Abstract
The Duffy binding protein is considered a leading vaccine candidate against asexual blood-stage Plasmodium vivax. The interaction of P. vivax merozoites with human reticulocytes through Duffy binding protein (DBP) and its cognate receptor is vital for parasite infection. The ligand domain of DBP (DBPII) is polymorphic, showing a diversity characteristic of selective immune pressure that tends to compromise vaccine efficacy associated with strain-specific immunity. A previous study resolved that a polymorphic region of DBPII was a dominant B-cell epitope target of human inhibitory anti-DBP antibodies, which we refer to as the DEK epitope for the amino acids in the SalI allele. We hypothesized that the polymorphic residues, which are not functionally important for erythrocyte binding but flank the receptor binding motif of DBPII, comprise variant epitopes that tend to divert the immune response away from more conserved epitopes. In this study, we designed, expressed, and evaluated the immunogenicity of a novel artificial DBPII allele, termed DEKnull, having nonpolar amino acids in the naturally occurring polymorphic charged residues of the DEK epitope. The DEKnull antigen retained erythrocyte-binding activity and elicited antibodies to shared epitopes of SalI DBPII from which it was derived. Our results confirmed that removal of the dominant variant epitope in the DEKnull vaccine lowered immunogenicity of DBPII, but inhibitory anti-DBPII antibodies were elicited against shared neutralizing epitopes on SalI. Focusing immune responses toward more conserved DBP epitopes may avoid development of a strain-specific immunity and enhance functional inhibition against broader range of DBPII variants.
INTRODUCTION
Merozoite invasion of erythrocytes involves a series of highly specific, sequential interactions between the merozoite and erythrocyte surface proteins and is a crucial step in the parasite's life cycle. In Plasmodium vivax, merozoite invasion of human reticulocytes is dependent on interaction of the Duffy binding protein (DBP) with its cognate receptor, the Duffy antigen receptor for chemokines (DARC) (5, 20, 28). It is believed that this molecule plays a critical role during the process of junction formation just before invasion, and unlike Plasmodium falciparum, there is no known alternate invasion pathway (3, 15). Merozoite proteins involved in the invasion process represent important candidates for development of malaria vaccines aimed at neutralizing blood-stage growth.
The DBP is a member of the Duffy binding-like erythrocyte binding protein (DBL-EBP) family from the erythrocyte-binding-like (ebl) genes, sequestered in the micronemes and released to the merozoite surface to bind receptors on erythrocytes during the early invasion process (1, 2). The receptor-binding domain is a conserved cysteine-rich motif known as region II and is the defining N-terminal extracellular domain of the DBP and related ligands where residues critical for receptor recognition are located (2, 10, 30, 38). Several studies have shown that anti-DBP antibodies that are either naturally or artificially induced can functionally inhibit invasion and DBPII binding to human erythrocytes in standard in vitro assays (12, 14, 17, 24, 25, 45). These data indicate that DBPII is a suitable target for an antibody-based vaccine against asexual blood-stage P. vivax. Therefore, the potential for effective antibody inhibition of merozoites, its biological importance, and the lack of apparent alternate ligands provide compelling reasons for development of DBPII as a vaccine.
Diversity in the ligand domain represents a potential problem that may compromise vaccine efficacy. Similar to other microbial ligands, alleles of this target domain have a very high ratio of nonsynonymous mutations to synonymous mutations, reflecting a mechanism consistent with a high selection pressure driving DBP allelic diversity as a mechanism of evasion for targets of inhibitory immunity (4, 13, 29, 35, 43). As a consequence, these polymorphisms pose a potentially serious challenge for development of a broadly effective vaccine based on the DBPII against diverse P. vivax strains.
In a recent study, we used naturally acquired human antibodies to identify dominant B-cell epitopes within DBPII that correlated with functional inhibition of the ligand domain (12). The dominant B-cell epitopes identified were surface-exposed motifs with clusters of polymorphic residues, which previous mutagenesis studies determined were not functionally important for erythrocyte binding but do flank residues critical for erythrocyte receptor recognition (7, 38). We refer to the epitope with the most variable residues as “DEK,” since these are the amino acids present in the SalI allele. Immune selection reported with other pathogens commonly involves variant residues adjacent to residues important for receptor recognition (6, 32, 39). As most naturally acquired infections with P. vivax tend to elicit weakly reactive and strain-specific antibodies, we concluded that the polymorphic dominant B-cell epitopes represent an evasion mechanism that misdirects the immune response away from more conserved less immunogenic epitopes that are potential targets for strain-transcending immunity. The goal of immunization is to accelerate the induction of protective immunity, but the presence of dominant variant epitopes can create an inherent bias toward a strain-specific response and limit induction of immune responses toward more conserved protective epitopes (12, 33, 41). To overcome this inherent bias, we have created a novel DBPII immunogen, referred to as DEKnull, lacking the polymorphic polar or charged residues that are normally present in the polymorphic DEK epitope (12). The design is to focus the immune response toward more conserved neutralizing epitopes that will have broader functional inhibition against allelic variants. In this proof-of-concept study, we present the design and characterize this novel synthetic antigen, comparing its immunogenicity to that of the naturally occurring allele product, SalI, which was used as a template.
MATERIALS AND METHODS
Design of DEKnull.
DBPII-SalI was used as a template to design the novel synthetic antigen DEKnull, in which the polar/highly charged residues 384D, 385E, 386K, 388Q, 389Q, 390R, 392K, and 393Q, which form part of the B-cell epitope (H3) identified from our previous study (12), were replaced by 384A, 385S, 386T, 388A, 389T, 390S, 392T, and 393S, respectively (Fig. 1). This epitope is the most polymorphic region within DBP region II.
Fig 1.
Alignment of the amino acid residues of the coding regions of DBPII-SalI and DEKnull. The gene coding for the ligand domain of DBPII-SalI was used as a template to create a novel synthetic DBPII allele (DEKnull). The most highly variant cluster of polar charged residues within the dominant B-cell epitope (underlined) identified in a previous study (12) was mutated to either alanine, threonine, or serine. Asterisks show different polymorphic sites within the DBP region II (39).
Production of recombinant antigens.
The coding sequences of DEKnull and SalI ligand domains were codon optimized for expression in Escherichia coli, and the DNA was commercially synthesized and cloned into pET21a+ (Novagen). The resulting plasmid was then used to transform E. coli BL21(DE3) LysE (Invitrogen). Pilot expression was used to identify bacterial clones expressing the recombinant proteins. Glycerol stocks were made of the positive clones and stored at −80°C until needed. For large-scale expression, an overnight culture was prepared from the frozen stock in LB medium containing 50 μg ml−1 ampicillin. The next day, the culture was diluted 1:10 in LB medium containing 50 μg ml−1 ampicillin and maintained at 37°C in a shaker flask with shaking at 250 rpm. Cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) with a final concentration at optical density at 600 nm [OD600] of 0.8 to 1.0. Cells were harvested 3 h postinduction and stored at −80°C until needed. Expressed proteins were purified from inclusion bodies under denaturing conditions as previously described (31, 34, 44) with modifications. Briefly, cells were resuspended in prechilled lysis buffer (10 mM Tris buffer [pH 8.0], 1 mM EDTA, 3% sucrose, 200 μg ml−1 lysozyme, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1 M NaCl, 20 μg ml−1 DNase), inclusion bodies were recovered by centrifugation, washed two times (10 mM Tris-HCl [pH 8.0], 3 M urea, 1 mM EDTA, 1 mM PMSF), and solubilized (20 mM phosphate buffer [pH 7.8] containing 8 M urea and 0.5 M NaCl), and recombinant proteins were purified by nickel Sepharose 6 affinity chromatography (GE Healthcare). Eluted fractions were checked for purity by separation by SDS-PAGE. Fractions containing pure protein were pooled and refolded by rapid dilution as previously described (31). The refolded product was dialyzed against 50 mM phosphate buffer (pH. 6.5) containing 1 M urea for 36 h, with two buffer changes every 12 h. The dialyzed product was concentrated down, filtered through a 0.2-μm-pore filter, dialyzed, adjusted to 1 mg ml−1 in phosphate-buffered saline (PBS), and stored at −80°C.
Functional analysis of refolded antigens.
Refolded antigens were tested for their ability to bind Duffy-positive human erythrocytes in an erythrocyte-binding assay similar to that previously described (8, 19, 31, 34, 42). Duffy-positive erythrocytes treated with chymotrypsin were used as a control. Twenty micrograms of refolded antigens was incubated with previously washed erythrocytes or erythrocytes treated with chymotrypsin in PBS with 1% bovine serum albumin (BSA) for 4 h at room temperature. The reaction mixture was layered over silicone oil (Dow Corning) and centrifuged for 30 s at 500 × g. Bound antigen was eluted from cells with 300 mM NaCl. Eluted proteins in ∼40 μl of buffer were diluted to 100 μl with SDS-PAGE buffer, heated at 65°C for 3 min, and separated on SDS-PAGE. Bound antigens were detected by immunoblot analysis with MAb-3D10, an anti-DBPII specific monoclonal antibody (MAb). Binding of the antigens to human erythrocytes was also tested by flow cytometry as previously reported (17, 34). Briefly, 3 μg of either antigen was incubated with 1 μl of Duffy-positive erythrocytes in 100 μl PBS with 1% BSA for 2 h at room temperature. Unbound antigen on red blood cells was washed off, and erythrocytes were incubated with a mouse anti-His monoclonal antibody conjugated to Alexa Fluor 488 (Qiagen). After washing, cells were suspended in 300 μl of PBS with 1% BSA, and 100,000 events were analyzed using an Accuri C6 flow cytometer on the FL1 channel. Binding was quantified by measuring the mean fluorescent intensity (MFI) of each sample relative to cells without bound antigen serving as a control.
Antibody production.
Polyclonal antibodies were produced in rats by immunizing with purified refolded recombinant SalI and recombinant DEKnull. Rats in each group received 0.1 mg of either recombinant DEKnull or SalI emulsified in TiterMax Gold (TiterMax) three times 3 weeks apart. Preimmune sera were collected on day 0, and immune sera were collected 3 weeks after the last immunization. Serum samples were pooled within each group for experiment analysis.
Immunoassays.
The enzyme-linked immunosorbent assay (ELISA) was used to quantify the anti-DBPII total IgG antibody titers in each serum sample for recombinant SalI and recombinant DEKnull. Wells of 96-well microtiter plates were coated and incubated overnight at 4°C with 200 ng of either recombinant DEKnull or recombinant SalI diluted in PBS. After washing away unbound antigens with PBS–0.05% (vol/vol) Tween 20 (PBS-T), antigen-coated wells were blocked with 5% skim milk in wash solution. In triplicate, half-log serial dilutions of each serum sample were added to wells and incubated with shaking for 2 h at room temperature. Plates were again washed, and wells were incubated for 90 min with a phosphatase-conjugated goat anti-rat monoclonal antibody (KPL, Inc., Gaithersburg, MD), diluted 0.5 μg ml−1 in wash buffer. Wells were washed, bound antibodies were detected with an alkaline phosphatase substrate (Phospho Blue; KPL, Inc.), and absorbance readings were measured at 630 nm on an automated microplate reader (Synergy II; Biotek, Inc.). The antibody titer for each serum sample was determined as the reciprocal of the dilution required to give an absorbance of 1 (OD630, 1.0) in the same assay. To determine whether recombinant DEKnull was recognized by naturally acquired anti-DPBII antibodies, recombinant SalI and DEKnull precoated and blocked plates were incubated with anti-SalI and anti-DEKnull specific antibodies affinity purified from Papua New Guinea (PNG) human sera, which was previously shown to contain high-titer anti-DBPII inhibitory antibodies (12, 21). After washing and incubating wells in phosphatase-conjugated goat anti-human monoclonal antibody (KPL, Inc.), bound antibody was detected as described above. The antibody titer was determined as concentration of antibody required to give an absorbance of 2 (OD630, 2).
For immunoblot analysis, antigens were separated by SDS-PAGE and electrophoretically transferred onto nitrocellulose membranes (Millipore). Membranes were rinsed in wash buffer (PBS–0.05% Tween 20) and blocked in 5% skim milk in wash buffer overnight at 4°C. The membranes were rinsed in wash buffer and reacted with either 2.5 μg ml−1 MAb-3D10 or rat antisera diluted 1:4,000. After three washes, membranes were incubated in either (HRP)-conjugated goat anti-rat or goat anti-mouse monoclonal antibody (KPL, Inc.) at 0.5 μg ml−1 in wash buffer. Bound antibody was then detected with ECL (enhanced chemiluminescence) substrate (GE Healthcare).
Inhibition of DBPII binding to human erythrocytes by COS7 assay.
Functional DBPII is expressed on the surface of COS7 cells, using herpes simplex virus signal and transmembrane targeting sequences, and with a C-terminal tag of green fluorescent protein (GFP), as reported previously (11, 24, 25). The cloning of DBPII-SalI coding sequence into pEGFP-N1 plasmid (Clontech) was previously described, and DEKnull was cloned in an identical manner (11, 16, 25, 26). DNA of each plasmid was purified using an endotoxin-free plasmid maxi-kit (Qiagen). COS7 cells were transfected with 125 ng DNA per well in a 24-well culture dish using Lipofectamine 2000 reagent (Invitrogen), and inhibition assays were carried out 42 h posttransfection. Each well was incubated with a different dilution of antiserum for 1 h at 37°C before human erythrocytes (2% hematocrit) were added and incubated for another 2 h. Nonadherent cells were washed off with PBS containing Ca2+ and Mg2+, and binding was quantified by counting rosettes observed in 30 microscope fields at magnification 200× as previously described (23, 25, 38). Binding inhibition of each antiserum was determined by accessing the percentage of rosettes in wells of transfected cells in the presence of antisera relative to wells with transfected cells in the presence of preimmune sera.
RESULTS
Design and expression of recombinant DEKnull.
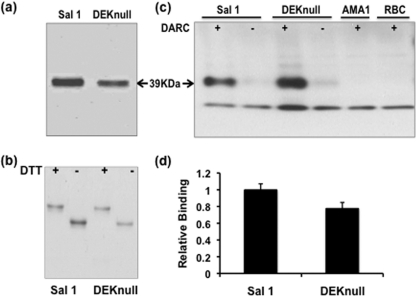
Analysis of coding sequences available in public databases for the P. vivax DBP ligand domain region (DBPII) revealed that most polymorphic residues were polar or charged. The most highly variant residues of DBPII occurred in the center of the ligand domain and were previously identified to be part of a dominant Bc epitope (DEKAQQRRKQWWNESK) that was also a target of inhibitory antibody (12). Using the DBPII-SalI as a template sequence, eight of the nine charged polar amino acids within the cluster of variant residues were mutated to small uncharged polar (serine or threonine) or small hydrophobic (alanine) amino acids to create a synthetic allele, termed DEKnull (Fig. 1). Other studies demonstrated that none of the residues mutated were critical for DBPII binding to erythrocytes (18, 38). Recombinant proteins of DEKnull and SalI were expressed in bacteria, purified from inclusion bodies, and refolded by rapid dilution by the same protocol. The expression level and stability of the DEKnull product were similar to those of SalI, and both refolded antigens migrated at the expected size of 39 kDa on SDS-PAGE (Fig. 2a). DEKnull and SalI had similar sensitivities to the reducing agent dithiothreitol (DTT), and a comparable mobility shift was observed with both refolded proteins (Fig. 2b).
Fig 2.
Purification and functional analysis of recombinant proteins. (a) SDS-PAGE gel of purified and refolded recombinant SalI and recombinant DEKnull. (b) SDS-PAGE showing differential mobility of reduced (+) and refolded (−) antigens. Mobility shift is a simple indicator of native conformation of recombinant antigens. (c) Western blot. A standard erythrocyte binding assay shows binding of purified proteins to Duffy antigen receptor on erythrocytes (+) and no binding to erythrocytes treated with chymotrypsin (−). Arrows indicate the 39-kDa size of the recombinant proteins. P. falciparum AMA-1 (PfAMA-1) and erythrocytes alone served as controls. (d) Recombinant proteins were tested for binding to erythrocytes by flow cytometry. Bars indicate average relative binding of recombinant DEKnull to human erythrocytes compared to recombinant SalI for two independent experiments, and error bars represent ± standard deviation.
DEKnull erythrocyte binding activity.
The purified refolded DEKnull was tested by two standard in vitro assays for binding specificity to Duffy antigen surface receptor on human erythrocytes. In a modified version of the original erythrocyte-binding assay used to validate erythrocyte receptor specificity of malaria parasite ligands (19), purified refolded DEKnull and SalI in isotonic buffer were tested for specificity of binding to Duffy-positive erythrocytes and Duffy-negative erythrocytes. Both recombinant proteins strongly bound to Duffy-positive erythrocytes, while only trace amounts of each recombinant protein could be detected as having been eluted from Duffy-negative erythrocytes. No bands of expected size were observed in lanes with erythrocytes incubated with recombinant P. falciparum AMA1 or control lanes with erythrocytes alone (Fig. 2c). Demonstration of binding of refolded DEKnull and SalI to erythrocytes was repeated by a flow cytometry erythrocyte-binding assay that can quantify binding (34). With this assay, we confirmed there was no significant quantitative difference in binding to Duffy-positive erythrocytes between recombinant DEKnull and SalI (Fig. 2d).
Testing immunogenicity of DEKnull.
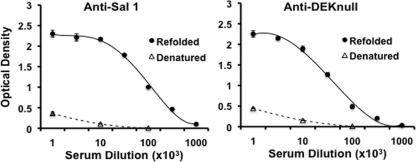
Serologic response to recombinant DEKnull was compared to that of recombinant SalI by laboratory immunizations of rats to evaluate the effect of the loss of the dominant Bc epitope in DEKnull. Immunogenicity was determined by ELISA endpoint titration reactivity against each homologous recombinant protein (Fig. 3). DEKnull appeared equally immunogenic to SalI in the rats, inducing high levels of anti-DBPII antibodies (Fig. 3). In the same way, each antiserum had a strong bias toward reactivity to conformational epitopes. Comparing the reactivities of anti-DEKnull and anti-SalI sera to refolded and reduced homologous antigens demonstrated ∼86% of the antibodies reacted to refolded antigens versus reduced homologous antigens (Fig. 3). Anti-DEKnull serum reacted to the same level with both antigens at an endpoint antibody titer of 1:65,000, while in contrast, anti-SalI reactivity was significantly different (P < 0.005) for the two antigens at 1:100,000 and 1:68,000 for recombinant SalI and recombinant DEKnull, respectively, suggesting SalI DBPII was more immunogenic (Fig. 4) as a result of the presence of the dominant B-cell epitope, which is absent in DEKnull.
Fig 3.
Reactivity of rat anti-SalI and anti-DEKnull sera with recombinant antigens. Antisera raised in rats against recombinant DBPII-SalI and recombinant DEKnull were tested by endpoint dilution for reactivity with homologous refolded (solid lines) and denatured (broken lines) antigens. Antigen preparations were adsorbed onto wells of a microtiter plate and allowed to react with different dilutions of the antisera. Each point on the curves represents the mean of triplicate wells, while error bars represent ± standard deviation.
Fig 4.
Reactivity profiles of anti-SalI and anti-DEKnull sera with recombinant antigens. Each antiserum was tested by endpoint dilution for cross-reactivity with homologous and heterologous refolded antigens. Antigen preparations were adsorbed onto wells of a micro titer plate and allowed to react with different dilutions of the antisera. Solid and broken lines represent recombinant SalI and recombinant DEKnull, respectively. Each point on the curves represents the mean of triplicate wells, while error bars represent ±standard deviation.
Further analysis was conducted to determine whether naturally acquired anti-DBPII antibodies can recognize recombinant DEKnull. Affinity-purified human sera from PNG previously shown to have high titer anti-DBPII inhibitory antibodies were highly reactive to both recombinant SalI and DEKnull (Fig. 5). Similar to the rat anti-DBPII antibodies, the human anti-DEKnull antibodies showed the same levels of reactivity to both antigens (P = 0.042), while anti-SalI reactivity was significantly higher against recombinant SalI than DEKnull (P < 0.001), confirming the immunogenicity of the DEK epitope.
Fig 5.
Reactivity of human sera against recombinant SalI and DEKnull. A pool of high-titer anti-DBPII inhibitory human serum from PNG was affinity purified and tested by endpoint dilution for reactivity with recombinant SalI and DEKnull. Recombinant SalI or DEKnull was adsorbed onto wells of the microtiter plate and probed with tripling dilutions of either affinity-purified anti-SalI or anti-DEKnull. Each point on the curves represents the mean OD values of each dilution tested in triplicate.
Anti-DBPII inhibition.
A standardized in vitro cell-based assay, which is used to evaluate erythrocyte-binding activity of DBP and other Plasmodium ligands, provides a useful platform to measure anti-DBP inhibition. Comparative effects of different antibodies can be evaluated against DBPII-erythrocyte binding by calculating the 50% inhibitory concentration (IC50) of each test antiserum, which is achieved by determining the inhibitory effects of serial dilutions ranging from 0 to 100% inhibition. Preimmune sera tested at 1:1,000 showed no inhibition of binding and served as a control for the experiments. As anticipated, the antisera to DEKnull and SalI were observed to inhibit DBPII-erythrocyte binding in a concentration-dependent manner (Fig. 6). The IC50 titer for anti-SalI at 1:5,000 was significantly more inhibitory than anti-DEKnull at 1:3,500 (P < 0.006). This result confirms that the DEKnull epitope comprises a significant target of anti-DBP inhibitory antibody, but other more conserved epitopes also exist on DBPII.
Fig 6.
Inhibition of erythrocyte binding to DBPII-SalI expressed on the surface of COS7 cells by rat anti-SalI and anti-DEKnull. A monolayer culture of COS7 cells expressing recombinant DBPII-SalI on the cell surface was tested for binding to Duffy blood group antigen on human erythrocytes in the presence of anti-DEKnull and anti-SalI sera. Antibodies were tested by endpoint dilution, and percent inhibition was quantified by comparing the number of rosettes bound to COS7 cells in 30 microscope fields (×200) in the presence of test sera and preimmune sera. Curves represent means of three independent experiments done in duplicate. Error bars represent ±standard deviation.
DISCUSSION
Plasmodium vivax preferentially infects human reticulocytes expressing the surface DARC blood group antigen (27). The critical nature of the host cell specificity for parasite survival is evident from the virtual absence of P. vivax malaria from populations with high levels of DARC negativity and the difficulty culturing this parasite in blood not enriched for reticulocytes (27, 36, 40). Unlike P. falciparum, alternate invasion pathways have not been identified. Consequently, the DBPII represents an ideal potential target for vaccine development against P. vivax asexual blood-stage infection. In spite of its biologic importance, potential confounding factors that may compromise its use as a vaccine have been revealed in studies on the naturally occurring serologic responses to DBP after infection. As with other blood-stage malaria vaccine candidates, DBP appears to be a relatively poor immunogen, since functional anti-DBP activity is mostly weak and development of a broadly inhibitory strain-transcending immunity is a rather uncommon occurrence (12, 21).
The relative bias toward a weak, strain-specific DBP antibody response following human infection is likely a consequence of the polymorphic nature of epitopes in the DBP ligand domain. Most polymorphisms identified in DBP are clustered within the central portion of the ligand domain, especially in the major variant cluster we refer to as the DEK epitope (12). This region of the ligand domain is also identified as being responsible for determining receptor specificity, and in a recent study, we mapped to this region the epitopes that are the primary targets for antibody inhibition. In the present study, we wanted to evaluate the immunogenic quality of a DBPII immunogen lacking this Bc epitope and whether the absence of the dominant “DEK” epitope adversely affects induction of anti-DBP immunity.
In order to resolve this issue, we created a synthetic gene referred to as DEKnull based on the SalI ligand domain that lacks only the highly charged, polar residues in the dominant B-cell epitope (12). We expressed, purified, and refolded recombinant DEKnull into its native conformation and determined its immunological characteristics. The expressed antigen was shown to be correctly refolded and functionally active as it was able to bind human erythrocytes expressing DARC but not DARC-negative erythrocytes (31, 44, 45). The synthetic DEKnull antigen lacking all of those polymorphic residues bound to human erythrocytes with specificity similar to that of the native protein and confirms previous data demonstrating that these polymorphic residues within the hypervariable segment of DBPII are not critical for erythrocyte binding activity (18, 38). Furthermore, the functionality of the altered residues is consistent with similar mutagenesis of the homologous DBL domain of P. falciparum EBA-175, which also has naturally occurring polymorphisms that do not affect erythrocyte-binding properties (22, 43).
For an antigen to be a good vaccine candidate, it should be able to induce high-titer, functional antibodies. Immunizations with recombinant DEKnull and SalI proteins induced high-titer immune sera in rats with antibodies reactive against both DEKnull and SalI. As expected, the SalI antiserum displayed a higher level of reactivity to the SalI versus the DEKnull protein, whereas the anti-DEKnull antiserum had nearly identical reactivity profiles to both homologous SalI and heterologus DEKnull antigens. This confirms the immunogenicity of the DEK epitope and also the presence of other immunogenic epitopes on native DBPII. Consistent with the presence of other target epitopes, we found that naturally acquired, inhibitory immune sera (12, 21) were highly reactive with DEKnull (Fig. 5), indicating that the synthetic DEKnull antigen contained conserved epitopes associated with a protective immune response despite lacking a dominant epitope. Functional in vitro assays demonstrated both antisera completely inhibited DBPII-erythrocyte binding at a dilution up to 1:500. The functional importance of the DEK epitope as a target of protective anti-DBPII immunity was confirmed by differences in the IC50 values for anti-SalI and anti-DEKnull inhibition of SalI-erythrocyte binding.
In previous studies, our group and others have demonstrated that DBP is naturally immunogenic; naturally acquired or artificially induced anti-DBPII antibodies can effectively block DBPII binding to human erythrocytes and identified epitopes' targets of inhibition in the ligand domain (17, 31, 45). However, anti-DBPII antibodies tend to be weak or unstable (12), as well as short-lived and strain specific (9, 37). In an effort to design a DBP immunogen to overcome these challenges, we designed and evaluated an artificial DBPII immunogen referred to as DEKnull, lacking the strongly charged residues of a dominant B-cell epitope that s a target of functional antibody. Serologic studies of this synthetic antigen in laboratory animals demonstrated that it was highly immunogenic and induced the production of functional antibodies that inhibited erythrocyte-binding activity of the natural SalI allele. It is expected that this type of immunogen may be necessary to avoid induction of strain-specific responses to the dominant variant epitopes and focus the immune response to more conserved neutralizing epitopes that are potential targets for strain-transcending immunity.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R01AI064478.
We have no commercial or other association that poses a conflict of interest.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Adams JH, et al. 1990. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63:141–153. [DOI] [PubMed] [Google Scholar]
- 2. Adams JH, et al. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 89:7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aikawa M, Miller LH, Johnson J, Rabbege J. 1978. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J. Cell Biol. 77:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. 1996. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol. Biochem. Parasitol. 78:269–272 [DOI] [PubMed] [Google Scholar]
- 5. Barnwell JW, Wertheimer SP. 1989. Plasmodium vivax: merozoite antigens, the Duffy blood group, and erythrocyte invasion. Prog. Clin. Biol. Res. 313:1–11 [PubMed] [Google Scholar]
- 6. Barouch DH. 2007. HIV-1 vaccine candidate ineffective. AIDS Clin. Care 19:93. [PubMed] [Google Scholar]
- 7. Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat. Struct. Mol. Biol. 18:908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camus D, Hadley TJ. 1985. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230:553–556 [DOI] [PubMed] [Google Scholar]
- 9. Ceravolo IP, et al. 2009. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin. Exp. Immunol. 156:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184:1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chootong P, et al. 2010. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect. Immun. 78:1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole-Tobian J, King CL. 2003. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol. Biochem. Parasitol. 127:121–132 [DOI] [PubMed] [Google Scholar]
- 14. Dutta S, Daugherty JR, Ware LA, Lanar DE, Ockenhouse CF. 2000. Expression, purification and characterization of a functional region of the Plasmodium vivax Duffy binding protein. Mol. Biochem. Parasitol. 109:179–184 [DOI] [PubMed] [Google Scholar]
- 15. Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. 1975. Invasion of erythrocytes by malaria merozoites. Science 187:748–750 [DOI] [PubMed] [Google Scholar]
- 16. Fraser T, et al. 1997. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect. Immun. 65:2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimberg BT, et al. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hans D, et al. 2005. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol. Microbiol. 55:1423–1434 [DOI] [PubMed] [Google Scholar]
- 19. Haynes JD, et al. 1988. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J. Exp. Med. 167:1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horuk R, et al. 1993. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261:1182–1184 [DOI] [PubMed] [Google Scholar]
- 21. King CL, et al. 2008. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc. Natl. Acad. Sci. U. S. A. 105:8363–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang H, Sim BK. 1997. Conservation of structure and function of the erythrocyte-binding domain of Plasmodium falciparum EBA-175. Mol. Biochem. Parasitol. 84:241–245 [DOI] [PubMed] [Google Scholar]
- 23. Mayer DC, et al. 2004. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc. Natl. Acad. Sci. U. S. A. 101:2518–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McHenry AM, Barnes SJ, Ntumngia FB, King CL, Adams JH. 2011. Determination of the molecular basis for a limited dimorphism, N417K, in the Plasmodium vivax Duffy-binding protein. PLoS One 6:e20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Michon P, Fraser T, Adams JH. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michon P, et al. 2001. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett. 495:111–114 [DOI] [PubMed] [Google Scholar]
- 27. Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295:302–304 [DOI] [PubMed] [Google Scholar]
- 28. Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. 1975. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189:561–563 [DOI] [PubMed] [Google Scholar]
- 29. Ntumngia FB, et al. 2009. Genetic variation among Plasmodium vivax isolates adapted to non-human primates and the implication for vaccine development. Am. J. Trop. Med. Hyg. 80:218–227 [PMC free article] [PubMed] [Google Scholar]
- 30. Ranjan A, Chitnis CE. 1999. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 96:14067–14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh S, et al. 2001. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax Duffy-binding protein. J. Biol. Chem. 276:17111–17116 [DOI] [PubMed] [Google Scholar]
- 32. Smith DJ, et al. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376 [DOI] [PubMed] [Google Scholar]
- 33. Tobin GJ, et al. 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26:6189–6199 [DOI] [PubMed] [Google Scholar]
- 34. Tran TM, et al. 2005. Detection of a Plasmodium vivax erythrocyte binding protein by flow cytometry. Cytometry A 63:59–66 [DOI] [PubMed] [Google Scholar]
- 35. Tsuboi T, et al. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect. Immun. 62:5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Udomsangpetch R, Kaneko O, Chotivanich K, Sattabongkot J. 2008. Cultivation of Plasmodium vivax. Trends Parasitol. 24:85–88 [DOI] [PubMed] [Google Scholar]
- 37. VanBuskirk KM, et al. 2004. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J. Infect. Dis. 190:1556–1562 [DOI] [PubMed] [Google Scholar]
- 38. VanBuskirk KM, Sevova E, Adams JH. 2004. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. U. S. A. 101:15754–15759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker BD, Burton DR. 2008. Toward an AIDS vaccine. Science 320:760–764 [DOI] [PubMed] [Google Scholar]
- 40. Welch SG, McGregor IA, Williams K. 1977. The Duffy blood group and malaria prevalence in Gambian West Africans. Trans. R. Soc. Trop. Med. Hyg. 71:295–296 [DOI] [PubMed] [Google Scholar]
- 41. Welsh RM, Fujinami RS. 2007. Pathogenic epitopes, heterologous immunity and vaccine design. Nat. Rev. Microbiol. 5:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickramarachchi T, Devi YS, Mohammed A, Chauhan VS. 2008. Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion. PLoS One 3:e1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xainli J, Adams JH, King CL. 2000. The erythrocyte binding motif of Plasmodium vivax Duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111:253–260 [DOI] [PubMed] [Google Scholar]
- 44. Yazdani SS, Shakri AR, Chitnis CE. 2004. A high cell density fermentation strategy to produce recombinant malarial antigen in E. coli. Biotechnol. Lett. 26:1891–1895 [DOI] [PubMed] [Google Scholar]
- 45. Yazdani SS, Shakri AR, Mukherjee P, Baniwal SK, Chitnis CE. 2004. Evaluation of immune responses elicited in mice against a recombinant malaria vaccine based on Plasmodium vivax Duffy binding protein. Vaccine 22:3727–3737 [DOI] [PubMed] [Google Scholar]