PTEN nuclear entry driven by ubiquitination is mediated by the ligase-interacting protein Ndfip1 and is essential for neuronal survival in mice after cerebral ischemia.
Abstract
PTEN (phosphatase and tensin homologue deleted on chromosome TEN) is the major negative regulator of phosphatidylinositol 3-kinase signaling and has cell-specific functions including tumor suppression. Nuclear localization of PTEN is vital for tumor suppression; however, outside of cancer, the molecular and physiological events driving PTEN nuclear entry are unknown. In this paper, we demonstrate that cytoplasmic Pten was translocated into the nuclei of neurons after cerebral ischemia in mice. Critically, this transport event was dependent on a surge in the Nedd4 family–interacting protein 1 (Ndfip1), as neurons in Ndfip1-deficient mice failed to import Pten. Ndfip1 binds to Pten, resulting in enhanced ubiquitination by Nedd4 E3 ubiquitin ligases. In vitro, Ndfip1 overexpression increased the rate of Pten nuclear import detected by photobleaching experiments, whereas Ndfip1−/− fibroblasts showed negligible transport rates. In vivo, Ndfip1 mutant mice suffered larger infarct sizes associated with suppressed phosphorylated Akt activation. Our findings provide the first physiological example of when and why transient shuttling of nuclear Pten occurs and how this process is critical for neuron survival.
Introduction
PTEN (phosphatase and tensin homologue deleted on chromosome TEN) is the major negative regulator of signaling by phosphatidylinositol 3-kinase (PI 3-K), thereby playing a central role in controlling many important cellular activities regulated by this pathway, including cell division, cell growth, cell survival, and DNA damage (Chalhoub and Baker, 2009). PTEN exerts its negative effect through its phosphatase activity on the plasma membrane lipid phosphatidylinositol 3,4,5-triphosphate (PIP3), reducing levels of phosphorylated Akt (pAkt; Maehama and Dixon, 1998; Stambolic et al., 1998). Thus, loss of PTEN, as demonstrated by genetic inactivation in human cancer or mouse knockout (KO) models, causes constitutive activation of Akt in cells, resulting in dysregulated cell proliferation, growth, and survival, which are hallmarks of tumorigenesis (Hobert and Eng, 2009; Nardella et al., 2010). PTEN can be found in both the cytoplasm and nucleus of many cell and tissue types, and its aberrant localization has been implicated in disease. The nucleocytoplasmic distribution of PTEN has been proposed to affect its tumor-suppressive function both within and outside the PI 3-K pathway (Planchon et al., 2008). However, it has remained unclear what physiological stimulus can drive PTEN into the nucleus and under what in vivo circumstances this can occur.
In the brain, PTEN is required for multiple aspects of neuronal function and development, including maintenance of neuron structure, size, synaptic plasticity, and survival (Endersby and Baker, 2008). Thus, conditional deletion of Pten in the brain increases astrocyte proliferation and neuron hypertrophy that is associated with increased dendrites and synapses and aberrant cerebellar development (Backman et al., 2001; Kwon et al., 2001, 2006). Although these phenotypes may be attributable to increased Akt signaling, the lack of tumor formation in these brains emphasizes multifaceted roles for Pten in neurons (Endersby and Baker, 2008; Chalhoub and Baker, 2009). So although Pten status might not dictate proliferation in neurons, it still appears to be important for apoptosis during cerebral ischemia (Ning et al., 2004; Lee et al., 2009). What is entirely unclear is the molecular mechanism underscoring Pten function in neuronal ischemia, despite studies advocating Pten inhibition as a possible therapeutic route (Chang et al., 2007; Li et al., 2009).
In the current study, we demonstrate that cerebral ischemia is the stimulus for trafficking of Pten to the nucleus, leading to neuron survival. This nuclear trafficking of Pten is downstream of Nedd4 family–interacting protein 1 (Ndfip1), an adaptor for Nedd4-mediated ubiquitination (Shearwin-Whyatt et al., 2006). Interestingly, Ndfip1 up-regulation and neuronal survival were not associated with Pten degradation. Instead, Ndfip1 directly increases the rate of Pten translocation to cell nuclei, and, without Ndfip1 in vivo, Pten fails to accumulate in neuronal nuclei, resulting in larger infarct sizes in ischemia. Therefore, Pten ubiquitination and nuclear import, previously shown to be antioncogenic in the colon (Trotman et al., 2007; Wang et al., 2007), serve the unexpected function of protecting neurons from death after ischemia in the brain.
Results and discussion
Nuclear trafficking of Pten in neurons is stimulated by cerebral ischemia and requires Ndfip1
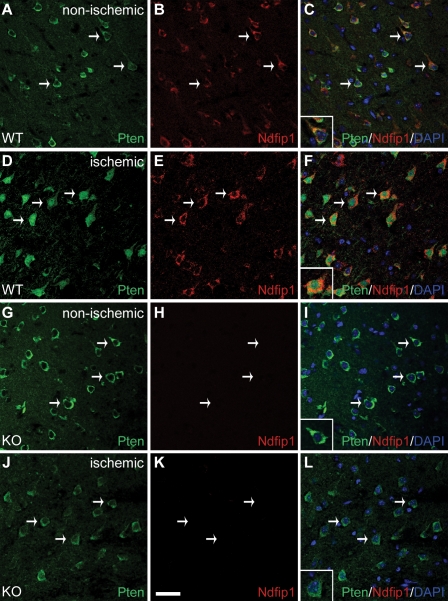
Under normal homeostasis, Pten is found predominantly in the cytoplasm of neurons in the cerebral cortex (Fig. 1, A–C). After ischemia, we observed a change in the cellular location of Pten in neurons of the periinfarct region, from the cytoplasm to the nucleus (Fig. 1, D–F; Pten antibody validation is shown in Fig. S1). This relocalization of Pten is strongly correlated with neurons up-regulating Ndfip1, an adaptor protein for the Nedd4 family of ubiquitin ligases (Fig. 1, A–F). Previously, we reported that Ndfip1 is rapidly up-regulated in surviving neurons after brain trauma (Sang et al., 2006); here, we also show that Ndfip1 is up-regulated in surviving neurons after brain ischemia (Fig. S2). The tight correlation of Pten nuclear localization with Ndfip1 up-regulation in the same neurons suggested that Ndfip1 could play a role in nuclear trafficking of Pten during ischemic stress. To determine whether Pten nuclear accumulation was Ndfip1 dependent, we conditionally deleted Ndfip1 in neurons using the Emx1-Cre transgene (Ndfip1loxP/loxP;Emx1-Cre+; Fig. S2; Iwasato et al., 2000). Mutant mice were viable and exhibited normal body weights and brain sizes despite Emx1-specific loss of Ndfip1 protein in neurons of the cortex (unpublished data). Similar to the wild-type (WT) animals, mutant neurons in a comparable cortical region of the nonischemic hemisphere showed a predominantly cytoplasmic distribution pattern for Pten (Fig. 1, G–I and inset). In contrast to the WT animals, the ischemic hemisphere of KO animals maintained a predominantly cytoplasmic Pten distribution (Fig. 1, J–L and inset). Further confirmation of nuclear Pten accumulation was obtained in vitro using nuclear/cytoplasmic cell fractionation to demonstrate that hypoxic primary neurons accumulate Pten in the nuclei of Ndfip1+/+ but not Ndfip1−/− mice (Fig. S3). Therefore, Pten distribution is cytoplasmic in both WT and KO cortical neurons, but, in ischemic cortices, Pten nuclear accumulation requires the presence of Ndfip1. These in vitro and in vivo observations point to Ndfip1 as a key regulator of Pten nuclear import in response to this adverse physiological stimulus.
Figure 1.
Pten nuclear import in neurons is triggered by cerebral ischemia and requires Ndfip1 up-regulation. (A–C) Confocal microscopy of a brain section in the nonischemic hemisphere of WT mice (Ndfip1loxP/loxP;Emx1-Cre−) showing low levels of basal Ndfip1 expression together with cytoplasmic Pten in these neurons (arrows). The inset in C shows coextensive staining for both proteins in the cytoplasm, whereas their nuclei (DAPI) are relatively devoid of both proteins. (D–F) In the ischemic cortex of WT mice (24 h after HI), Pten is predominantly localized in the nucleus of neurons (arrows) that are up-regulated for Ndfip1. The inset in F shows nuclear localization of Pten with double labeling for both Ndfip1 and Pten (orange) in the cytoplasm. (G–I) In Ndfip1 conditional KO mice (Ndfip1loxP/loxP;Emx1-Cre+) 24 h after HI, Pten in the nonischemic cortex is localized only in the cytoplasm of neurons (arrows and inset). (J–L) In contrast to WT littermates in D–F, Pten in the ischemic cortex of Ndfip1 conditional KO mice remains in the cytoplasm of neurons (arrows). The inset in L shows the absence of Ndfip1 and nuclear exclusion of Pten after ischemia. Bar, 30 µm.
Ndfip1 binds to and enhances ubiquitination of Pten
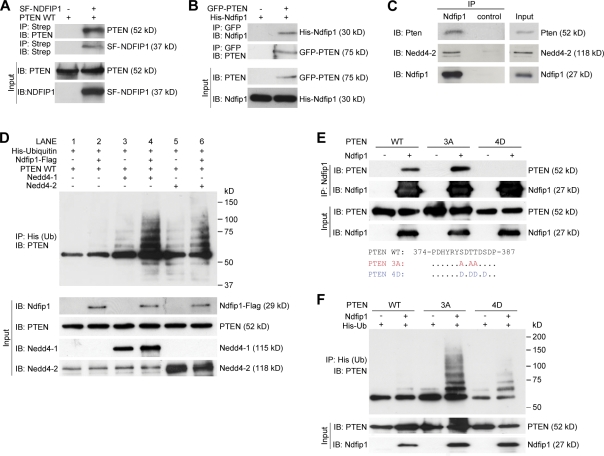
Next, we looked for a direct interaction between PTEN and Ndfip1. Ndfip1 acts by recruiting target proteins that lack PY motifs and facilitates their ubiquitination by Nedd4 E3 ligases (Shearwin-Whyatt et al., 2006). One member of the Nedd4 family, Nedd4-1, has been shown to be important for PTEN ubiquitination (Trotman et al., 2007; Wang et al., 2007; Drinjakovic et al., 2010), but there is controversy regarding this interaction (Fouladkou et al., 2008; Wang et al., 2008). As PTEN lacks the classical PY motifs required for Nedd4 binding, we investigated whether Ndfip1 may mediate the interaction between PTEN and the ubiquitin ligases. To determine whether Ndfip1 binds to PTEN, coimmunoprecipitation experiments were performed in human embryonic kidney (HEK-293T) cells (Fig. 2, A and B) and brain tissue (Fig. 2 C). In cells, Ndfip1 and PTEN can mutually coprecipitate each other (Fig. 2, A and B), and, in brain lysates, Ndfip1 can pull down both Pten and Nedd4-2, indicating endogenous interaction (Fig. 2 C).
Figure 2.
Ndfip1 binds to PTEN and enhances PTEN ubiquitination by Nedd4 E3 ligases. (A) PTEN (PTEN WT) coprecipitates with Strep-Flag-NDFIP1 (SF-NDFIP1) after expression in HEK-293T cells. IB, immunoblotted; IP, immunoprecipitated. (B) Ndfip1 coprecipitates with GFP-PTEN after expression in HEK-293T cells. (C) Mouse cortical lysates show coimmunoprecipitation of both Pten and Nedd4-2 with Ndfip1. (D) PTEN ubiquitin (Ub) assay using Ndfip1, Nedd4-1, and Nedd4-2 in HEK-293T cells. Lanes without Ndfip1 coexpression (lanes 1, 3, and 5) show decreased PTEN ubiquitination compared with lanes expressing Ndfip1 (lanes 2, 4, and 6). The presence of Ndfip1 with Nedd4-1 (lane 4) or Nedd4-2 (lane 6) produced the characteristic monoubiquitinated ladder together with higher molecular mass–polyubiquitinated smear. (E) Phosphorylation status of PTEN changes Ndfip1 interaction. PTEN WT and the PTEN 3A phosphorylation-defective mutant interact strongly with Ndfip1, but PTEN 4D phosphomimic mutant interacts weakly with Ndfip1. (F) Ndfip1-mediated ubiquitination of PTEN is dependent on the phosphorylation status of PTEN. In the absence of Ndfip1, little PTEN ubiquitination occurs. Coexpression with Ndfip1 results in strong ubiquitination of the PTEN 3A phosphorylation-defective mutant, but both PTEN WT and the PTEN 4D phosphomimic mutant are weakly ubiquitinated.
The aforementioned results suggest that in the brain, Nedd4-2 may be the E3 ligase required for Pten ubiquitination. To further investigate this, we tested the role of Ndfip1 in mediating PTEN ubiquitination by either Nedd4-1 or Nedd4-2 (Fig. 2 D). Transfected PTEN alone (Fig. 2 D, lane 1) in HEK-293T cells showed a lack of ubiquitination. Overexpression of either Nedd4-1 (Fig. 2 D, lane 3) or Nedd4-2 (Fig. 2 D, lane 5) revealed multiple PTEN ladders corresponding to multimonoubiquitinated forms of PTEN (Trotman et al., 2007). These discrete forms of monoubiquitinated PTEN were significantly enhanced when Ndfip1 was cotransfected with either Nedd4-1 (Fig. 2 D, lane 4) or Nedd4-2 (Fig. 2 D, lane 6), indicating increased substrate specificity or increased ligase efficiency as a result of Ndfip1. In the presence of Ndfip1, the adduct forms that correspond to monoubiquitinated bands of PTEN (Trotman et al., 2007; Wang et al., 2007) appear stronger, although the uniform smear representing polyubiquitinated PTEN (Wang et al., 2007; Maccario et al., 2010) is also increased. Ndfip1 alone (Fig. 2 D, lane 2) also produced basal levels of ubiquitinated PTEN, likely assisted by endogenous levels of NEDD4-2 in HEK-293T cells (Fig. 2 D, bottom). To test this further, we conducted RNAi experiments to knock down either NEDD4-1 or NEDD4-2 (Fig. S3). In the presence of Ndfip1, knockdown of either NEDD4-1 or NEDD4-2 decreased the overall level of PTEN ubiquitination, reaffirming the pivotal role of Ndfip1 in conjunction with both E3 ligases (Fig. S3, lanes 2, 4, and 6). Phosphorylation of PTEN at C-terminal residues is known to stabilize the enzyme by preventing its ubiquitination but reduces its catalytic activity; conversely, dephosphorylated PTEN is active but rapidly degraded (Vazquez et al., 2001; Raftopoulou et al., 2004). To ascertain whether or not Ndfip1-mediated binding and ubiquitination have a preferred form of PTEN substrate, experiments were conducted between three types of PTEN: WT, a phosphorylation-defective PTEN 3A mutant (S380A/T382A/T383A), and a phosphomimic PTEN 4D mutant (S380D/T382D/T383D/S385D; Vazquez et al., 2000). We found that PTEN 3A was able to interact with Ndfip1 at a level similar to or increased compared with PTEN WT, but PTEN 4D interaction was weak (Fig. 2 E). In the presence of Ndfip1, PTEN 3A was ubiquitinated more than PTEN WT, generating the characteristic multimonoubiquitinated bands as well the polyubiquitinated smear (Fig. 2 F, fourth lane). Interestingly, the PTEN 4D mutant is capable of being ubiquitinated despite low binding to Ndfip1 (Fig. 2 F, sixth lane).
In vitro nuclear import of PTEN is accelerated by Ndfip1
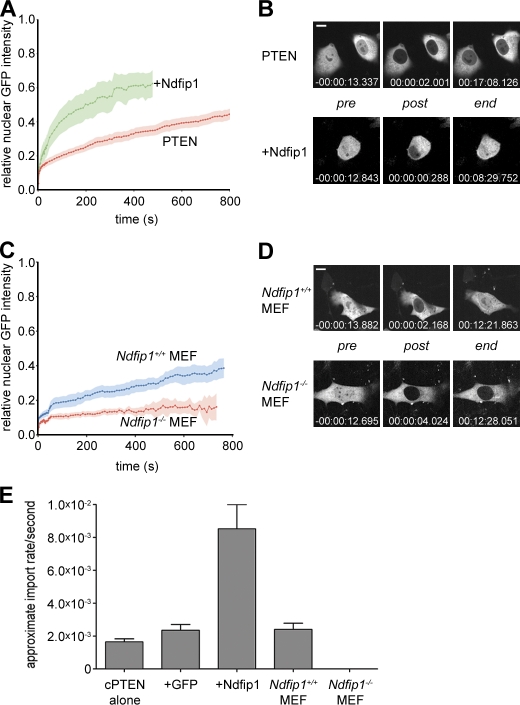
NEDD4-1 has been shown to control PTEN monoubiquitination and nuclear import (Trotman et al., 2007). To test whether Ndfip1 is a mediator of this process, we measured nuclear PTEN transport as a function of Ndfip1 status. To this end, we used nuclear FRAP of cherryFP-PTEN (cherry-PTEN) in the PTEN-deficient PC3 prostate cancer cell line, as previously described (Trotman et al., 2007). We first tested whether overexpression of GFP-Ndfip1 could affect cherry-PTEN nuclear transport. As shown in Fig. 3 A, recovery of nuclear PTEN fluorescence typically proceeds >800 s after nuclear bleaching. In contrast, cooverexpression of GFP-Ndfip1 strongly accelerated this process such that a plateau was reached within <500 s, with an apparent import rate greater than fourfold higher than what was observed in GFP-expressing cells (Fig. 3 E). Moreover, cells coexpressing both plasmids were readily able to reestablish the prebleach nuclear cytoplasmic signal ratios, indicating that upon Ndfip1 expression, a high fraction of PTEN in the cell is nuclear transport competent (Fig. 3 B). Next, we made use of the genetic system to test whether mouse Ndfip1 is an essential component of the PTEN import machinery and transfected Ndfip1+/+ and Ndfip1−/− primary mouse embryonic fibroblasts (MEFs) with GFP-PTEN for subsequent FRAP analysis. As shown in Fig. 3 (C and D), Ndfip1+/+ MEFs showed PTEN import curves and apparent import rates comparable with those observed in the PC3 cells. Yet, the Ndfip1−/− cells showed only negligible signal recovery. As a result (Fig. 3 E), the KO MEFs displayed only 0.1% of the normal PTEN transport rate (2.4 × 10−6/s vs. 2.4 × 10−3/s for KO and WT cells, respectively; Fig. 3, D and E). Collectively, our in vitro analysis shows that Ndfip1 is essential for PTEN import, which can be enhanced by Ndfip1 overexpression, in full agreement with our in vivo findings using Ndfip1 KO mice.
Figure 3.
Ndfip1 regulates PTEN nuclear import. (A) Nuclear FRAP analysis of cherry-PTEN shows enhanced transport upon coexpression of Ndfip1-GFP after photobleaching. Errors (SEM) are shown as shaded areas. (B) Images of typical cells from A at pre- or postbleach times and at experimental end points. Relative time points are indicated. (C) Nuclear FRAP analysis of GFP-PTEN shows the absence of transport in Ndfip1−/− MEFs compared with Ndfip1+/+ MEFs. Errors (SEM) are shown as shaded areas. (D) Images of typical cells from C at pre- or postbleach times and at experimental end points. Relative time points are indicated. (B and D) Bars, 10 µm. (E) Quantification of FRAP analyses after expression of cherry-PTEN (cPTEN) alone, with GFP (+GFP) or Ndfip1-GFP (+Ndfip1), or in the indicated cell type (cPTEN, n = 11; GFP, n = 4; Ndfip1, n = 5; Ndfip1+/+ and Ndfip1−/− MEF experiments, n = 10). Values are mean ± SEM.
Mice lacking Ndfip1 in the brain suffer larger infarct sizes after cerebral ischemia
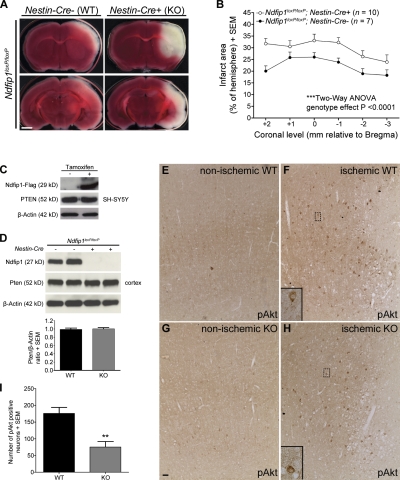
The aforementioned results indicate a causal relationship between Ndfip1 up-regulation and Pten nuclear localization in surviving neurons after ischemia. If Ndfip1 induction protects neurons from ischemic death (Sang et al., 2006), elimination of Ndfip1 in neurons would be expected to increase cell death after cerebral ischemia. To test this, we quantitated the effect of neuron-specific ablation of Ndfip1 (Ndfip1loxP/loxP;Nestin-Cre+). These mutant mice were compared with littermate controls (Ndfip1loxP/loxP;Nestin-Cre−) 24 h after ischemia, using 2,3,5-triphenyltetrazolium chloride (TTC) staining of coronal slices to map the extent of infarction between the two groups (Fig. 4 A). Results show increased infarct areas in all brain regions examined of Ndfip1loxP/loxP;Nestin-Cre+ mice compared with Ndfip1loxP/loxP;Nestin-Cre− controls (Fig. 4 B), suggesting a neuroprotective role for Ndfip1.
Figure 4.
Ndfip1 deletion increases neuron death after HI and reduces the number of pAkt-positive neurons. (A) Conditional KO of Ndfip1 increases total infarct size 24 h after HI. TTC-stained brain slices show a larger infarct area (unstained white tissue) in Ndfip1 KO (Ndfip1loxP/loxP;Nestin-Cre+) versus control littermates (Ndfip1loxP/loxP;Nestin-Cre−) for all ischemic regions examined (cortex, striatum, hippocampus, and thalamus). Bar, 2 mm. (B) The quantification of total infarct area indicates that Ndfip1 KO mice sustain larger infarct size compared with control littermates (significant genotype effect; two-way analysis of variance [ANOVA]). (C) PTEN levels in neuronal cell lines (SH-SY5Y) are unaffected by tamoxifen-induced overexpression of Ndfip1. (D) Western blot densitometry reveals deletion of Ndfip1 (Ndfip1loxP/loxP;Nestin-Cre+) does not alter Pten levels in the mouse cortex compared with littermate controls (Ndfip1loxP/loxP;Nestin-Cre−; P = 0.7392; two-tailed unpaired Student’s t test; n = 5 per genotype). (E and F) Immunostaining for pAkt in WT mice (Ndfip1loxP/loxP;Nestin-Cre−) 24 h after HI (nonischemic and ischemic cortex) shows increased numbers of pAkt-positive neurons in the periinfarct region. (G and H) In Ndfip1 KO mice (Ndfip1loxP/loxP;Nestin-Cre+), fewer pAkt-positive neurons are observed in the periinfarct region after HI. (E–H) Insets show magnifications of the boxed areas. Bar, 30 µm. (I) Quantification of pAkt levels in the ischemic cortex 24 h after HI shows that fewer neurons activate Akt in the periinfarct region in Ndfip1 KO mice. **, P < 0.005 compared with WT (two-tailed unpaired Student’s t test; n = 5 per genotype). Values are mean ± SEM throughout.
Ndfip1-mediated ubiquitination of Pten can lead to either monoubiquitination promoting nuclear accumulation or polyubiquitination promoting Pten degradation in the cytoplasm. Our results show that cytoplasmic Pten levels in normal (nonischemic) neurons appear comparable between the two Ndfip1 genotypes (Fig. 1, A and G), suggesting that the absence of Ndfip1 might not compromise levels of cytoplasmic Pten. In ischemic neurons, some cytoplasmic Pten also remains present despite Pten nuclear accumulation (Fig. 1 D), suggesting that the Ndfip1 surge in these neurons mediates Pten trafficking rather than Pten degradation. In addition, overall Pten levels appear unaltered by Ndfip1 status; this was confirmed by Ndfip1 overexpression in SH-SY5Y neuronal cells (Fig. 4 C) and also in brain lysates comparing the two Ndfip1 genotypes (Fig. 4 D). Hence, the absence of Ndfip1 in ischemia abolishes nuclear transport rather than increasing cytoplasmic Pten levels (compare Fig. 1, A and G). To examine the effects of Pten misregulation in neurons after the loss of Ndfip1 in ischemia, pAkt immunostaining was performed. We found that pAkt-positive neurons are increased in periinfarct areas of WT mice after ischemia (Fig. 4 F), indicating neuron survival (Zhao et al., 2006), but this number is significantly reduced in mice lacking Ndfip1 (Fig. 4, H and I). Thus, the loss of Ndfip1 does not totally abolish pAkt up-regulation in neurons after ischemia (Fig. 4 H) but attenuates it (Fig. 4 I), suggesting that the loss of Pten nuclear transport as a result of the absence of Ndfip1 reduces pAkt levels in neurons, a result of persistent cytoplasmic Pten. In turn, this may lead to a greater degree of cell death found in Ndfip1 mutants. Additionally, loss of nuclear Pten may also deprive neurons of other survival benefits provided by nuclear Pten, including repairing DNA and promoting genome stability (Shen et al., 2007; Song et al., 2011).
In the present study, we have uncovered an altogether unexpected facet of Pten regulation in the brain after ischemia besides its known roles in normal brain development and cancer (Chalhoub and Baker, 2009). We show that in the normal brain, the role of Pten as a guardian against proliferation is recast as a defense against neuronal apoptosis, using almost identical protein modification pathways involving Nedd4 ubiquitination. Importantly, this novel Ndfip1-mediated system equips the cell with a dynamic and reversible Pten shuttling module that is geared toward improving neuron survival. It is intriguing that nuclear Pten import, rather than cytoplasmic Pten degradation, is the preferred response to this physiological stimulus, even though similar ubiquitination pathways can mediate both processes. However, we are unable to resolve how Pten nuclear entry can elicit neuron survival responses at such rapid intervals, given that the known functions of PTEN in tumor suppression are quite delayed. It may be that cerebral ischemia/hypoxia represents a unique physiological context that accelerates PTEN-mediated signaling for cell survival (Ning et al., 2004). Nonetheless, our evidence demonstrating that Pten is not degraded in the cytoplasm but is imported into the nucleus for neuron survival presents a conceptual challenge to the existing dogma of promoting Pten inhibition for the treatment of stroke (Ning et al., 2004; Hong et al., 2006; Liu et al., 2010).
Our findings also provide the first in vivo example of an acute stimulus that can regulate nuclear trafficking of Pten. This has powerful implications for PTEN-mediated mechanisms that are known to operate widely in the organism under normal and disease settings, including development and cancer. Because lack of PTEN import has been demonstrated to be tumor promoting (Trotman et al., 2007), it will be of interest to determine whether Ndfip1 alteration is implicated in neoplasia. However, it remains to be seen how ischemic stimuli and tumorigenic stimuli can both be interpreted by the cell to initiate nuclear trafficking of Pten and how, in postmitotic neurons, this leads to survival; but in dividing cancer cells, this results in tumor suppressive function. Given that PTEN nuclear accumulation can occur under a variety of conditions, including cell cycle states and hormonal changes (Mutter et al., 2000; Ginn-Pease and Eng, 2003), the downstream consequence of nuclear PTEN is likely to be cell and context specific. In the current instance, we demonstrate that in neurons, ischemic damage is a trigger for subcellular distribution of Pten with survival benefit. However, it will be of interest to determine whether PTEN nuclear import in all other scenarios is also regulated by the Ndfip1/Nedd4 system.
Materials and methods
Animals
All procedures were approved by the Florey Neuroscience Institutes Animal Ethics Committee. C57BL/6J mice were obtained from the Australian Research Council. Emx1-Cre (C57BL/6) mice were provided by T. Iwasato (RIKEN Brain Science Institute, Saitama, Japan; Iwasato et al., 2000), and Nestin-Cre mice (B6.Cg-Tg(Nes-cre)1Kln/J) were obtained from The Jackson Laboratory.
Generation of Ndfip1 conditional KO mice
The targeting construct was produced using homologous recombineering of bacterial artificial chromosome clone RP23-65L6 in Escherichia coli according to the method of Liu et al. (2003). Using this approach, a 10-kb fragment of RP23-65L6, which included exon 1 of Ndfip1, was recombineered into the targeting plasmid pL253. A loxP site was inserted 962 bp upstream of exon 1 and the Neo selection cassette flanked by FRT sites, and a single loxP site was inserted 1,059 bp 3′ of exon 1 (Fig. S2). After electroporation into BRUCE 4 cells (a C57BL/6 embryonic stem cell line), homologously recombined embryonic stem cell clones were selected for production of germline chimeras. Conditional KO Ndfip1loxP/loxP;Emx1-Cre+ or Ndfip1loxP/loxP;Nestin-Cre+ mice were derived by crossing Ndfip1loxP/loxP (C57BL/6) homozygous mice with those carrying the Emx1-Cre transgene (Iwasato et al., 2000) or the Nestin-Cre transgene, respectively. Mice were genotyped by PCR of tail biopsies with the Ndfip1 floxed allele primers across the 5′ loxP site (forward 5′-TCATCCATGATGGATGTGTGC-3′ and reverse 5′-TTTACTCACAGACCCTGAGCC-3′) and the Cre transgene primers (forward 5′-TTTGCCTGCATTACCGGTCGATGCAAC-3′ and reverse 5′-TGCCCCTGTTTCACTATCCAGGTTACGGA-3′).
Unilateral cerebral hypoxia-ischemia (HI)
HI was induced in 4-wk-old male mice using a protocol adapted from Vannucci et al. (2001), with modifications as described. Mice were anesthetized by inhalation of isoflurane (4% induction and 2% maintenance) in a mixture of 0.05–0.1% O2 and balanced room air. A neck incision was performed, and the right common carotid artery (CCA) was exposed and dissected free of surrounding tissue and nerve before permanent double ligation with 5–0 surgical silk. The first ligation was made immediately proximal to the bifurcation of the CCA into the internal and external carotid arteries with the second ligation made ∼3 mm proximal to the bifurcation. After ligation, the CCA was inspected to verify that blood flow had ceased and the wound was sutured, and the animal was allowed to recover for 3 h with free access to food and water. To induce HI, conscious mice were subsequently exposed to 40 min of systemic hypoxia in a humidified incubator containing 8% O2/92% N2 (model 3130; Thermo Fisher Scientific). The incubator was equipped with an internal O2 sensor, and O2 concentration was continuously monitored throughout the hypoxic period. The incubator temperature was maintained between 35.5 and 35.7°C to preserve core body temperature during hypoxia (Vannucci et al., 2001). Mice were removed from the incubator after hypoxia and allowed to recover in a normoxic environment before returning to their cages with free access to food and water.
Immunohistochemistry
For Pten and pAkt immunostaining, mice were killed under deep anesthesia after HI by transcardial perfusion of PBS, pH 7.4, followed by 4% PFA in 0.1 M phosphate buffer. Brains for Pten staining were postfixed for 1 h in 4% PFA in 0.1 M phosphate buffer and then cryoprotected for 24 h in 20% sucrose in 0.1 M phosphate buffer at 4°C before coronal sectioning. Brains for pAkt staining were postfixed for 24 h in 4% PFA in 0.1 M phosphate buffer at 4°C before being paraffin embedded. For Ndfip1/TUNEL staining, mice were anesthetized with isoflurane and killed by decapitation after HI before brains were fresh frozen in Tissue-Tek (Sakura). 14 µm of perfusion-fixed sections was permeabilized with 0.3% Triton X-100 in 0.1 M phosphate buffer and blocked with 10% FBS in 0.1% Triton X-100 with 0.1 M phosphate buffer. 8 µm of paraffin-embedded coronal sections was deparaffinized and microwaved in 10 mM citrate buffer, pH 6.0, to unmask epitopes and treated with 0.03% hydrogen peroxide for 5 min to block endogenous peroxidase. Tissue was then washed in PBST (0.1 M PBS and 0.1% Triton X-100) and blocked with 10% normal horse serum in PBST. 14 µm of fresh frozen coronal sections was fixed with 4% PFA in 0.1 M phosphate buffer, blocked, and permeabilized in 10% normal goat serum with 0.3% Triton X-100 in 0.1 M phosphate buffer. Sections were incubated with primary antibodies overnight followed by appropriate secondary antibodies for 1 h at room temperature. Cell death was detected using terminal deoxynucleotidyl transferase–mediated biotinylated UTP nick-end labeling (TUNEL) as per the manufacturer’s instructions (Roche). Sections were counterstained with DAPI (1:10,000; Dako) before mounting under glass coverslips with antifade mounting reagent. For detection of pAkt, the VECTASTAIN ABC kit was used according to manufacturer’s instructions (Vector Laboratories). Fluorescent images of Pten/Ndfip1 staining were obtained at room temperature with a 40× objective (oil immersion NA 1.3) on a laser-scanning confocal microscope (FluoView FV1000; Olympus) using FV100-ASW software (Olympus). For Ndfip1/TUNEL staining, fluorescent images were captured at room temperature with a 10× objective (NA 0.03) on a fluorescent microscope (BX51; Olympus) equipped with a SPOT 2.3.1 camera (Diagnostic Instruments) and using Image-Pro Plus software (Media Cybernetics). Brightfield images of pAkt staining were obtained at room temperature using 10× (NA 0.30) and 40× (NA 1.00–0.50) objectives on a fluorescent microscope (DMLB2; Leica) equipped with a charge-coupled device camera (Optronics ORCA-R2; Hamamatsu Photonics) and using Picture Frame software (MBF Bioscience). All images were taken at 22°C. The primary antibodies used were purified rat monoclonal anti-Ndfip1 (clone 1G5; 1:1,000 or 1:200 for fresh frozen sections), rabbit monoclonal anti-PTEN (clone Y184; 1:500; Abcam), mouse monoclonal PTEN (clone 6H2.1; 1:500; Cascade BioScience), and rabbit monoclonal anti-pAkt Ser 473 (1:50; Cell Signaling Technology). The secondary antibodies used were Alexa Fluor 594–conjugated goat anti–rat IgG (1:500; Invitrogen) and Alexa Fluor 488–conjugated goat anti–rabbit IgG (1:500; Invitrogen).
Evaluation of infarct area in TTC-stained sections
Mice were anesthetized by isoflurane inhalation after HI and decapitated, and brains were removed and sectioned in a mouse coronal brain matrix at predefined 1-mm intervals. Sections were incubated in 2% TTC (Merck & Co., Inc.) for 30 min, rinsed in PBS, and then fixed in 4% PFA in 0.1 M phosphate buffer for 24 h before imaging sections on a flatbed color scanner. Infarct area was measured by planimetry of scanned sections using ImageJ software (National Institutes of Health), and the infarct area was calculated and corrected for edema according to the method of Swanson et al. (1990).
Cell culture, transfection, and plasmid constructs
HEK-293T cells were cultured in DME (Invitrogen), 10% FCS, 4 mM l-glutamine, and 50 µg/ml PenStrep. Human SH-SY5Y neuroblastoma cells were cultured in RPMI medium (Invitrogen), 15% FCS, 2 mM l-glutamine, and 50 µg/ml PenStrep. HEK-293T cells were transiently transfected with appropriate constructs using the Effectene Transfection Reagent kit according to the manufacturer’s instructions (QIAGEN). PC3 cells or primary MEFs were cultured in DME (Mediatech, Inc.), 10% FCS, 50 U/ml penicillin, and 100 µg/ml PenStrep and were transfected with appropriate constructs using Lipofectamine 2000 (for PC3; Invitrogen) or Effectene (for MEFs). Lentiviral infection was used to make stable SH-SY5Y cell lines of inducible Ndfip1-Flag. In brief, lentiviral particles were made through transfection of HEK-293T cells with packaging constructs plasmid cytomegalovirus ðR8.2 and vesicular stomatitis virus glycoprotein and the Ndfip1-Flag lentiviral plasmid using Effectene. Supernatants were harvested, and SH-SY5Y cells were infected with virus supernatant for 24 h. Successful infection was selected for with 2 mg/ml puromycin (Sigma-Aldrich) and 100 mg/ml hygromycin B (Sigma-Aldrich) to form a stable cell line for inducible Ndfip1-Flag (Howitt et al., 2009). To create N terminus Strep-Flag–tagged NDFIP1, human NDFIP1 cDNA flanked with Nhe1 and Xho1 restriction sites was generated by PCR with the primers forward 5′-GTGCTAGCATGGCGTTGGCGTTG-3′ and reverse 5′-GTCTCGAGATAAATAAAGAGAACTCTGGTCCT-3′. The purified NDFIP1 PCR fragment was digested with Nhe1 and Xho1 restriction enzymes and cloned into N-SF-TAPpcDNA3 (provided by C.J. Gloeckner, Institute of Human Genetics, Munich-Neuherberg, Germany), generating the Strep-Flag-NDFIP1 (SF-TAP-NDFIP1pcDNA3) construct. Other constructs used were GFP-PTEN in pcDNA3 (plasmid 13039; obtained from the Addgene plasmid repository), His-Ndfip1 in pcDNA3, Ndfip1-Flag in pcDNA3, PTEN WT in pcDNA3, PTEN 3A in pcDNA3, PTEN 4D in pcDNA3, His-Ubiquitin in pcDNA5, and Nedd4-1 and Nedd4-2 in pcDNA3. Cells were lysed 20 h after transfection for immunoprecipitation assays as described in the next section. PTEN WT, PTEN 3A, and PTEN 4D were provided by H. Zhu (Royal Melbourne Hospital, The University of Melbourne, Parkville, Victoria, Australia).
Protein lysate preparation and immunoprecipitation assays
Mouse brain cortices or HEK-293T and SH-SY5Y cells were lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.2, 0.15 M NaCl, 2 mM EDTA, 1% NP-40, and 0.1% SDS) with protease inhibitor cocktail (Complete, Mini; Roche) for 20 min at 4°C. Brain homogenates and cell lysates were cleared of insoluble debris by centrifugation at 13,000 rpm for 15 min at 4°C. Protein concentration of lysates was measured using the detergent-compatible protein assay according to the manufacturer’s instructions (Bio-Rad Laboratories). For precipitation experiments, protein G beads (Thermo Fisher Scientific) were used for the precipitation of antibodies. Other beads used include Strep-Tactin (QIAGEN) for Strep-Flag–tagged Ndfip1 and GFP-Trap (ChromoTek) for GFP-PTEN. For each experiment, beads were washed four times with radioimmunoprecipitation assay buffer before elution using the manufacturer’s instructions. For the ubiquitination assays, HEK-293T cells were transfected with His-Ubiquitin, Ndfip1-Flag or Ndfip1, PTEN WT, PTEN 3A, PTEN 4D, Nedd4-1, and Nedd4-2. For RNAi knockdown of Nedd4 ligases, DharmaFECT transfection reagent was used with siGENOME SMART pool NEDD4 (M-007178-02), NEDD4L (D-007187-04), and the control nontargeting siRNAi Pool #1 (D-001206-13; Thermo Fisher Scientific). PTEN, Ndfip1-Flag, and His-Ubiquitin were transfected 24 h after RNAi transfection, and ubiquitination assay was performed 48 h later. Lysates were immunoprecipitated with HisLink (Promega) under denaturing condition using 6 M guanidine HCl and N-ethylmaleimide for inhibiting the deubiquitinase enzyme. Beads were washed three times before ubiquitinated proteins were eluted using 300 mM imidazole. Eluted fractions were suspended in Laemmli buffer for SDS-PAGE and analyzed by Western blotting.
Nuclear and cytoplasmic fractionation of mouse primary neurons
Mouse primary neurons from Ndfip1loxp/loxp;Nestin-Cre− and Ndfip1loxp/loxp;Nestin-Cre+ embryonic brains (embryonic day 16 [E16]) were cultured for 21 d before being exposed to hypoxia (<1% O2 and 5% CO2 at 37°C) for 8 h in a humidified chamber (model 3130; Thermo Fisher Scientific). Fractionation of primary neurons was performed using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Western blotting
Lysates or immunoprecipitates were resolved on 10% SDS-PAGE gels followed by transfer onto Hybond-C nitrocellulose membrane (GE Healthcare). Membranes were blocked for 1 h at room temperature in 5% nonfat milk in TBS and 0.05% Tween 20. Blots were incubated overnight with primary antibodies at 4°C followed by appropriate HRP-conjugated secondaries for 1 h at room temperature. Proteins were detected using ECL reagent according to the manufacturer’s instructions (GE Healthcare) and visualized by exposure to x-ray film. The primary antibodies used were rat monoclonal anti-Ndfip1 (clone 1G5; 1:2,000), rabbit monoclonal anti-PTEN (clone Y184; 1:2,000; Abcam), mouse monoclonal anti-PTEN (clone 6H2.1; 1:500; Cascade BioScience), mouse monoclonal anti–StrepMAB-Classical (1:1,000; IBA GmbH), purified rabbit polyclonal anti–Nedd4-2 (1:1,000), mouse monoclonal anti–Nedd4-1 (clone 15; 1:1,000; BD), mouse monoclonal anti–β-actin (clone AC-40; 1:5,000; Sigma-Aldrich), and mouse monoclonal anti–Flag M2 (1:1,000; Sigma-Aldrich). The HRP-conjugated secondary antibodies used were goat polyclonal anti–rat (1:5,000 or 1:10,000; Millipore), goat polyclonal anti–rabbit (1:10,000; Millipore), and goat polyclonal anti–mouse (1:15,000; Millipore).
Photobleaching nuclear transport assays
PC3 or primary MEF cells were plated, transfected, and observed in glass-bottom dishes (MatTek Corporation) at 24 h after transfection (MEF cells) or between 12 and 14 h after transfection (PC3 cells; Trotman et al., 2007). Note that cotransfection of PC3 cells with cherry-PTEN and GFP-Ndfip1 results in cell death after 14 h. Cotransfection was validated using GFP-Ndfip1 fluorescence. FRAP assays were performed on a spinning-disk confocal microscope (UltraVIEW VoX; PerkinElmer) using the 488- and 561-nm laser lines (set between 3 and 13% power). All experiments were performed with cells growing on a heated stage at 37°C in phenol red–free DME (Invitrogen), and imaging was performed with a 60× objective (NA 1.49 oil immersion). The FRAP protocol involved 12 s of imaging at 3-s intervals of prebleach followed by bleaching and postbleach recording at 2-s intervals for the first 40 s and 10-s intervals for the remainder of the experiments, typically using between 50 and 250 ms of exposure per channel. Images were recorded at 512 × 672 pixels and a 12-bit depth. Bleaching was performed at 100% laser intensity on the entire nuclear signal, using between 8 and 20 cycles (total of >200 ms; step size 2). Mean nuclear fluorescence intensities were measured using either the FRAP Analysis or the region of interest functions of Volocity software (v. 5.3.2; PerkinElmer) for mean nuclear fluorescence calculation with background subtraction. Note that unspecific bleaching of fluorescence signal over the imaging period was not detectable. Nuclear flux rates were calculated using nonlinear curve fitting of Prism software (v. 5; GraphPad Software) to a pseudo-first order association kinetics curve using the model equation Y = Y0 + (Yindef − Y0) × (1 − e(−K × t)) to determine the apparent transport rate constant K for each series. Analyses were performed on at least 10 cells or on 4 cells for GFP control overexpression with cherry-PTEN and 5 cells for Ndfip1 coexpression, whereas experiments were reproduced in independent biological replicates.
Statistical analysis
All data are presented as mean ± SEM unless otherwise indicated. Where appropriate, data were tested for normality before statistical analysis using the D’Agostino–Pearson test. All data were analyzed using Prism software, and P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 demonstrates the specificity of the antibody Y184 for PTEN and compares it with the antibody 6H2.1 for immunostaining and Western blotting. Fig. S2 demonstrates that Ndfip1 is up-regulated in surviving neurons (TUNEL negative) of WT mice after cerebral ischemia, and the generation of Ndfip1 conditional KO mice along with a Western blot confirming the loss of Ndfip1 protein in Ndfip1loxP/loxP;Emx-Cre+ mice is also shown. Fig. S3 shows the nuclear and cytoplasmic fractionation of mouse primary neurons after treatment with hypoxia to determine Pten localization. The ubiquitination of PTEN after the knockdown of endogenous Nedd4-1 and Nedd4-2, with and without overexpression of Ndfip1, is also demonstrated. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105009/DC1.
Acknowledgments
This work was supported by the National Health and Medical Research Council and the Victorian government through the Operational Infrastructure Support Program.
Footnotes
Abbreviations used in this paper:
- CCA
- common carotid artery
- HI
- hypoxia-ischemia
- KO
- knockout
- MEF
- mouse embryonic fibroblast
- pAkt
- phosphorylated Akt
- RCCAo
- right carotid artery occlusion
- WT
- wild type
References
- Backman S.A., Stambolic V., Suzuki A., Haight J., Elia A., Pretorius J., Tsao M.S., Shannon P., Bolon B., Ivy G.O., Mak T.W. 2001. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29:396–403 10.1038/ng782 [DOI] [PubMed] [Google Scholar]
- Chalhoub N., Baker S.J. 2009. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 4:127–150 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., El-Hayek Y.H., Gomez E., Wan Q. 2007. Phosphatase PTEN in neuronal injury and brain disorders. Trends Neurosci. 30:581–586 10.1016/j.tins.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Drinjakovic J., Jung H., Campbell D.S., Strochlic L., Dwivedy A., Holt C.E. 2010. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 65:341–357 10.1016/j.neuron.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby R., Baker S.J. 2008. PTEN signaling in brain: Neuropathology and tumorigenesis. Oncogene. 27:5416–5430 10.1038/onc.2008.239 [DOI] [PubMed] [Google Scholar]
- Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., Rotin D. 2008. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. USA. 105:8585–8590 10.1073/pnas.0803233105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn-Pease M.E., Eng C. 2003. Increased nuclear phosphatase and tensin homologue deleted on chromosome 10 is associated with G0-G1 in MCF-7 cells. Cancer Res. 63:282–286 [PubMed] [Google Scholar]
- Hobert J.A., Eng C. 2009. PTEN hamartoma tumor syndrome: An overview. Genet. Med. 11:687–694 10.1097/GIM.0b013e3181ac9aea [DOI] [PubMed] [Google Scholar]
- Hong K.W., Lee J.H., Kima K.Y., Park S.Y., Lee W.S. 2006. Cilostazol: Therapeutic potential against focal cerebral ischemic damage. Curr. Pharm. Des. 12:565–573 10.2174/138161206775474323 [DOI] [PubMed] [Google Scholar]
- Howitt J., Putz U., Lackovic J., Doan A., Dorstyn L., Cheng H., Yang B., Chan-Ling T., Silke J., Kumar S., Tan S.S. 2009. Divalent metal transporter 1 (DMT1) regulation by Ndfip1 prevents metal toxicity in human neurons. Proc. Natl. Acad. Sci. USA. 106:15489–15494 10.1073/pnas.0904880106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T., Datwani A., Wolf A.M., Nishiyama H., Taguchi Y., Tonegawa S., Knöpfel T., Erzurumlu R.S., Itohara S. 2000. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 406:726–731 10.1038/35021059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.H., Zhu X., Zhang J., Knoop L.L., Tharp R., Smeyne R.J., Eberhart C.G., Burger P.C., Baker S.J. 2001. Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat. Genet. 29:404–411 10.1038/ng781 [DOI] [PubMed] [Google Scholar]
- Kwon C.H., Luikart B.W., Powell C.M., Zhou J., Matheny S.A., Zhang W., Li Y., Baker S.J., Parada L.F. 2006. Pten regulates neuronal arborization and social interaction in mice. Neuron. 50:377–388 10.1016/j.neuron.2006.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Zhao H., Maier C.M., Steinberg G.K. 2009. The protective effect of early hypothermia on PTEN phosphorylation correlates with free radical inhibition in rat stroke. J. Cereb. Blood Flow Metab. 29:1589–1600 10.1038/jcbfm.2009.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qu Y., Mao M., Zhang X., Li J., Ferriero D., Mu D. 2009. Involvement of the PTEN-AKT-FOXO3a pathway in neuronal apoptosis in developing rat brain after hypoxia-ischemia. J. Cereb. Blood Flow Metab. 29:1903–1913 10.1038/jcbfm.2009.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li L., Zhang Q., Chang N., Wang D., Shan Y., Li L., Wang H., Feng H., Zhang L., et al. 2010. Preservation of GABAA receptor function by PTEN inhibition protects against neuronal death in ischemic stroke. Stroke. 41:1018–1026 10.1161/STROKEAHA.110.579011 [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccario H., Perera N.M., Gray A., Downes C.P., Leslie N.R. 2010. Ubiquitination of PTEN (phosphatase and tensin homolog) inhibits phosphatase activity and is enhanced by membrane targeting and hyperosmotic stress. J. Biol. Chem. 285:12620–12628 10.1074/jbc.M109.072280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Dixon J.E. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273:13375–13378 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Mutter G.L., Lin M.C., Fitzgerald J.T., Kum J.B., Eng C. 2000. Changes in endometrial PTEN expression throughout the human menstrual cycle. J. Clin. Endocrinol. Metab. 85:2334–2338 10.1210/jc.85.6.2334 [DOI] [PubMed] [Google Scholar]
- Nardella C., Carracedo A., Salmena L., Pandolfi P.P. 2010. Faithfull modeling of PTEN loss driven diseases in the mouse. Curr. Top. Microbiol. Immunol. 347:135–168 10.1007/82_2010_62 [DOI] [PubMed] [Google Scholar]
- Ning K., Pei L., Liao M., Liu B., Zhang Y., Jiang W., Mielke J.G., Li L., Chen Y., El-Hayek Y.H., et al. 2004. Dual neuroprotective signaling mediated by downregulating two distinct phosphatase activities of PTEN. J. Neurosci. 24:4052–4060 10.1523/JNEUROSCI.5449-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon S.M., Waite K.A., Eng C. 2008. The nuclear affairs of PTEN. J. Cell Sci. 121:249–253 10.1242/jcs.022459 [DOI] [PubMed] [Google Scholar]
- Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. 2004. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 303:1179–1181 10.1126/science.1092089 [DOI] [PubMed] [Google Scholar]
- Sang Q., Kim M.H., Kumar S., Bye N., Morganti-Kossman M.C., Gunnersen J., Fuller S., Howitt J., Hyde L., Beissbarth T., et al. 2006. Nedd4-WW domain-binding protein 5 (Ndfip1) is associated with neuronal survival after acute cortical brain injury. J. Neurosci. 26:7234–7244 10.1523/JNEUROSCI.1398-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin-Whyatt L., Dalton H.E., Foot N., Kumar S. 2006. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 28:617–628 10.1002/bies.20422 [DOI] [PubMed] [Google Scholar]
- Shen W.H., Balajee A.S., Wang J., Wu H., Eng C., Pandolfi P.P., Yin Y. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 128:157–170 10.1016/j.cell.2006.11.042 [DOI] [PubMed] [Google Scholar]
- Song M.S., Carracedo A., Salmena L., Song S.J., Egia A., Malumbres M., Pandolfi P.P. 2011. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 144:187–199 10.1016/j.cell.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V., Suzuki A., de la Pompa J.L., Brothers G.M., Mirtsos C., Sasaki T., Ruland J., Penninger J.M., Siderovski D.P., Mak T.W. 1998. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 95:29–39 10.1016/S0092-8674(00)81780-8 [DOI] [PubMed] [Google Scholar]
- Swanson R.A., Morton M.T., Tsao-Wu G., Savalos R.A., Davidson C., Sharp F.R. 1990. A semiautomated method for measuring brain infarct volume. J. Cereb. Blood Flow Metab. 10:290–293 10.1038/jcbfm.1990.47 [DOI] [PubMed] [Google Scholar]
- Trotman L.C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N.P., Carver B.S., Cordon-Cardo C., Erdjument-Bromage H., et al. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 128:141–156 10.1016/j.cell.2006.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucci S.J., Willing L.B., Goto S., Alkayed N.J., Brucklacher R.M., Wood T.L., Towfighi J., Hurn P.D., Simpson I.A. 2001. Experimental stroke in the female diabetic, db/db, mouse. J. Cereb. Blood Flow Metab. 21:52–60 10.1097/00004647-200101000-00007 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Ramaswamy S., Nakamura N., Sellers W.R. 2000. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 20:5010–5018 10.1128/MCB.20.14.5010-5018.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Grossman S.R., Takahashi Y., Rokas M.V., Nakamura N., Sellers W.R. 2001. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 276:48627–48630 10.1074/jbc.C100556200 [DOI] [PubMed] [Google Scholar]
- Wang X., Trotman L.C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., et al. 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 128:129–139 10.1016/j.cell.2006.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi Y., Wang J., Huang G., Jiang X. 2008. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem. J. 414:221–229 10.1042/BJ20080674 [DOI] [PubMed] [Google Scholar]
- Zhao H., Sapolsky R.M., Steinberg G.K. 2006. Phosphoinositide-3-kinase/akt survival signal pathways are implicated in neuronal survival after stroke. Mol. Neurobiol. 34:249–270 10.1385/MN:34:3:249 [DOI] [PubMed] [Google Scholar]