Electron microscopy and crystallography studies of α4β7 integrin reveal the mechanism by which this atypical integrin enables rolling adhesion prior to integrin activation.
Abstract
The lymphocyte homing receptor integrin α4β7 is unusual for its ability to mediate both rolling and firm adhesion. α4β1 and α4β7 are targeted by therapeutics approved for multiple sclerosis and Crohn’s disease. Here, we show by electron microscopy and crystallography how two therapeutic Fabs, a small molecule (RO0505376), and mucosal adhesion molecule-1 (MAdCAM-1) bind α4β7. A long binding groove at the α4–β7 interface for immunoglobulin superfamily domains differs in shape from integrin pockets that bind Arg-Gly-Asp motifs. RO0505376 mimics an Ile/Leu-Asp motif in α4 ligands, and orients differently from Arg-Gly-Asp mimics. A novel auxiliary residue at the metal ion–dependent adhesion site in α4β7 is essential for binding to MAdCAM-1 in Mg2+ yet swings away when RO0505376 binds. A novel intermediate conformation of the α4β7 headpiece binds MAdCAM-1 and supports rolling adhesion. Lack of induction of the open headpiece conformation by ligand binding enables rolling adhesion to persist until integrin activation is signaled.
Introduction
Among adhesion molecule families, integrins are striking for the diversity of the ligands they recognize, and the types of adhesion that they mediate. This diversity is reflected not only in the large number of 18 α and 8 β subunits, but also in the low sequence identity of 22% among α subunits belonging to six different integrin families (Fig. S1 A). Despite crystal structures for three of the 24 different integrin heterodimers (Xiao et al., 2004; Zhu et al., 2008; Xiong et al., 2009; Xie et al., 2010), much of the structural basis for integrin functional diversity remains to be described.
The α4 integrin family includes one of the most broadly expressed integrins, α4β1, and two of the most specialized integrins, α4β7 and α9β1. Unique among integrins, α4β7 is a homing receptor that targets lymphocytes and specific leukocyte subsets in the bloodstream to mucosal tissues, especially the gut (Springer, 1994). The primary ligand for α4β7-mediated homing is mucosal adhesion molecule-1 (MAdCAM-1), an addressin with two immunoglobulin superfamily (IgSF) domains and a mucin-like stalk that is specifically expressed on Peyer’s patch high endothelial venules and postcapillary venules in lamina propria (Streeter et al., 1988; Berlin et al., 1993; Briskin et al., 1993). In the multistep model of leukocyte binding to endothelium and migration into tissues, it is generally selectins that mediate tethering and rolling on the vessel wall and integrins that mediate subsequent firm adhesion and migration (Springer, 1994; von Andrian and Engelhardt, 2003). The largest exception to this rule is integrin α4β7, which clearly mediates rolling as well as firm adhesion in vivo as it functions as a homing receptor (Bargatze et al., 1995).
Integrin α4β1 is expressed on many connective tissue cells, as well as on lymphocytes and monocytes. α4β1 and its ligand, vascular cell adhesion molecule-1 (VCAM-1), have roles in vascular and heart development (Bouvard et al., 2001), atherosclerosis (Cybulsky and Gimbrone, 1991), co-stimulation of immune responses, and lymphocyte and monocyte trafficking in inflammation (von Andrian and Engelhardt, 2003). VCAM-1 has seven IgSF domains, the first two of which are 25% identical to those of MAdCAM-1. α4β1 and α4β7 also bind fibronectin through an LDV motif in the alternatively spliced connecting III segment (IIICS) distinct from the RGD motif in Fn3 domain 10 of fibronectin recognized by integrins αVβ3, αIIbβ3, and α5β1. α4β1 binds better to VCAM-1 and fibronectin than to MAdCAM-1, and α4β7 binds to MAdCAM-1 better than or similarly to VCAM-1 and fibronectin (Andrew et al., 1994; Strauch et al., 1994; Newham et al., 1997; Day et al., 2002).
Antagonists of α4 integrins represent an important new class of therapeutics for autoimmune diseases (Davenport and Munday, 2007). The humanized antibody natalizumab to the integrin α4 subunit is approved for both relapsing multiple sclerosis (MS) and Crohn’s disease (Miller et al., 2003; Sandborn et al., 2005). However, use of natalizumab is limited by progressive multifocal leukoencephalopathy, caused by a polyoma-like virus that is latent in the majority of adults and can emerge after suppression of the adaptive immune response (Major, 2010).
Greater selectivity may be achieved by only targeting α4β7 or α4β1. Act-1 antibody (Lazarovits et al., 1984), which binds α4β7 and not α4β1 or αEβ7 (Schweighoffer et al., 1993), disrupts the interaction between α4β7 and MAdCAM-1, and rapidly resolves chronic colitis in an animal model (Hesterberg et al., 1996). Humanized Act-1, vedolizumab, demonstrates efficacy in clinical trials for ulcerative colitis and Crohn’s disease (Hesterberg et al., 1996; Feagan et al., 2005; Davenport and Munday, 2007; Feagan et al., 2008; Soler et al., 2009). Interestingly, Act-1/vedolizumab selectively blocks α4β7 binding to MAdCAM-1 and fibronectin but not to VCAM-1 (Soler et al., 2009).
Small molecule antagonists of α4β1/α4β7 are in trials for inflammatory bowel disease, asthma, and multiple sclerosis (Davenport and Munday, 2007). The most widely developed class of antagonist is based on a phenylalanine moiety with a free carboxyl group, e.g., RO0505376 (Davenport and Munday, 2007). A structure of a small molecule antagonist bound to α4β7 would be enormously useful for development of α4β7-selective and α4β1-selective antagonists for inflammatory bowel and central nervous system diseases, respectively.
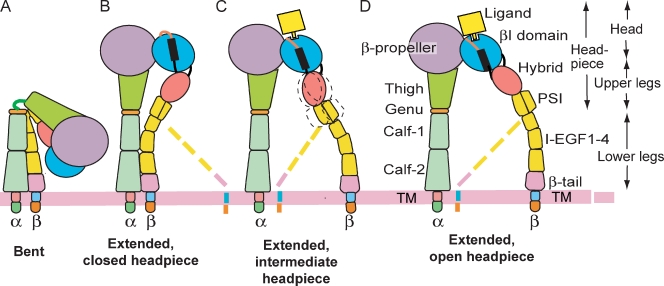
Structural studies on integrins have revealed three overall conformational states, and correlated them with ligand binding or affinity state (Fig. 1, A, B, and D; Luo et al., 2007). Crystal, small-angle x-ray scattering (SAXS), as well as EM studies on liganded and unliganded integrin ectodomains or headpieces, and on function-perturbing Fab-bound integrins using αVβ3, αIIbβ3, α5β1, and αXβ2, have shown that the bent and the extended-closed headpiece conformations have low affinity for ligand, whereas the extended-open headpiece conformation has high affinity (Takagi et al., 2002, 2003; Mould et al., 2003; Luo et al., 2004; Xiao et al., 2004; Iwasaki et al., 2005; Springer et al., 2008; Zhu et al., 2008, 2010; Chen et al., 2010; Xie et al., 2010). Although not all groups have seen integrin extension (Adair et al., 2005; Xiong et al., 2009), extension and headpiece opening are being increasingly described as important in activation by multiple independent groups (Iwasaki et al., 2005; Askari et al., 2010; Kamata et al., 2010; Ye et al., 2010).
Figure 1.
Schematic of integrin domain organization and conformational states. The pink loop and black bar in B–D represent the β6-α7 loop and α7-helix of the βI domain, respectively. Broken lines symbolize lower β leg flexibility. The intermediate headpiece state is shown here for the first time.
It has been hypothesized that rolling adhesion is mediated by a closed-headpiece, low-affinity state of α4β7, whereas firm adhesion is mediated by an open-headpiece, high-affinity state of α4β7 (Chen et al., 2003). Both types of adhesion require an intact metal ion–dependent adhesion site (MIDAS) in the βI domain that is hypothesized to hold an Mg2+ ion that coordinates to a critical Asp in IgSF domain 1 of MAdCAM-1 (Newham et al., 1997; Green et al., 1999). Metal binding sites on either side of the MIDAS, the synergistic metal ion binding site (SyMBS), and adjacent to MIDAS (ADMIDAS), have opposing, bipolar effects (Chen et al., 2003). SyMBS mutations result in rolling adhesion, and ADMIDAS mutations result in firm adhesion, no matter whether Ca2+, Ca2+/Mg2+, Mg2+, or Mn2+ are present.
Studies on RGD-binding integrins αVβ3, αIIbβ3, and α5β1 have shown that ligand binding induces headpiece opening (Takagi et al., 2002, 2003; Mould et al., 2003; Xiao et al., 2004; Zhu et al., 2010). This is hypothesized to be incompatible with rolling adhesion; if MAdCAM-1 binding induced the open-headpiece conformation of α4β7, this would be predicted to give firm adhesion. Here, we investigate the specializations that enable α4β7 to mediate both rolling adhesion and firm adhesion. We describe EM and crystal structures of the α4β7 headpiece in complex with Act-1 and natalizumab Fab and a ligand-mimetic antagonist. The shape of the ligand-binding pocket differs from previously described integrins, and is specialized for binding IgSF domains. Other specialized structural features include a novel intermediate conformation of the headpiece that is important in rolling adhesion, and an auxiliary MIDAS residue that is important for binding to MAdCAM-1 in Mg2+.
Results
Overall α4β7 headpiece structure
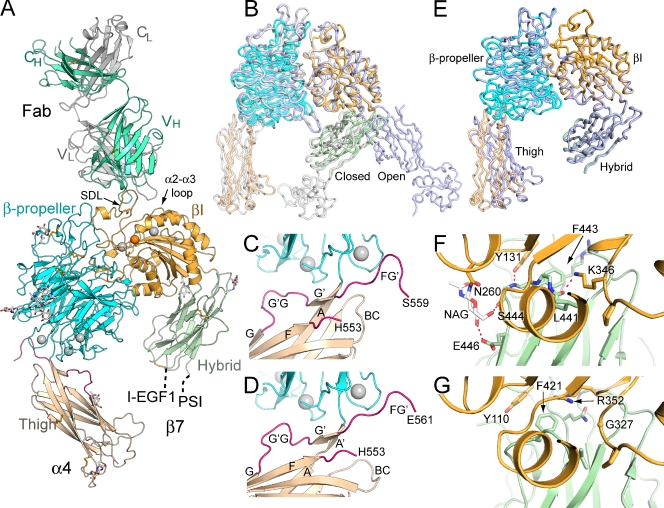
A headpiece fragment of α4β7 co-crystallized with Act-1 Fab and diffracted to 3.15 Å (Fig. 2 and Table 1). The structure was solved by multi- and single-wavelength anomalous diffraction at 3.5 and 3.6 Å. Phases were improved by multicrystal averaging using lattices with two or four headpiece–Fab complexes per asymmetric unit (Table 1). Final structures at 3.15 and 3.1 Å resolutions are well refined with R free of 25.1 and 23.1%, respectively.
Figure 2.
The α4β7 headpiece crystal structure. (A) Overall structure of the α4β7 headpiece complex with Act-1 Fab. Ca2+ and Mg2+ ions are silver and orange spheres, respectively. Disulfide bonds are shown in orange and glycans with white carbons are shown as sticks. (B) Superposition of the closed (white; Zhu et al., 2008) and open (light blue; Springer et al., 2008) headpiece of αIIbβ3 on α4β7 (colored domains). (C and D) Two distinct examples of the β-propeller domain (cyan) interface with the thigh domain (wheat), shown in the same orientation. FG′ and G′G loops are hot pink. (E) Superposition of the two most dissimilar α4β7 molecules. (F and G) The interface between the βI (orange) and hybrid (green) domains in closed conformations of α4β7 (F) and αIIbβ3 (G).
Table 1.
X-ray diffraction data and refinement
| Criteria | Nativelattice A | Se-SADlattice A | Se-MAD lattice B | RO0505376lattice C | ||
| Se peak | Se inflection | Se remote | ||||
| Wavelength (Å) | 1.00695 | 0.97945 | 0.97945 | 0.9796 | 0.94957 | 1.03337 |
| Space group | P21 | P21 | P21 | P21 | ||
| Cell a, b, c (Å) | 137.8, 122.7, 158.1 | 136.9, 121.8, 157.8 | 158.4, 123.2, 240.6 | 127.6, 123.5, 154 | ||
| α, β, γ (°) | 90, 115.4, 90 | 90, 115.4, 90 | 90, 101.8, 90 | 90, 112.6, 90 | ||
| Resolution (Å) | 50-3.15 | 50-3.6 | 50-3.5 | 50-3.1 | ||
| Reflections (total/unique) | 310,175/82,079 | 410,287/105,656 | 873,259/114,453 | 433,115/114,453 | 433,092/114,412 | 286,405/79,083 |
| Completeness (%) | 99.3/99.5a | 98.9/99.0a | 99.9/99.9a | 99.9/99.9a | 99.9/99.8a | 98.8/97.8a |
| I/s (I) | 10.3/1.9a | 7.7/2.1a | 15.6/3.5a | 10.1/1.9a | 10.0/2.2a | 5.4/1.6a |
| Rmerge (%)b | 8.9/91.5a | 13.0/70.2a | 11.5/64.0a | 12.0/73.1a | 10.7/60.2a | 16.1/84.5a |
| Refinement | ||||||
| Rworkc | 0.220 | 0.197 | ||||
| Rfreed | 0.251 | 0.231 | ||||
| RMSD | ||||||
| Bond (Å) | 0.003 | 0.005 | ||||
| Angle (Å) | 0.689 | 0.791 | ||||
| Complex/asymmetric unit | 2 | 2 | 4 | 2 | ||
| Ramachandran (percentage favored/allowed/outlier)e | 91.4/8.0/0.6 | 91.8/7.6/0.6 | ||||
| PDB accession no. | 3V4P | 3V4V |
Numbers correspond to the last resolution shell.
Rmerge = Σh Σi |Ii (h) − < I(h) > |/Σh Σi Ii (h), where Ii (h) and < I(h) > are the ith and mean measurement of the intensity of reflection h.
Rwork = Σh || Fobs (h)| − | Fcalc(h)||/Σ h | Fobs (h)|, where Fobs (h) and Fcalc (h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 5% subset of the data set excluded from refinement.
Residues in favorable, allowed, and outlier regions of the Ramachandran plot as reported by MOLPROBITY (Davis et al., 2007).
The crystal structure contains the α4 β-propeller and thigh domains, the β7 βI and hybrid domains, and Fab (Figs. 2 A and S2). The β7 PSI and I-EGF1 domains are present in the crystallization construct and in crystals, as confirmed by the molecular weight of the β7 subunit in SDS-PAGE. However, weak density for the PSI domain and lack of density for the I-EGF1 domain suggest flexibility at their connections to the N and C termini of the β7 hybrid domain, respectively (Fig. 2 A). Superposition of αIIbβ3 headpieces in the closed and open conformations demonstrates that the α4β7 headpiece is in the closed conformation (Fig. 2 B).
Whereas many integrins that lack αI domains contain a furin cleavage site in the α subunit calf-2 domain, α4 is unusual for lacking such a site and containing a furin cleavage site in its thigh domain. The furin site was deleted in the crystallization construct with an α4 R558A mutation, and lack of proteolytic processing of α4 was confirmed by SDS-PAGE.
Despite mutational abolition of cleavage of the FG loop, different examples of α4β7 molecules in crystal lattices show up to a 17° difference in orientation at the β-propeller–thigh interface (Fig. 2 E). The FG loop forms a long, flexible bumper-like interface with the β-propeller domain, allowing it to move in concert with change in orientation between the β-propeller and thigh domains that may be important in rolling adhesion (Fig. 2, C and D). During reorientation of the β-propeller and thigh domains by torsion of the thigh A strand (Fig. 2 C), which can result in separation into A and A′ strands (Fig. 2 D), remodeling of the FG′ and G′G loop segments prevents clashes between the two domains and enables maintenance of the hydrogen bonds between the A and G′ (Fig. 2 C) or A′ and G′ β strands (Fig. 2 D).
The interface between the βI and hybrid domains appears unusually stable in α4β7. Interdomain orientation varied only 4 ± 2° (mean and SD) among four independent α4β7 molecules (Fig. 2 E). This rigid orientation was enforced by hydrogen bonds that were invariant among the four examples of α4β7 molecules (Fig. 2 F). Lys-346 has a key role in the interface. This βI residue hydrogen bonds to the backbone of hybrid domain residue 441 and forms a π-cation bond to residue Phe-443. Furthermore, the aliphatic portion of the βI Lys-346 side chain is buried by the side chain of Leu-441 in the hybrid domain. Moreover, the side chain of βI residue Tyr-131 hydrogen bonds to the backbone of residue 444, and the Tyr-131 and Phe-443 side chains pack at right angles to one another, optimizing interaction of their quadrupoles (Chakrabarti and Bhattacharyya, 2007). An interesting interaction with the hybrid domain is mediated by the N-linked glycan attached to βI residue Asn-260 (Fig. 2 F). This N-linked glycan covers one flank of the hybrid domain, and its first N-acetyl glucosamine residue hydrogen bonds to Ser-444 and Glu-446 (Fig. 2 F).
In αIIbβ3, Arg-352, the last residue of the βI domain, is key in the βI hybrid domain interface and gives it a flatter shape than in β7 (Fig. 2 G). β7 Lys-346 and β3 Arg-352 approach the same part of the βI hybrid interface from different directions and give it a different shape. Another difference is the lack of a glycosylation site in β3 equivalent to that at N260 in β7. The direction of hybrid domain swing-out is such that even upon partial opening, the hybrid domain moves away from βI residues N260 and K346, isolating them from the interdomain interface. In contrast, in swing-out in the β3 subunit, Arg-352 as the last βI residue moves along with the hybrid domain, and reorients (Xiao et al., 2004).
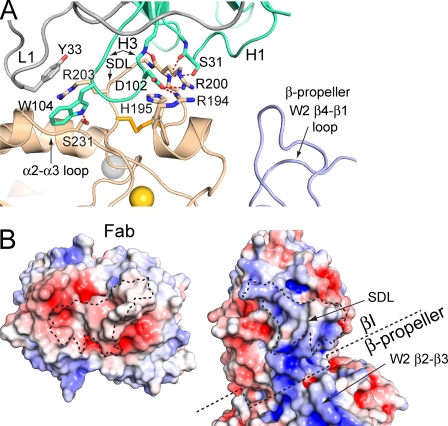
Act-1 Fab complex
Act-1 recognizes an extended, disulfide-bonded loop known as the specificity-determining loop (SDL) that projects from the β7 βI domain near its MIDAS at the interface with the α4 β-propeller domain (Fig. 2 A). The Fab contacts just the β7 subunit and buries a total solvent-accessible surface area of 1,300 Å2. Most of the area on β7 is contributed by the SDL (88%) with the remainder centered at Ser-231 in the βI domain α2-α3 loop (Figs. 2 A and 3 A). Residues 194–205 of the SDL are buried in a cleft formed by complementarity-determining regions (CDRs) H1 and H3 of the heavy chain (Fig. 3 A). CDR L1 of the light chain forms much of the contact with the βI domain α2-α3 loop. The Fab interface is rich with hydrogen bonds to both integrin side chains and backbone. SDL residue Arg-200 has a prominent role at the center of the interface (Fig. 3 A). Furthermore, the β7 Arg-203 guanido group stacks between H3 residue Trp-104, which protrudes from CDR H3, and L1 residue Tyr-33 to form two π-cation bonds (Fig. 3 A). Moreover, a basic patch on the SDL including residues Arg-194, His-195, Arg-200, and Arg-203 is complemented by a negatively charged binding interface on the Fab, as shown by electrostatic surfaces (Fig. 3 B).
Figure 3.
The Act-1 Fab complex. (A) The integrin–Fab interface showing the most significant side chains and their hydrogen bonds. (B) Open-book view of the Act-1–integrin complex. The contact areas have dashed outlines. The approximate β-propeller domain boundary with the βI domain is also shown with dashed lines. The electrostatic surface (−5 [red] to +5 [blue] kT/e) was calculated using the Adaptive Poisson-Boltzmann Solver software plug-in for PyMOL (Baker et al., 2001).
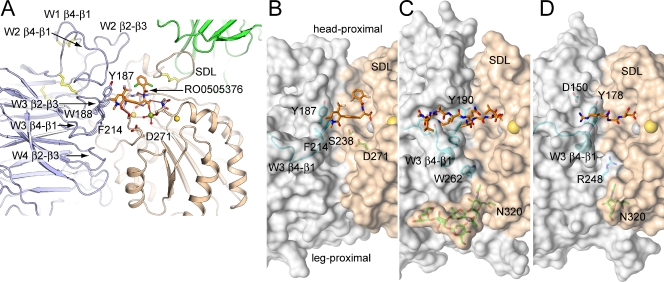
Ligand-binding site
The β-propeller domain of α4 differs from those of the previously characterized αIIb, αV, and αX integrins, particularly in the loops on the face of the β-propeller domain that bind the βI domain and form the ligand-binding site (Fig. 4). These loops are named after the β strands they connect, and the β-sheet, i.e., propeller blade, in which they are present. Each blade has four antiparallel β strands in a W topology, where each stroke of the letter W corresponds to one β strand. The propeller has seven blades, W1–W7. The α4 β-propeller W1 β4-β1 loop, which connects W1 and W2, contains a disulfide bond that is absent in αIIb, αV, and αX (Fig. 4 A). The W1 β4-β1 loop forms important contacts with two adjacent loops that help form the α4β7 ligand-binding pocket, the W2 β2-β3 loop, and the W2 β4-β1 loop (Fig. 4 A). The lengths, conformations, and locations on the β-propeller surface of these three loops differ greatly among α4, αV, αIIb, and αX (Fig. S1, B–D; and Fig. S2 A). The short W3 β2-β3 loop, in the center of the ligand-binding site (Fig. 4 A), is the only loop with a well-conserved sequence and backbone (Figs. S1 and S2). However, because of its surroundings, its Tyr-187 side chain differs from Tyr-190 in αIIb in both being higher on the α4 ligand-binding wall and further from the β subunit wall, helping to create a wider crevice in α4β7 (Fig. 4, B and C). The W3 β4-β1 loop also differs markedly in conformation between α4 and other known integrin subunits. In αV and αIIb, three residues in the W3 β4-β1 loop form a 310 helix, and the loop moves ∼6 Å toward the ligand-binding pocket and narrows the crevice in the middle (Fig. 4, C and D). In α4, this loop contains no secondary structure and locates far from the binding interface (Fig. 4 B). Finally, the α4 W4 β2-β3 loop (Fig. 4 A) is one residue shorter than in αIIb and αV and lacks the Trp-262 in αIIb or Arg-248 in αV that protrudes into the crevice (Fig. 4, B–D).
Figure 4.
Integrin ligand-binding sites. (A) The loops in the α4 β-propeller and β7 I domain that build the ligand-binding site. (B–D) Transparent molecular surfaces are shown with key residues in stick and loops in cartoon. The surfaces are light gray (α subunit β-propeller domains) or wheat (β subunit βI domains). Ligands are shown in their entirety (α4β7), or key residues of peptides are shown (αIIbβ3 and αVβ3). Metal ions are shown as spheres. (B) α4β7 bound to small molecule antagonist. (C) αIIbβ3 bound to fibrinogen γ C-terminal peptide (PDB ID 2VDO) (Springer et al., 2008). (D) αVβ3 bound to an RGD-mimetic (Xiong et al., 2009).
The net effect of the α4 β-propeller substitutions is to create a ligand-binding site with an overall different shape than those of αIIbβ3 and αVβ3 (Fig. 4, B–D). The α4β7 ligand-binding site has an approximately straight ∼40 Å–long crevice running along the α–β subunit interface that is ∼10 Å wide and ∼10 Å deep (Fig. 4 B). In contrast, the crevices in αIIbβ3 and αVβ3 are narrowed by the specializations described in the proceeding paragraph. Furthermore, the α4 wall completely lacks a groove cut through it, which in αIIb forms the binding site for the N-terminal portion of the fibrinogen γC LGGAKQAGDV peptide, which thus binds in a crevice essentially at a right angle to the crevice in α4β7 (Fig. 4 C). The ligand-binding wall of αV has a groove cut through it that is similar to that in αIIb, and the W3 β4-1 loop protrudes into the αV–β3 interface near the RGD-binding site (Fig. 4 D).
The “head-proximal” end of the groove near the W2 β2-β3 loop and “leg-proximal” end near the W4 β2-β3 loop are also wider in α4β7 (Fig. 4, A–D). The β7 SDL creates a wide head-proximal end because, compared with β3, its disulfide-bonded loop with a Pro lacks a 310 helix, and thus extends away from α (Fig. 4 B) rather than toward α (Fig. 4, C and D). Moreover, together with the protruding Trp-262 residue in αIIb and Arg-248 residue in αV, N-glycosylation of Asn-320 in the α6-helix of β3 blocks the leg-proximal end of the groove site in αVβ3 and αIIbβ3 integrins (Fig. 4, C and D).
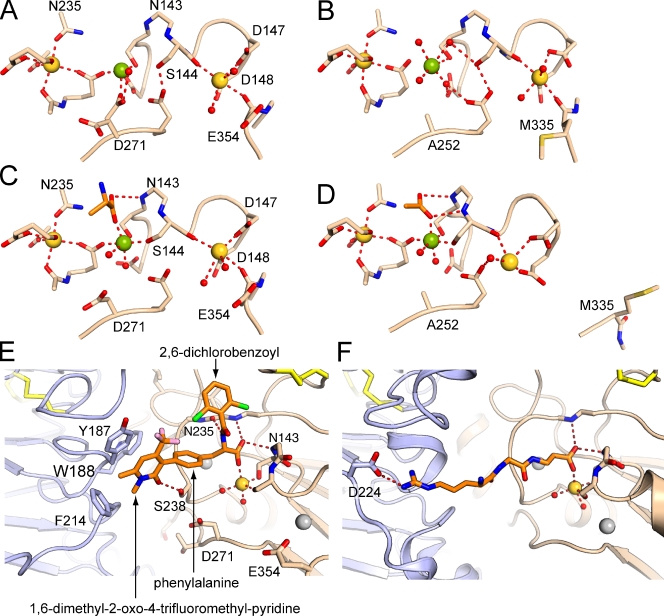
An extra MIDAS-coordinating residue in β7
Prominent densities at the SyMBS, MIDAS, and ADMIDAS in the β7 I domain show that all three sites are occupied by metal ions (Figs. 5 A and S3). These derive from the protein preparation that contained 1 mM Mg2+ and 1 mM Ca2+. The coordination at the MIDAS and ADMIDAS, and the positions in the βI domain of the β1-α1 loop, α1 helix, β6-α7 loop, and α7 helix are similar to those in closed αIIbβ3 (Fig. 5 B) and not in open αIIbβ3 (Fig. 5 D). These findings agree with the closed position of the β7 hybrid domain, and confirm that the β7 I domain adopts the closed conformation.
Figure 5.
βI domain metal-binding sites and rearrangements upon small molecule antagonist binding. (A and C) α4β7 metal-binding sites in the absence (A) or presence (C) of a small molecule antagonist (with orange carbons). (B and D) Metal-binding sites in αIIbβ3 in the closed conformation (B; PDB accession no. 3NID; Zhu et al., 2010) and open conformation (C; PDB accession no. 2VDL; Springer et al., 2008). A–D were superimposed on the βI domain and then displaced vertically or horizontally on the page. (E) RO0505376 binding to α4β7. (F) RGD peptide binding to αIIbβ3 (PDB accession no. 2VDR; Springer et al., 2008). E and F are shown in the same orientation.
Most interestingly, the β7 subunit, unlike β3, has an extra Asp residue at the MIDAS (Fig. 5, A and B). Density for the side chain of Asp-271 clearly shows two conformations, one of which forms a direct coordination to the MIDAS Mg2+ with an O–Mg2+ distance of 2.2 Å (Figs. 5 A and S3). An extra Glu side chain is also nearby the ADMIDAS, Glu-354, but does not appear to be close enough to form a direct coordination.
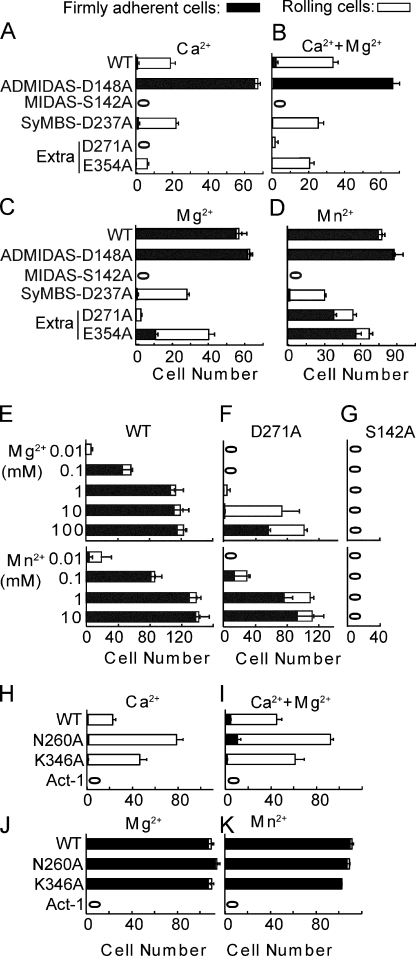
The functional significance of residues Asp-271 and Glu-354 was tested mutationally and compared with representative SyMBS, MIDAS, and ADMIDAS mutations (Fig. 6). As described previously (Chen et al., 2003) in Ca2+ or Ca2+ + Mg2+, wild-type α4β7 mediates rolling adhesion (Fig. 6, A and B), whereas in Mg2+ alone and Mn2+ it mediates firm adhesion (Fig. 6, C and D). The MIDAS S142A mutation completely abolishes rolling and firm adhesion in all metal ion conditions (Fig. 6, A–D). Compared with wild type, the ADMIDAS D148A mutation in Ca2+ and Ca2+ + Mg2+ mediates firm adhesion instead of rolling adhesion (Fig. 6, A and B). In contrast, compared with wild type, the SyMBS D237 mutation in Mg2+ and Mn2+ mediates rolling adhesion instead of firm adhesion (Fig. 6, C and D; Chen et al., 2003). Mutation of the extra ADMIDAS residue Glu-354 had a marked effect, decreasing rolling adhesion in Ca2+ and Ca2+ + Mg2+, decreasing firm adhesion in Mg2+ and Mn2+, and partially converting firm adhesion to rolling adhesion (Fig. 6, A–D). Mutation of extra MIDAS residue Asp-271 had an even more striking phenotype. The D271A mutation eliminated rolling adhesion in Ca2+ (Fig. 6 A), greatly diminished rolling in Ca2+ + Mg2+ (Fig. 6 B), and greatly diminished adhesion in 1 mM Mg2+ (Fig. 6 C). However, in 0.5 mM Mn2+, D271A had only a small effect (Fig. 6 D).
Figure 6.
Effects of β7 mutations on rolling and firm adhesion of α4β7 transfectants on MAdCAM-1 substrates in shear flow. (A–D) Adhesion of βI domain metal-binding site mutants in 1 mM Ca2+ (A), 1 mM Ca2+ + 1 mM Mg2+ (B), 1 mM Mg2+ (C), and 0.5 mM Mn2+ (D). (E–G) Metal ion concentration dependence of adhesion. (H–K) Adhesion of mutants in the βI domain interface with the hybrid domain in the indicated metal ions. Bars show cumulative numbers of firmly adherent and rolling cells at 2 dyn/cm2 wall shear stress and error bars show the standard deviation between three experiments.
We hypothesized that the concentrations of Mg2+ and Mn2+ used in these experiments might be sufficient for Mn2+ but not Mg2+. To test this hypothesis, adhesion in shear flow was assayed over a wide range of Mg2+ and Mn2+ concentrations (Fig. 6, E–G). Adhesion of the D271A mutant was nearly absent in 1 mM Mg2+ and nearly wild type in 1 mM Mn2+ (Fig. 6 F). However, adhesion was almost completely recovered in 100 mM Mg2+. Furthermore, adhesion was greatly diminished in 0.01 mM and 0.1 mM Mn2+ compared with wild type (Fig. 6, E and F). The titrations demonstrate that Asp-271 regulates the metal ion concentration required for adhesion. The D271A mutation raises the EC50 ∼1,000-fold for Mg2+, from 0.1 to 100 mM, and raises the EC50 ∼10-fold for Mn2+, from 0.1 to 1 mM (Fig. 6, E and F). This function of Asp271 differs qualitatively from MIDAS residue Ser-142, which is indispensible for adhesion even at high metal ion concentrations (Fig. 6 G).
Complex with a small antagonist molecule
We subjected the α4β7 headpiece Fab complex to crystallization in the presence of a small molecule α4β7 antagonist, RO0505376. Crystals grew under similar conditions as in the absence of ligand, although with different lattice dimensions (Table 1), and, remarkably, revealed a complex with an antagonist with the closed headpiece. This contrasts with the αIIbβ3 headpiece, which yields only the open headpiece when co-crystallized with ligand-mimetic compounds (Xiao et al., 2004; Zhu et al., 2010).
The antagonist binds in the wide cleft between α4 and β7 with one oxygen of its carboxyl group coordinating the MIDAS Mg2+ (Fig. 4, A and B; and Fig. 5, C and E). The other oxygen of the carboxyl hydrogen bonds to the backbone NH group of residue Asn-143, in the β1-α1 loop in the second X position of the DXSXS MIDAS motif (Fig. 5 C). The antagonist belongs to the most common class of α4 antagonists, the phenylalanine class, and its carboxyl corresponds to the phenylalanine α-carboxyl group. The phenylalanine α-amino group hydrogen bonds to the Asn-235 backbone carbonyl, and is linked through an amide to a 2,6-dichlorobenzoyl group (Fig. 5 E). This group occupies the spacious half of the ligand-binding cleft toward the SDL (Fig. 4 B). The phenylalanine ring is joined at its para position to a third aromatic group, 1,6-dimethyl-2-oxo-4-trifluoromethyl-pyridine, which locates to the middle of the ligand-binding cleft. This positively charged aromatic ring makes both van der Waals and π- cation bonds with α4 residues Tyr-187 and Phe-214, which form an important portion of the α4 wall of the binding pocket (Fig. 4 B). Furthermore, the 2-oxo group of the pyridine moiety forms a strong 2.6 Å hydrogen bond to the side chain of Ser-238 (Fig. 5 E). Ser-238 projects prominently from the base of the ligand-binding groove, and, together with the adjacent depression at the MIDAS Mg2+, separates head-proximal and leg-proximal halves of the cleft (Fig. 4 B). The antagonist has few rotatable bonds, and its conformation is further stabilized by van der Waals contact between chloro and trifluoromethyl groups of the two distal aromatic rings (Fig. 5 E).
Although α4β7 remains closed after small molecule antagonist binding, some readjustment occurs in the ligand-binding site (Fig. 5, A and C). These changes are significant because they are very similar when the two independent α4β7–Fab complexes in antagonist-liganded and unliganded crystals are compared and are greater than the estimated coordinate error. First, the tip of the β1-α1 loop centered at Ser-144 moves 1 Å closer to the MIDAS Mg2+ ion. This brings the Ser side chain oxygen within 2.1–2.2 Å of the Mg2+, an optimal distance for Mg2+ coordination (Fig. 5 C). A similar 1 Å movement of the β1-α1 backbone brings the nitrogen of residue 143 within hydrogen-bonding distance (3.1 Å) of the nonmetal coordinating oxygen of the ligand carboxyl group (Fig. 5, A and C). The amount of backbone movement gradually diminishes to 0.7 or 0.5 Å at the backbones of Asp-147 and Asp-148, the side chains of which coordinate the ADMIDAS. The backbone carbonyl of Ser-144 also coordinates the ADMIDAS Ca2+, and thus the ADMIDAS Ca2+ also moves 0.6 Å (Fig. 5, A and C).
Furthermore, the MIDAS-coordinating alternate conformation of auxiliary MIDAS residue Asp-271 is lost, and now it occupies the same position as the noncoordinating alternate conformation in the unliganded structure, 4 Å further away from the Mg2+ (Fig. 5, A and C). The Asp-271 side chain thus moves close to Ser-238, and their two side chains together form the projection in the middle of the ligand-binding cleft (Fig. 4 B). Although there is electrostatic repulsion between the ligand carboxyl group and the metal-coordinating conformation of Asp-271, there is no steric clash, and they bind to opposite rather than neighboring positions in the octahedral Mg2+ coordination shell (Fig. 5, A and C).
EM studies on α4β7 conformation and ligand binding
EM allowed our studies to be extended to the complete α4β7 ectodomain and complexes with MAdCAM-1 and further Fab (Fig. S4). The full α4β7 ectodomain was expressed with the extracellular portions of α4 and β7 (Fig. 1) fused at their C termini to flexible linkers followed by disulfide-linked ACID-BASE α-helical coiled coils that mimic the α and β subunit transmembrane domain association. The complete α4β7 ectodomain had a compact rather than extended conformation (Fig. 7 A); however, it appeared not as compact and did not give class averages with as much detail as αVβ3, αIIbβ3, or αXβ2 (Takagi et al., 2002; Nishida et al., 2006; Zhu et al., 2008). Cross-correlations with the αIIbβ3 or αVβ3 ectodomain crystal structures fit markedly less well than with EM class averages of αIIbβ3 or αVβ3. Furthermore, although αXβ2 has a different orientation between the αX calf-1 and calf-2 domains than in αIIb or αV (Xie et al., 2010), cross correlation with the αXβ2 crystal structure lacking the αI domain fit the α4β7 EM class averages no better. Thus, although it clearly has a compact conformation, the orientation between domains in α4β7 appears distinct from that in the bent integrins thus characterized in crystal structures.
Figure 7.
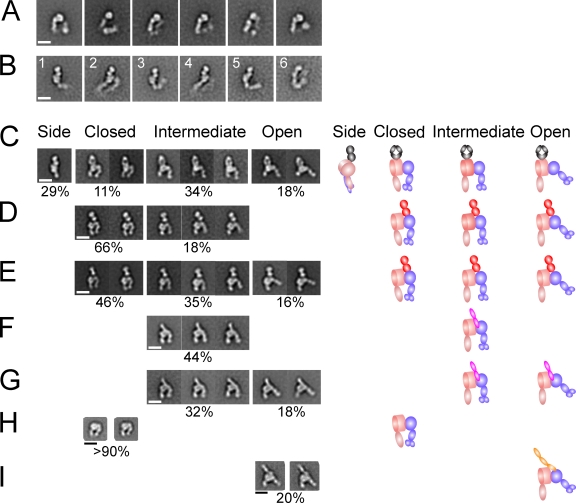
EM of the complete ectodomain and headpiece of α4β7, and complexes with Fab and ligands. (A and B) C-terminally clasped α4β7 ectodomain wild-type (A) or α4 R558A mutant ectodomain complexed with natalizumab Fab (B). (C–G) The α4β7 R558A mutant headpiece construct complexed with Fab, MAdCAM-1, and a small molecule antagonist as indicated, in buffer with 1 mM Ca2+ and 1 mM Mg2+, except for the indicated sample in 1 mM Mn2+. The percentage of α4β7 particles in each state is shown below the representative class averages, and does not sum to 100% because headpiece class averages without bound Fab or MAdCAM-1, or that were poorly resolved, were omitted. (C) Natalizumab. (D) Act-1 Fab. (E) Act-1 Fab and RO0505376. (F) MAdCAM-1. (G) MAdCAM-1 and Mn2+. Schematics depicting orientations in representative class averages are shown to the right. (H and I) Previously published (Takagi et al., 2003) representative EM class averages of the α5β1 headpiece (H) and its complex in Mn2+ with fibronectin Fn3 domains 7–10 (I). Integrins or complexes were isolated by Superdex S200 gel filtration except the complex between α4β7 headpiece (0.02 mg/ml) and MAdCAM-1 (0.2 mg/ml) in Ca2+ + Mg2+, which was formed by mixing the two at room temperature for 30 min and diluting 10-fold just before grid preparation. Bars, 10 nm.
Ectodomain complexes with natalizumab Fab revealed distinct orientations on the substrate (Fig. 7 B) compared with absence of Fab and also appeared to reveal differences in the amount of bending at the knees (Fig. 7 B, compare panels 3 and 5). Complexes between natalizumab and only the α4β7 headpiece revealed two different orientations on the substrate with the headpiece seen on its side, with the Fab also seen on its side (Fig. 7 C, side), or with the headpiece lying flat on the substrate and the Fab seen as a flat oval (Fig. 7 C, closed, intermediate, or open). Comparison of the side view of the natalizumab Fab headpiece complex (Fig. 7 C) to averages of the natalizumab Fab ectodomain complex (Fig. 7 B, panels 1, 3, and 5) suggests that the latter represents similar side views, and emphasizes that density seen in the ectodomain and not headpiece complex class averages represent the α4β7 legs, which appear to vary in orientation with respect to the headpiece.
The α4β7 headpiece was examined with Fab and with or without the antagonist RO0505376 (Fig. 7, C–E) and in the absence of Fab (Fig. S4). Because Act-1 Fab helped orient the headpiece on the substrate, its complexes provided more accurate assessment of headpiece conformation than the headpiece alone; however, class averages of the headpiece alone also revealed the intermediate conformation described below (Fig. S4). The α4 β-propeller domain and β7 I domain in the integrin head were resolved as separate densities that were large and small, respectively (Fig. 7, D and E). Furthermore, Act-1 Fab bound to the smaller β I domain density (Fig. 7, D and E). Two densities in the β-leg correspond to (1) the hybrid domain and (2) the PSI and I-EGF1 domains, which are side-by-side in the β-leg (Fig. 7, D and E). A single density in the α-leg corresponds to the thigh domain.
We assigned headpiece conformation based on cross-correlation with the closed headpiece crystal structure and a model of the open α4β7 headpiece, and by visual inspection of class averages. By both criteria, α4β7 can also adopt an intermediate conformation of the headpiece that is clearly distinct from the previously described integrin closed and open headpiece conformations. In the absence of RO0505376, the majority of the α4β7 particles had a closed headpiece (Fig. 7 D). Almost all remaining particles had an intermediate conformation of the headpiece: cross-correlation scores with the open and closed headpiece were similar, and the β-leg showed a position intermediate between closed and open (Fig. 7 D). Few if any particles exhibited an open headpiece. In the presence of RO0505376, a substantial proportion of headpieces remained closed, demonstrating the representativeness of the small molecule antagonist-bound closed crystal structure (Fig. 7 E). Additionally, a substantial proportion of intermediate headpieces were present. Moreover, the truly open headpiece conformation was also seen in presence of RO0505376 (Fig. 7 E). Distinct densities in the β-leg for the hybrid and PSI/EGF-1 domains were more evident for the closed and open headpieces than for the intermediate headpiece (Fig. 7, D and E), which is consistent with a less well-defined orientation between the βI and hybrid domains in the intermediate opening of the headpiece.
Natalizumab binds to the α4 β-propeller domain, close to its ligand-binding interface with the β7 βI domain (Fig. 7 C). All three headpiece conformations were seen with natalizumab. Natalizumab appears to bind more out of the major plane formed by the headpiece domains than Act-1, because its Fab caused more variation in headpiece orientations on the substrate, including side views (Fig. 7 C), as described three paragraphs earlier.
MAdCAM-1 bound to the α4β7 headpiece with sufficient strength in Mn2+ for the complex to be isolated by gel filtration; binding was weaker in Ca2+ and Mg2+, and therefore freshly isolated α4β7 headpiece and MAdCAM-1 were mixed together before EM grid preparation. In each metal ion condition, a substantial proportion of headpieces did not bind MAdCAM-1, resulting in the percentage of complexes shown below each panel (Fig. 7). MAdCAM-1 bound at the interface between the α4 β-propeller and β7 I domain, with one and possibly a portion of a second IgSF domain clearly projecting out of the interface (Fig. 7, F and G). MAdCAM-1 was aligned in the same orientation as the 40 Å–long α4β7 ligand-binding crevice. The MAdCAM-1–bound α4β7 headpiece was in the intermediate conformation in Ca2+ and Mg2+ (Fig. 7 F); 100% of α4β7–MAdCAM-1 complexes were in the intermediate conformation. Mn2+ substantially increased the proportion of headpieces with the open conformation (Fig. 7 G). Among α4β7–MAdCAM-1 complexes in Mn2+, 64% had the intermediate conformation and 36% had the open conformation.
Previously published data on the α5β1 headpiece (Takagi et al., 2003; Luo et al., 2004) provide a direct comparison (Fig. 7, H and I). The α5β1 headpiece alone, either in Ca2+ or in Mn2+, was found uniformly in a closed conformation (Fig. 7 H). When the headpiece was mixed in Mn2+ with an excess of a fragment of fibronectin containing Fn3 domains 7–10 and isolated by gel filtration, 20% of the α5β1 headpieces bound to Fn3 7–10, and 100% of the particles were in the open conformation. In contrast, all nonliganded α5β1 headpieces were in the closed conformation. Among integrins studied to date, α4β7 is unique not only for its ability to access both the closed and intermediate headpiece conformations in the absence of ligand, but also for its ability to access both the intermediate and open conformations in the presence of ligand.
The comparison to α5β1 also emphasizes the distinct ligand-binding modalities of α4β7 and RGD-binding integrins. The complexes with α5β1 show the RGD-bearing Fn3 domain 10 as a distinct density, which in the orientation on the page lies above the α5 β-propeller domain and β1 βI domain, at their interface (Fig. 7 I). In contrast, no such separate density is seen for domain 1 of MAdCAM-1, which is consistent with it binding to a long groove between the α4 β-propeller domain and the β7 βI domain, and the merging of its density with the β-propeller and βI domains (Fig. 7, F and G).
If the closed headpiece mediated rolling, destabilizing the closed βI domain–hybrid domain interface should decrease rolling. In contrast, if an intermediate headpiece conformation mediated rolling, destabilizing the closed headpiece should increase rolling. We destabilized the closed headpiece with a K346A mutation to remove the Lys in the interface, and a N260A mutation to remove the interacting N-glycan. Each mutation increased rolling adhesion, without affecting firm adhesion (Fig. 6, H–K). These data support the hypothesis that the intermediate headpiece conformation, seen so far only for α4β7, mediates rolling adhesion.
Discussion
Four independent examples in crystals of the α4β7 headpiece yield insights into its flexibility. The β-propeller–thigh interface enables substantial flexibility, yet maintains close association between these neighboring domains. Whereas αIIbβ3 headpiece structures show well-defined density for the β3 I-EGF1 and PSI domains (Springer et al., 2008; Zhu et al., 2010), in α4β7, density for I-EGF1 was lacking and density for PSI was weak. Overall, flexibility at the hybrid domain interface with the I-EGF and PSI domains, and at the β-propeller–thigh domain interface, may make important contributions to encountering ligands in rolling adhesion, which requires fast on-rates. In an interesting contrast, we found little flexibility, and well-defined noncovalent bonds, at the interface in β7 between the βI and hybrid domains.
We identified a novel auxiliary MIDAS residue, Asp-271. It can form a direct coordination to the MIDAS Mg2+ in one of two alternate conformations in the absence of ligand. In the presence of RO0505376, it swings away from the Mg2+, perhaps because of electrostatic repulsion by the ligand carboxyl group. This auxiliary MIDAS residue is present only in the β2, β5, and β7 integrin subunits. Previous 3.5-Å resolution αXβ2 crystal structures showed that the homologous residue was near the MIDAS, but lacked metal at the βI MIDAS, and thus could not define whether it was a MIDAS residue (Xie et al., 2010).
Our functional studies of rolling and firm adhesion in shear flow demonstrate that the auxiliary MIDAS residue makes a profound contribution to binding MAdCAM-1 by lowering the effective concentration of Mg2+ required from ∼100 mM to ∼0.1 mM. Thus, Asp-271 is indispensable for ligand binding at physiological 1 mM Mg2+ concentrations. Asp-271 is in the ligand-binding pocket and may make a direct contribution to binding of macromolecular ligands. Residue Glu-354 near the ADMIDAS also markedly contributed to rolling adhesion in Ca2+ and firm adhesion in Mg2+. Functionally, it might be considered an auxiliary ADMIDAS residue; structurally, it did not unambiguously coordinate the ADMIDAS. These findings provide insights into specializations of β7 for its function in rolling and firm adhesion.
In α4β7, the ligand-binding site is a long, wide groove that is open at both ends and is appropriate for lengthwise binding of one or two protein domains in which the integrin-binding motif is tightly embedded. In contrast, the binding sites at the αIIb–β3 and αV–β3 interfaces are pockets that are closed at one end by projections of αIIb Trp or αV Arg residues and a β3 N-linked glycan, and at the other end by αV and αIIb loops and the β3 SDL. These differences correlate with recognition by αVβ3 and αIIbβ3 of peptide motifs, such as RGD, largely independent of their macromolecular context. The wall lining the binding groove is relatively straight and even in height in α4, whereas the αIIb and αV walls are crooked and notched. These notches correlated with binding to peptide fragments in a groove that is at right angles to the groove in α4β7.
Our study provides the first example of how a small molecule antagonist binds across an integrin α–β interface outside of RGD-recognizing integrins. Early studies showed that an I/L-D-T/S motif in domain 1 of MAdCAM-1 and VCAM-1 (Vonderheide et al., 1994; Viney et al., 1996; Newham et al., 1997; Green et al., 1999), and a similar LDV motif in the alternatively spliced type IIICS segment in fibronectin (Komoriya et al., 1991), was important for binding α4 integrins. Notably, both RGD and I/L-D-T/S/V motifs contain an Asp that is recognized by the βI domain MIDAS Mg2+ ion. The structure of VCAM-1 revealed that the integrin-binding motif is in a loop between the C and D β strands of IgSF domain 1 (Jones et al., 1995; Wang et al., 1995). The conformation of the integrin-binding CD loop in VCAM-1 is strongly supported by hydrogen bonds to its backbone, and is identical in independent VCAM-1 structures. In contrast, the integrin-binding loop in MAdCAM-1 does not have a well-defined orientation (Tan et al., 1998; Dando et al., 2002), and further structures have revealed additional backbone conformations (unpublished data).
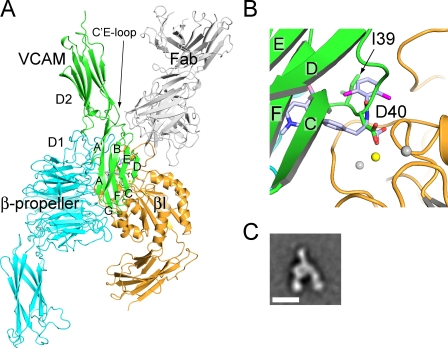
Therefore, we manually docked domains 1 and 2 of VCAM-1 with its rigid integrin-binding loop to α4β7, and were able to obtain an unambiguous overall binding orientation (Fig. 8). The key Asp side chain carboxyl must lie on the β7 subunit side of the ligand-binding groove, which fixes the N-to-C orientation of D1 in the groove. Unlike the other loops in domain 1 of VCAM-1, the CD loop locates near the middle rather than at one of the two ends of the domain; furthermore, although it links edge strands in the A′GFC and BED β-sheets, the CD loop is bent so that its Asp is on the GFC side (Fig. 8 B). Docking to place the Asp side chain in a similar orientation to the RO0505376 carboxyl group places the GFC sheet of domain 1 of VCAM-1 in the α4-β7 groove (Fig. 8, A and B). D2 projects from the head-proximal end of the groove, with its C′E loop contacting α4 at this end. The finding by mutagenesis that the residues in domain 2 important in VCAM-1 binding to α4β7 are in the C′E loop confirms the docking model (Newham et al., 1997). MAdCAM-1 domains 1 and 2 can readily be superimposed on those of VCAM-1. The docking orientation is strongly supported by the EM projection averages of α4β7 headpiece complexes with MAdCAM-1, which show one IgSF domain of MAdCAM-1 (which docking suggests is domain 2) projecting from the headpiece, on the opposite side from the integrin legs, and aligned with the ligand-binding groove (Fig. 8, A and C).
Figure 8.
VCAM-1 docked to α4β7. (A) VCAM-1 domains 1 and 2 (PDB accession no. 1VCA; Jones et al., 1995) were manually placed in an approximate binding orientation to the α4β7 head (β-propeller and βI domain only, no Fab). The orientation was optimized by RosettaDock (Lyskov and Gray, 2008). The α4β7 headpiece/Act-1 Fab/RO0505376 structure was superimposed, and the pose of VCAM-1 was slightly adjusted to align the putative MIDAS-coordinating carboxyl group in VCAM-1 to that in RO0505376. (B) Zoomed-in view of ligand-binding site in A. (C) A representative class average of MAdCAM-1 bound to the α4β7 headpiece from Fig. 7 H. Note the similar orientations between the docked VCAM-1 in the model and MAdCAM-1 bound to α4β7. The only difference is that the headpiece is closed in the crystal structure and in an intermediate conformation when bound to MAdCAM-1.
In the docking orientation, the CAM domains that link to the membrane—IgSF domains 3–7 in VCAM-1 or a mucin-like domain in MAdCAM-1—extend from IgSF domain 2 in one direction, and the integrin legs extend away in the other direction. This geometry is not only ideal for cell adhesion but is also force-resistant because tensile force applied to the complex as it mediates adhesion in shear flow is aligned with the binding groove, and a large number of interactions along the groove would have to be broken at once for receptor–ligand dissociation to occur.
Our structure of α4β7 and the proposed docking model are compatible with previous mutational studies on α4. In the α4 W3 β2-β3 loop, which is central in the ligand-binding site, Tyr-187, which forms the ligand-binding wall opposite the MIDAS and contacts RO0505376, is required for α4β1 binding to VCAM-1 and the IIICS segment of fibronectin (Irie et al., 1995). Moreover, exchange of loops between the α4 and α5 subunits at the head end of the α-β groove (W2 β2-β3) and leg end of the groove (W4 β2-β3) blocked α4β1 binding to VCAM-1 and a fibronectin IIICS peptide (Irie et al., 1997). These results are compatible with contact of protein ligands with an extensive interface along the α–β groove as proposed here, and suggest extension of this concept to the α4β1 interface and fibronectin.
Alignment of VCAM-1 Asp-40 with the α-carboxyl group of RO0505376 allows assessment of how the other functional groups of the phenylalanine class of α4 integrin antagonists mimic biological ligands (Fig. 8). Interestingly, the amide-linked hydrophobic aromatic ring extending toward the head end of the groove falls precisely in the position of the key hydrophobic Ile-39 side chain that precedes Asp-40 in VCAM-1 (Fig. 8 B); this is conserved as Ile or Leu in VCAM-1, MAdCAM-1, and IIICS of fibronectin. Furthermore, a pocket-like depression here in the α4-β7 groove accommodates this hydrophobic group (Fig. 4 B). Thus, a key difference between I/L-D and RGD motifs is that while the Arg of RGD extends across the α–β interface toward α, the Leu of I/L-D extends perpendicularly to the Arg, along the α-β groove. In contrast, the function of the phenylalanine side chain of phenylalanine-based α4 integrin antagonists is to extend across the α–β interface, and to bring the aromatic ring to which it is connected into contact with the α4 wall of the binding groove. Thus, these pharmacophores mimic the thickness of an IgSF domain as it is buried in a hydrophobic binding groove.
The RO0505376 antagonist cocrystallized here is closely related to the phenylalanine class antagonist firategrast, which is currently in clinical trials for multiple sclerosis (Davenport and Munday, 2007). The α4β7 structure and the binding pose of cocrystallized antagonist will be invaluable for further development of antagonist potency and selectivity for α4βl or α4β7. For example, the structure demonstrates that the amide-linked benzoyl ring could be extended toward the SDL at the head end of the groove, and that other substituents could interact with β7 Asp-271. Because β7 and β1 differ markedly in their SDL loops, and the β7 auxiliary MIDAS residue Asp-271 is an Ala in β1, such modifications could improve selectivity for α4β1, as desired for treatment of multiple sclerosis, or selectivity for α4β7, as desired for treatment of Crohn’s disease.
Humanized Act-1 antibody, vedolizumab, is in clinical trials for treatment of ulcerative colitis (Feagan et al., 2005) and Crohn’s disease (Feagan et al., 2008). The Act-1 Fab has a concave binding site that wraps around the β7 SDL. Interestingly, Ser-231 in the β7 α2-α3 loop, which hydrogen bonds to H3 Trp-104 (Fig. 3 A), is Asn in mouse β7, which introduces an N-linked glycosylation site at residue 231. The S231N mutation in human β7 loses Act-1 reactivity, and the N231S mutation in mouse β7 gains reactivity (Tidswell et al., 1997). Thus, lack of glycosylation at this site creates a unique human epitope recognized by a mouse antibody. However, this glycosylation site is distal from the α4-β7 groove, and does not affect binding of MAdCAM-1 (Tidswell et al., 1997).
Act-1 binds α4β7 but not α4β1 or αEβ7 (Schweighoffer et al., 1993). This finding, together with our finding that Act-1 binds only to β7, is likely to be explained by the presence of an αI domain in αE. The αI domain is inserted in the β-propeller W2 β4-β1 loop, opposite the SDL on the α4β7 groove (Figs. 3 A and 4 A). Superpositions of an αI integrin, αXβ2 (Xie et al., 2010), show a clash of the αI domain with Act-1 Fab (Fig. S5).
A further interesting property of Act-1/vedolizumab is that it blocks binding of MAdCAM-1 and fibronectin to α4β7, but does not block (Soler et al., 2009), or synergistically enhances (Schweighoffer et al., 1993), binding to VCAM-1. Consistent with this finding, our model of docked VCAM-1 shows that it fits in the α4-β7 groove without clashing with, but is in close proximity to, Act-1 Fab (Fig. 8). In contrast to the C′E loop in domain 2 of VCAM-1, MAdCAM-1 contains a much longer “charged antenna” DE loop in domain 2 (Tan et al., 1998; Dando et al., 2002), which is disordered in crystal structures, and important in binding to α4β7 (Green et al., 1999). Superposition of MAdCAM-1 on docked VCAM-1 locates the charged antenna near the β7 SDL, and explains selective blocking by Act-1/vedolizumab. Basic patches of residues on the SDL of β7 and in the W2 β2-β3 loop of the α4 β-propeller domain (Fig. 3 B) are present in the α4-β7 groove for interaction with the MAdCAM-1 charged antenna.
Natalizumab to the α4 integrin subunit is in clinical use for treatment of relapsing multiple sclerosis and Crohn’s disease (Miller et al., 2003; Sandborn et al., 2005). EM shows that this antibody binds to the α4 β-propeller domain close to the α4-β7 groove. The mouse precursor antibody to natalizumab, termed an100226m (Kent et al., 1995) or TY21/6 (Newham et al., 1998), is similar to α4 antibodies to B1 and B2 epitopes (Pulido et al., 1991; Newham et al., 1998) that require human α4 residues 152–203 in β-propeller blade W3 for recognition (Schiffer et al., 1995). This is consistent with localization by EM of the natalizumab Fab-binding site adjacent to the α4-β7 groove. EM further demonstrated that neither Act-1 nor natalizumab inhibited ligand binding by stabilizing the integrin headpiece in the closed conformation, in contrast to some β1 and β2 antibodies (Luo et al., 2004; Chen et al., 2010).
The α4β7 ectodomain has a less compact bent conformation than seen for αVβ3, αIIbβ3, αXβ2, and αLβ2 integrins (Takagi et al., 2002; Nishida et al., 2006; Zhu et al., 2008), and has a novel intermediate conformation of the headpiece. Previously, αVβ3, αIIbβ3, αXβ2, αLβ2, and α5β1 integrins have shown only closed and open headpiece conformations (Takagi et al., 2002, 2003; Luo et al., 2004; Xiao et al., 2004; Nishida et al., 2006; Xiong et al., 2009; Chen et al., 2010; Zhu et al., 2010). Closed, intermediate, and open conformations of the α4β7 headpiece were all seen in the presence of saturating concentrations of a small molecule antagonist. Moreover, the same small molecule cocrystallized with Act-Fab and α4β7 headpiece in the closed conformation. These results strongly contrast with the αIIbβ3 headpiece, which crystallizes in the open conformation in the presence of RGD-like antagonists, and in the closed conformation in their absence (Xiao et al., 2004; Zhu et al., 2010). Furthermore, when complexed with MAdCAM-1, α4β7 displayed intermediate as well as open headpiece conformations. This contrasts with the α5β1 headpiece, which was always open when bound to a fibronectin fragment (Takagi et al., 2003).
Previously, the rolling and firm phases of α4β7 adhesion were proposed to, respectively, correlate with the low-affinity, closed-headpiece, and high-affinity, open-headpiece states seen with other integrins (Chen et al., 2003, 2004). The observation that α4β7 is either intermediate or open and not closed when complexed with MAdCAM-1, together with mutational studies, now strongly suggests that rolling is mediated by the intermediate headpiece conformation. If rolling were mediated by the closed headpiece, mutational destabilization of the closed headpiece would decrease rolling, whereas the opposite was found. Furthermore, if there were no intermediate state, destabilization of the closed headpiece would increase firm adhesion; however, firm adhesion was unaffected. Thus, mutational enhancement of rolling and lack of effect on firm adhesion provide strong support for the concept that the intermediate headpiece conformation mediates rolling adhesion. We also found that the α4β7 headpiece could be crystallized in the closed headpiece conformation in the presence of a small molecule antagonist, which is consistent with EM results. However, the antagonist induced movements in the MIDAS β1-α1 loop, which may in part mimic movement toward the intermediate headpiece conformation.
Thus, data now suggest that the rolling phase of adhesion mediated by the MAdCAM-1 complex with α4β7 corresponds to an intermediate state of the α4β7 headpiece, and the firm phase corresponds to the open state of the headpiece. Detailed understanding of these states must await crystal structures of complexes of MAdCAM-1 and α4β7. The important point biologically is that ligand binding does not induce the open headpiece conformation with high affinity for ligands, as seen with RGD-binding integrins that mediate firm adhesion; therefore, the intermediate conformation can persist after ligand binding, so that α4β7 can mediate rolling adhesion. This specialization of α4β7 enables rolling versus firm adhesion to be regulated by signals from within the cell. Thus, physiological signals encountered by a leukocyte rolling through α4β7 on endothelium, such as those received by G protein–coupled chemokine receptors, can activate inside-out signals that cause α4β7 headpiece opening, and activate firm adhesion through α4β7 as a prelude to lymphocyte transendothelial migration.
Materials and methods
Protein expression, purification, and crystallization
Soluble α4β7 headpiece and ectodomain were prepared similarly to αXβ2 (Xie et al., 2010). In brief, the α4 subunit cloned in pcDNA3.1/Hygro(−) was followed by a tobacco etch virus (TEV) site, the ACID coiled-coil, and a Strep-II tag. The β7 subunit cloned in pEF1-puro was followed by a TEV site, the BASE coiled-coil, and a His tag. Proteins expressed in CHO-Lec3.2.8.1 cells were purified using Ni-NTA and Strep-Tactin affinity chromatography. Natalizumab was from S. Goelz (Biogen Idec, Cambridge, MA). Antibody Act-1 and further natalizumab were from E. Fedyk (Millenium: The Takeda Oncology Co., Cambridge, MA). The headpiece was stabilized with Act-1 Fab (1:3 molar ratio) and cleaved with TEV protease and EndoH at room temperature overnight. The mixture was passed through Ni-NTA resin and further purified by gel filtration. The α4β7–Act-1 Fab complex was concentrated to 3.6 mg/ml in 10 mM Tris, pH 7.5, 0.15 M NaCl, 1 mM CaCl2, and 1 mM MgCl2. RO0505376 was added to a final concentration of 0.5 mM. Crystals were grown in 0.1 M Tris, pH 8.5, 0.1 M NaCl, and 8–10% polyethylene glycol (PEG) 20,000 at 4C°. To incorporate Se-Met in protein, cells were washed with PBS supplemented with 1% dialyzed FCS, incubated with methionine-free α-MEM (SAFC Biosciences) supplemented with 50 mg/l l-Se-Met (Sigma-Aldrich) and 10% dialyzed FCS, replaced with the same medium after 6 h, and cultured for 4 d (Xie et al., 2010). Crystals were cryoprotected by first increasing the PEG 20,000 to 15%. Then PEG 400 or 2-methyl-2,4-pentaneidiol for native or RO0505376 crystals, respectively, was added in 5% increments to 30%. RO0505376 was from J. Tilley (Roche, Nutley, NJ).
Structure determination
Structures were determined in three lattices (Table 1). Phases for a 3.5-Å MAD dataset in lattice B were solved with SHARP (Vonrhein et al., 2007). Phases for a 3.6 Å single-wavelength anomalous dispersion (SAD) dataset in lattice A were first obtained by molecular replacement using the Fab complex model from lattice B. PHASER (McCoy et al., 2007) was then used to calculate experimental phases for lattice A based on 24 Se atoms. The SAD phases were transferred to the 3.15 Å native data in this lattice. Multicrystal averaging improved and extended the phases. A better model was built, and new domain masks calculated. Multicrystal, multidomain averaging was performed once more. The resulting model was used with molecular replacement to solve the complex with RO0505376 in lattice C. Models were iteratively improved by building with COOT (Emsley and Cowtan, 2004), refinement with PHENIX (Adams et al., 2002), and validation with MOLPROBITY (Davis et al., 2007).
Negative stain EM
α4β7 ectodomain or headpiece proteins, with or without MAdCAM-1, Fab, or RO0505376, were loaded on a gel filtration column (Superdex 200 HR; GE Healthcare) pre-equilibrated with Tris-buffered saline, 1 mM CaCl2 and 1 mM MgCl2, or 1 mM MnCl2. When present, RO0505376 was also added to the running buffer (0.5 mM). Peak fractions were adsorbed to glow-discharged carbon-coated copper grids and stained with uranyl formate, and images acquired with an electron microscope (Tecnai 12; FEI) at 120 kV and a nominal magnification of 67,000. Particles were interactively picked using BOXER in EMAN (Ludtke et al., 1999). Grid location coordinates were converted to SPIDER (Frank et al., 1996) format and used in windowing particles to create a boxed image stack. Each image in the stack was normalized to a mean intensity of 0 and SD of 50. Normalized images were subjected to 10 iterations of centering, multivariate statistical analysis, K-means classification, class averaging, and multireference alignment, with the class averages used as input references for multireference alignment in the next iteration (Mi et al., 2011).
Flow chamber experiments
β7 mutations were generated using KOD-Plus-Mutagenesis (Toyobo Co.). Wild-type human β7 cDNA in vector pcDNA3.1/Hygro(–) (Invitrogen) was used as the template. All mutations were confirmed by DNA sequencing. 293T cells were transiently transfected with α4 and β7 cDNA using calcium phosphate precipitates. Adhesion in a parallel wall flow chamber on a plastic substrate coated with a human MAdCAM-1 Fc chimera was performed exactly as described previously (Chen et al., 2003). In brief, cells were pumped into a flow chamber mounted on a Petri dish coated with 10 µg/ml huMAdCAM-1/Fc, and allowed to accumulate for 30 s at 0.3 dyn/cm2 and 10 s at 0.4 dyn/cm2. Then, shear stress was increased every 10 s from 1 dyn/cm2 up to 32 dyn/cm2, in twofold increments. The number of cells remaining bound at the end of each 10-s interval and the rolling velocity were determined.
Online supplemental material
Fig. S1 shows the six integrin α subunit families, and variation in known ligand binding loops in their β-propellers. Fig. S2 shows structure-based sequence alignment of structurally defined integrins with α4β7. Fig. S3 shows density for βI domain metal ions and Asp-271. Fig. S4 shows all class averages from negative stain EM. Fig. S5 shows superimposition of αXβ2 on the α4β7 headpiece–Act-1 Fab complex. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110023/DC1.
Acknowledgments
We thank Dr. Jing Song for early contributions to this project, and the staff at GM/CA Collaborative Access Team beamline at the Advanced Photon Source, Argonne, IL.
This paper was supported by National Institutes of Health grant HL48675 and HHMI.
Footnotes
Abbreviations used in this paper:
- ADMIDAS
- adjacent to MIDAS
- CDR
- complementarity-determining region
- IgSF
- immunoglobulin superfamily
- MIDAS
- metal ion–dependent adhesion site
- SAD
- single-wavelength anomalous dispersion
- SDL
- specificity-determining loop
- SyMBS
- synergistic metal ion binding site
References
- Adair B.D., Xiong J.P., Maddock C., Goodman S.L., Arnaout M.A., Yeager M. 2005. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J. Cell Biol. 168:1109–1118 10.1083/jcb.200410068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Andrew D.P., Berlin C., Honda S., Yoshino T., Hamann A., Holzmann B., Kilshaw P.J., Butcher E.C. 1994. Distinct but overlapping epitopes are involved in α4β7-mediated adhesion to vascular cell adhesion molecule-1, mucosal addressin-1, fibronectin, and lymphocyte aggregation. J. Immunol. 153:3847–3861 [PubMed] [Google Scholar]
- Askari J.A., Tynan C.J., Webb S.E., Martin-Fernandez M.L., Ballestrem C., Humphries M.J. 2010. Focal adhesions are sites of integrin extension. J. Cell Biol. 188:891–903 10.1083/jcb.200907174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 98:10037–10041 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargatze R.F., Jutila M.A., Butcher E.C. 1995. Distinct roles of L-selectin and integrins α4β7 and LFA-1 in lymphocyte homing to Peyer’s patch-HEV in situ: the multistep model confirmed and refined. Immunity. 3:99–108 10.1016/1074-7613(95)90162-0 [DOI] [PubMed] [Google Scholar]
- Berlin C., Berg E.L., Briskin M.J., Andrew D.P., Kilshaw P.J., Holzmann B., Weissman I.L., Hamann A., Butcher E.C. 1993. α 4 β 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 74:185–195 10.1016/0092-8674(93)90305-A [DOI] [PubMed] [Google Scholar]
- Bouvard D., Brakebusch C., Gustafsson E., Aszódi A., Bengtsson T., Berna A., Fässler R. 2001. Functional consequences of integrin gene mutations in mice. Circ. Res. 89:211–223 10.1161/hh1501.094874 [DOI] [PubMed] [Google Scholar]
- Briskin M.J., McEvoy L.M., Butcher E.C. 1993. MAdCAM-1 has homology to immunoglobulin and mucin-like adhesion receptors and to IgA1. Nature. 363:461–464 10.1038/363461a0 [DOI] [PubMed] [Google Scholar]
- Chakrabarti P., Bhattacharyya R. 2007. Geometry of nonbonded interactions involving planar groups in proteins. Prog. Biophys. Mol. Biol. 95:83–137 10.1016/j.pbiomolbio.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Chen J.F., Salas A., Springer T.A. 2003. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat. Struct. Biol. 10:995–1001 10.1038/nsb1011 [DOI] [PubMed] [Google Scholar]
- Chen J.F., Takagi J., Xie C., Xiao T., Luo B.-H., Springer T.A. 2004. The relative influence of metal ion binding sites in the I-like domain and the interface with the hybrid domain on rolling and firm adhesion by integrin α4β7. J. Biol. Chem. 279:55556–55561 10.1074/jbc.M407773200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xie C., Nishida N., Li Z., Walz T., Springer T.A. 2010. Requirement of open headpiece conformation for activation of leukocyte integrin alphaXbeta2. Proc. Natl. Acad. Sci. USA. 107:14727–14732 10.1073/pnas.1008663107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulsky M.I., Gimbrone M.A., Jr 1991. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science. 251:788–791 10.1126/science.1990440 [DOI] [PubMed] [Google Scholar]
- Dando J., Wilkinson K.W., Ortlepp S., King D.J., Brady R.L. 2002. A reassessment of the MAdCAM-1 structure and its role in integrin recognition. Acta Crystallogr. D Biol. Crystallogr. 58:233–241 10.1107/S0907444901020522 [DOI] [PubMed] [Google Scholar]
- Davenport R.J., Munday J.R. 2007. α4-integrin antagonism—an effective approach for the treatment of inflammatory diseases? Drug Discov. Today. 12:569–576 10.1016/j.drudis.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S., Richardson D.C. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day E.S., Osborn L., Whitty A. 2002. Effect of divalent cations on the affinity and selectivity of α4 integrins towards the integrin ligands vascular cell adhesion molecule-1 and mucosal addressin cell adhesion molecule-1: Ca2+ activation of integrin α4β1 confers a distinct ligand specificity. Cell Commun. Adhes. 9:205–219 10.1080/15419060216014 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Feagan B.G., Greenberg G.R., Wild G., Fedorak R.N., Paré P., McDonald J.W., Dubé R., Cohen A., Steinhart A.H., Landau S., et al. 2005. Treatment of ulcerative colitis with a humanized antibody to the α4β7 integrin. N. Engl. J. Med. 352:2499–2507 10.1056/NEJMoa042982 [DOI] [PubMed] [Google Scholar]
- Feagan B.G., Greenberg G.R., Wild G., Fedorak R.N., Paré P., McDonald J.W., Cohen A., Bitton A., Baker J., Dubé R., et al. 2008. Treatment of active Crohn’s disease with MLN0002, a humanized antibody to the α4β7 integrin. Clin. Gastroenterol. Hepatol. 6:1370–1377 10.1016/j.cgh.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Green N., Rosebrook J., Cochran N., Tan K., Wang J.-h., Springer T.A., Briskin M.J. 1999. Mutational analysis of MAdCAM-1/alpha4beta7 interactions reveals significant binding interfaces in the second immunuglobulin domain. Cell Adhes. Commun. 7:167–181 10.3109/15419069909010800 [DOI] [PubMed] [Google Scholar]
- Hesterberg P.E., Winsor-Hines D., Briskin M.J., Soler-Ferran D., Merrill C., Mackay C.R., Newman W., Ringler D.J. 1996. Rapid resolution of chronic colitis with an antibody to a gut homing integrin alpha 4 beta 7. Gastroenterology. 111:1373–1380 10.1053/gast.1996.v111.pm8898653 [DOI] [PubMed] [Google Scholar]
- Irie A., Kamata T., Puzon-McLaughlin W., Takada Y. 1995. Critical amino acid residues for ligand binding are clustered in a predicted beta-turn of the third N-terminal repeat in the integrin alpha 4 and alpha 5 subunits. EMBO J. 14:5550–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie A., Kamata T., Takada Y. 1997. Multiple loop structures critical for ligand binding of the integrin α4 subunit in the upper face of the β-propeller mode 1. Proc. Natl. Acad. Sci. USA. 94:7198–7203 10.1073/pnas.94.14.7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Mitsuoka K., Fujiyoshi Y., Fujisawa Y., Kikuchi M., Sekiguchi K., Yamada T. 2005. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J. Struct. Biol. 150:259–267 10.1016/j.jsb.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Jones E.Y., Harlos K., Bottomley M.J., Robinson R.C., Driscoll P.C., Edwards R.M., Clements J.M., Dudgeon T.J., Stuart D.I. 1995. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution. Nature. 373:539–544 10.1038/373539a0 [DOI] [PubMed] [Google Scholar]
- Kamata T., Handa M., Ito S., Sato Y., Ohtani T., Kawai Y., Ikeda Y., Aiso S. 2010. Structural requirements for activation in alphaIIb beta3 integrin. J. Biol. Chem. 285:38428–38437 10.1074/jbc.M110.139667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent S.J., Karlik S.J., Cannon C., Hines D.K., Yednock T.A., Fritz L.C., Horner H.C. 1995. A monoclonal antibody to α 4 integrin suppresses and reverses active experimental allergic encephalomyelitis. J. Neuroimmunol. 58:1–10 10.1016/0165-5728(94)00165-K [DOI] [PubMed] [Google Scholar]
- Komoriya A., Green L.J., Mervic M., Yamada S.S., Yamada K.M., Humphries M.J. 1991. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 266:15075–15079 [PubMed] [Google Scholar]
- Lazarovits A.I., Moscicki R.A., Kurnick J.T., Camerini D., Bhan A.K., Baird L.G., Erikson M., Colvin R.B. 1984. Lymphocyte activation antigens. I. A monoclonal antibody, anti-Act I, defines a new late lymphocyte activation antigen. J. Immunol. 133:1857–1862 [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Strokovich K., Walz T., Springer T.A., Takagi J. 2004. Allosteric β1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J. Biol. Chem. 279:27466–27471 10.1074/jbc.M404354200 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Carman C.V., Springer T.A. 2007. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25:619–647 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S., Gray J.J. 2008. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 36:W233–W238 10.1093/nar/gkn216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major E.O. 2010. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu. Rev. Med. 61:35–47 10.1146/annurev.med.080708.082655 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L.Z., Lu C., Li Z., Nishida N., Walz T., Springer T.A. 2011. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 18:984–989 10.1038/nsmb.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.H., Khan O.A., Sheremata W.A., Blumhardt L.D., Rice G.P., Libonati M.A., Willmer-Hulme A.J., Dalton C.M., Miszkiel K.A., O’Connor P.W. 2003. A controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 348:15–23 10.1056/NEJMoa020696 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Symonds E.J., Buckley P.A., Grossmann J.G., McEwan P.A., Barton S.J., Askari J.A., Craig S.E., Bella J., Humphries M.J. 2003. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J. Biol. Chem. 278:39993–39999 10.1074/jbc.M304627200 [DOI] [PubMed] [Google Scholar]
- Newham P., Craig S.E., Seddon G.N., Schofield N.R., Rees A., Edwards R.M., Jones E.Y., Humphries M.J. 1997. α4 integrin binding interfaces on VCAM-1 and MAdCAM-1. Integrin binding footprints identify accessory binding sites that play a role in integrin specificity. J. Biol. Chem. 272:19429–19440 10.1074/jbc.272.31.19429 [DOI] [PubMed] [Google Scholar]
- Newham P., Craig S.E., Clark K., Mould A.P., Humphries M.J. 1998. Analysis of ligand-induced and ligand-attenuated epitopes on the leukocyte integrin α4β1: VCAM-1, mucosal addressin cell adhesion molecule-1, and fibronectin induce distinct conformational changes. J. Immunol. 160:4508–4517 [PubMed] [Google Scholar]
- Nishida N., Xie C., Shimaoka M., Cheng Y., Walz T., Springer T.A. 2006. Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity. 25:583–594 10.1016/j.immuni.2006.07.016 [DOI] [PubMed] [Google Scholar]
- Pulido R., Elices M.J., Campanero M.R., Osborn L., Schiffer S., García-Pardo A., Lobb R., Hemler M.E., Sánchez-Madrid F. 1991. Functional evidence for three distinct and independently inhibitable adhesion activities mediated by the human integrin VLA-4. Correlation with distinct α 4 epitopes. J. Biol. Chem. 266:10241–10245 [PubMed] [Google Scholar]
- Sandborn W.J., Colombel J.F., Enns R., Feagan B.G., Hanauer S.B., Lawrance I.C., Panaccione R., Sanders M., Schreiber S., Targan S., et al. 2005. Natalizumab induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med. 353:1912–1925 10.1056/NEJMoa043335 [DOI] [PubMed] [Google Scholar]
- Schiffer S.G., Hemler M.E., Lobb R.R., Tizard R., Osborn L. 1995. Molecular mapping of functional antibody binding sites of α 4 integrin. J. Biol. Chem. 270:14270–14273 [DOI] [PubMed] [Google Scholar]
- Schweighoffer T., Tanaka Y., Tidswell M., Erle D.J., Horgan K.J., Luce G.E.G., Lazarovits A.I., Buck D., Shaw S. 1993. Selective expression of integrin α4β7 on a subset of human CD4+ memory T cells with Hallmarks of gut-trophism. J. Immunol. 151:717–729 [PubMed] [Google Scholar]
- Soler D., Chapman T., Yang L.L., Wyant T., Egan R., Fedyk E.R. 2009. The binding specificity and selective antagonism of vedolizumab, an anti-α4β7 integrin therapeutic antibody in development for inflammatory bowel diseases. J. Pharmacol. Exp. Ther. 330:864–875 10.1124/jpet.109.153973 [DOI] [PubMed] [Google Scholar]
- Springer T.A. 1994. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 76:301–314 10.1016/0092-8674(94)90337-9 [DOI] [PubMed] [Google Scholar]
- Springer T.A., Zhu J., Xiao T. 2008. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 182:791–800 10.1083/jcb.200801146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch U.G., Lifka A., Gosslar U., Kilshaw P.J., Clements J., Holzmann B. 1994. Distinct binding specificities of integrins α4β7 (LPAM-1), α 4 β 1 (VLA-4), and α IEL β 7. Int. Immunol. 6:263–275 10.1093/intimm/6.2.263 [DOI] [PubMed] [Google Scholar]
- Streeter P.R., Berg E.L., Rouse B.T.N., Bargatze R.F., Butcher E.C. 1988. A tissue-specific endothelial cell molecule involved in lymphocyte homing. Nature. 331:41–46 10.1038/331041a0 [DOI] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., Springer T.A. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–11 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., Walz T. 2003. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 22:4607–4615 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Casasnovas J.M., Liu J.H., Briskin M.J., Springer T.A., Wang J.H. 1998. The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition. Structure. 6:793–801 10.1016/S0969-2126(98)00080-X [DOI] [PubMed] [Google Scholar]
- Tidswell M., Pachynski R., Wu S.W., Qiu S.-Q., Dunham E., Cochran N., Briskin M.J., Kilshaw P.J., Lazarovits A.I., Andrew D.P., et al. 1997. Structure-function analysis of the integrin β7 subunit: identification of domains involved in adhesion to MAdCAM-1. J. Immunol. 159:1497–1505 [PubMed] [Google Scholar]
- Viney J.L., Jones S., Chiu H.H., Lagrimas B., Renz M.E., Presta L.G., Jackson D., Hillan K.J., Lew S., Fong S. 1996. Mucosal addressin cell adhesion molecule-1: a structural and functional analysis demarcates the integrin binding motif. J. Immunol. 157:2488–2497 [PubMed] [Google Scholar]
- von Andrian U.H., Engelhardt B. 2003. α4 integrins as therapeutic targets in autoimmune disease. N. Engl. J. Med. 348:68–72 10.1056/NEJMe020157 [DOI] [PubMed] [Google Scholar]
- Vonderheide R.H., Tedder T.F., Springer T.A., Staunton D.E. 1994. Residues within a conserved amino acid motif of domains 1 and 4 of VCAM-1 are required for binding to VLA-4. J. Cell Biol. 125:215–222 10.1083/jcb.125.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C., Blanc E., Roversi P., Bricogne G. 2007. Automated structure solution with autoSHARP. Methods Mol. Biol. 364:215–230 [DOI] [PubMed] [Google Scholar]
- Wang J.-h., Pepinsky R.B., Stehle T., Liu J.-h., Karpusas M., Browning B., Osborn L. 1995. The crystal structure of an N-terminal two-domain fragment of (VCAM-1): A cyclic peptide based on the domain 1 C-D loop can inhibit VCAM-1-akpha 4 integrin interaction. Proc. Natl. Acad. Sci. USA. 92:5714–5718 10.1073/pnas.92.12.5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., Springer T.A. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Zhu J., Chen X., Mi L., Nishida N., Springer T.A. 2010. Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 29:666–679 10.1038/emboj.2009.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Mahalingham B., Alonso J.L., Borrelli L.A., Rui X., Anand S., Hyman B.T., Rysiok T., Müller-Pompalla D., Goodman S.L., Arnaout M.A. 2009. Crystal structure of the complete integrin αVb3 ectodomain plus an α/β transmembrane fragment. J. Cell Biol. 186:589–600 10.1083/jcb.200905085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A.A., McLean M.A., Sligar S.G., Taylor K.A., Ginsberg M.H. 2010. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188:157–173 10.1083/jcb.200908045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., Springer T.A. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., Negri A., Provasi D., Filizola M., Coller B.S., Springer T.A. 2010. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 116:5050–5059 10.1182/blood-2010-04-281154 [DOI] [PMC free article] [PubMed] [Google Scholar]