High-resolution time-lapse imaging of meiosis in C. elegans reveals stage-specific, dynein-driven chromosome motion that accelerates homologue pairing and triggers synapsis.
Abstract
Meiotic chromosome segregation requires homologue pairing, synapsis, and crossover recombination, which occur during meiotic prophase. Telomere-led chromosome motion has been observed or inferred to occur during this stage in diverse species, but its mechanism and function remain enigmatic. In Caenorhabditis elegans, special chromosome regions known as pairing centers (PCs), rather than telomeres, associate with the nuclear envelope (NE) and the microtubule cytoskeleton. In this paper, we investigate chromosome dynamics in living animals through high-resolution four-dimensional fluorescence imaging and quantitative motion analysis. We find that chromosome movement is constrained before meiosis. Upon prophase onset, constraints are relaxed, and PCs initiate saltatory, processive, dynein-dependent motions along the NE. These dramatic motions are dispensable for homologous pairing and continue until synapsis is completed. These observations are consistent with the idea that motions facilitate pairing by enhancing the search rate but that their primary function is to trigger synapsis. This quantitative analysis of chromosome dynamics in a living animal extends our understanding of the mechanisms governing faithful genome inheritance.
Introduction
Meiosis, the specialized cell division process required for sexual reproduction, produces haploid cells from diploid progenitors by segregating homologous chromosomes to different daughter cells. To segregate properly, each chromosome must first pair with its homologous partner, culminating in intimate lengthwise association. In most organisms, homologous interactions are stabilized through formation of the synaptonemal complex and the covalent linkage of homologues through crossover recombination. The mechanisms that promote chromosome pairing and assess homology to selectively reinforce appropriate interactions remain among the most enigmatic aspects of the meiotic program.
Homologue pairing and synapsis occur during the stage of meiotic prophase classically defined as zygotene. At the onset of this phase, chromosomes are dramatically repositioned through a mechanism involving attachment to the nuclear envelope (NE). This is commonly accompanied by clustering of telomeres near the microtubule (MT)-organizing center (centrosome or spindle pole body) to establish the “meiotic bouquet” (Scherthan et al., 1996; Zickler and Kleckner, 1998; Bass et al., 2000; Vazquez et al., 2002). In Caenorhabditis elegans, a modified bouquet is mediated by association of special chromosome regions called pairing centers (PCs), which are located toward one end of each chromosome, with the NE and the MT cytoskeleton (Sato et al., 2009). This conformation corresponds to the transition zone (TZ) region of the gonad, in which the chromatin mass adopts a crescent-shaped morphology (MacQueen and Villeneuve, 2001).
PCs contain many dispersed copies of short DNA sequences recognized by a family of zinc finger proteins, HIM-8, ZIM-1, ZIM-2, and ZIM-3 (MacQueen et al., 2005; Phillips et al., 2005, 2009; Phillips and Dernburg, 2006). These proteins promote interaction between PCs and a conserved pair of SUN and KASH domain-containing proteins, SUN-1 and ZYG-12, which span the NE (Penkner et al., 2007; Sato et al., 2009). During early meiosis, cytoplasmic dynein concentrates with SUN-1 and ZYG-12 at patches along the NE (Sato et al., 2009). The interaction between PCs and the SUN–KASH complex, as well as functional dynein and MTs, is required for normal pairing and synapsis, leading to the suggestion that dynein-driven translocation along MTs regulates pairing and coordinates appropriate pairing with synapsis (Sato et al., 2009).
Here, we present an analysis of meiotic chromosome dynamics based on high-speed, high-resolution time-lapse imaging. Adult C. elegans hermaphrodites contain ∼500 meiotic prophase nuclei arranged in a temporal progression within each arm of the gonad (Fig. 1 A). Meiotic nuclei are ∼3.5 µm in diameter, which is small enough to allow rapid 4D imaging yet large enough that details such as individual chromosomes and PCs can be resolved. We uncover processive chromosome motions (PCMs) that depend on dynein and MTs and are regulated by meiotic progression. These rapid motions are dispensable for homologue pairing at the PCs, suggesting that they play alternate roles in homology assessment and/or synapsis. Our analysis also reveals constraints on premeiotic chromosome motion, the relaxation of which contributes to homologue pairing in a dynein-independent manner.
Figure 1.
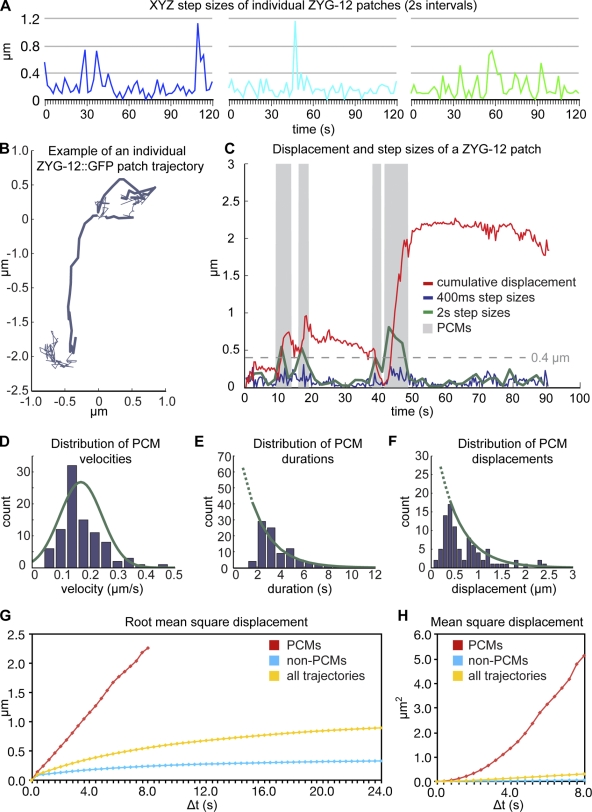
ZYG-12::GFP patches exhibit heterogeneous, independent trajectories along the NE. (A) Diagram of an adult C. elegans hermaphrodite indicating the temporospatial organization of germline nuclei. The rectangle indicates the TZ region imaged in C. ONM, outer nuclear membrane; INM, inner nuclear membrane. (B) Diagram of the organization of NE patches, showing a PC bound by HIM-8 and the associated aggregate of SUN-1 and ZYG-12. (C) Projection of one time point, comprising 33 optical sections (spanning a depth of 6 µm) showing ZYG-12::GFP in TZ nuclei. Meiosis progresses from left to right. (D) Selected projections showing a single TZ nucleus. In the bottom images, colored spheres mark two patches that merge and split (nuclear surface is indicated by a circle in the first image). (E) Colored tracks indicate all the steps for patches shown in D. (F) Selected frames showing a nucleus with six ZYG-12 patches (top) and overlays of the segmented patches (bottom). (G) Colored tracks indicate all the steps for patches shown in F over a 2-min time course. (H) Colored tracks representing all steps over a 2-min time course for all patches in three representative nuclei; each nucleus is shown from two orthogonal viewpoints to highlight the distribution of patches spanning the entire NE. Dotted lines indicate the nuclear surface. Times are given in minutes and seconds. Bars: (C) 5 µm; (D–H) 1 µm.
Results
PCs undergo independent processive motions during early meiotic prophase
At the onset of meiotic prophase, all PCs associate with the NE, where they induce the aggregation of the transmembrane proteins SUN-1 and ZYG-12 into patches ranging from 0.3 to 1.2 µm in diameter (Fig. 1, B and C; Penkner et al., 2007; Sato et al., 2009; Baudrimont et al., 2010; Harper et al., 2011). These patches persist until the completion of synapsis, at which point SUN-1 and ZYG-12 redistribute throughout the NE.
To analyze the motion of PCs, we first recorded images from animals expressing a zyg-12::gfp transgene (Malone et al., 2003). Initial observations revealed that the ZYG-12::GFP patches observed in early meiotic prophase were highly mobile (Sato et al., 2009). Using the OMX (Optical Microscope Experimental) high-speed wide-field imaging system (Carlton et al., 2010), we recorded single-wavelength 3D data stacks, with 2-s intervals between successive stacks (Fig. 1, C–G; and Video 1). These recordings were limited to a duration of ∼5 min before the signal/noise ratio was compromised by photobleaching. The resulting 4D datasets allowed segmentation and tracking of fluorescent patches along the nuclear surface using a semiautomated approach (see Materials and methods).
These recordings provided several insights into the motion of PCs during early prophase. First, the number of patches observed typically ranged from four to six per nucleus, fewer than the 12 individual chromosomes. This is consistent with evidence that homologous PCs pair early in prophase, based on immunofluorescence and FISH analysis (MacQueen et al., 2002; Phillips et al., 2005). The detection of fewer patches than chromosome pairs also suggests that chromosomes interact with both homologous and heterologous partners from the earliest stages at which patches are observed. Patches were frequently observed to merge and/or split over the course of a few minutes. An example of a nucleus in which two patches merge, remain together for 6 s, and subsequently separate can be seen in Fig. 1 (D and E) and Video 2. An example of a nucleus in which six patches are clearly moving independently is shown in Fig. 1 (F and G) and Video 2. Quantitative analysis of patch trajectories indicated that the motions were largely uncorrelated in their xyz direction and are therefore not a consequence of nuclear translation or rotation (Fig. S1). Although we observed many instances of multiple ZYG-12 patches in proximity, these clusters were transient and were not preferentially localized to one area of the NE, corroborating previous analysis of fixed samples, which indicated that attachment sites do not form a classical tightly clustered bouquet (Fig. 1 H).
The distribution of ZYG-12 step sizes was extremely heterogeneous, both within the population and for individual trajectories (Fig. 2 A). The mean xyz step size for all patches between adjacent data stacks acquired at 2-s intervals was 0.25 µm, corresponding to a mean apparent speed of 0.125 µm/s. However, we observed occasional jumps of >0.8 µm within a 2-s interval, indicating the occurrence of transient movements of ≥0.4 µm/s. At this 2-s sampling interval, we detected only a few instances in which the direction and rate of motion of individual patches were correlated over consecutive time points (Fig. S1). This weak correlation suggested that changes in direction and speed occur on a time scale more rapid than our sampling rate and that data collected at higher temporal resolution would enable a more complete description of patch motion.
Figure 2.
Rapid 2D imaging enables two modes of motion to be distinguished. (A) Plots depicting xyz step sizes between each time point for three patches shown in Fig. 1 (F and G). Plot colors correspond to patch color in time-lapse images. (B) Trajectory of a single ZYG-12 patch followed at 400-ms intervals. Bold segments denote processive chromosome motions (PCMs). (C) Cumulative distance plot for the single representative trajectory shown in B. Green line shows step sizes observed between every five frames (2-s intervals). (D) Speed distribution of ZYG-12 PCMs. Green line shows fitted normal distribution. n = 91 PCMs. (E and F) Distributions of durations (E) and total displacements (F) of ZYG-12 PCMs. Green lines show fitted exponential decay; dotted lines show extrapolation to shorter times. (G and H) RMSD and MSD plots for PCMs, non-PCMs, and all trajectories. The limited duration of PCMs allows RMSD analysis up to only 8 s.
We acquired data at a fivefold higher sampling rate by recording a single confocal optical section at 400-ms intervals, capturing multiple patches per nucleus (Video 3). In these 2D datasets, we detected a marked increase in directional correlation between adjacent patch movements (Fig. S1), indicating the occurrence of processive movements of a few seconds duration. This revealed that patch trajectories are composites of at least two distinct modes of displacement: PCMs, in which patches move continuously in the same direction for up to several seconds, interspersed with periods in which patches exhibit constant changes in direction and remain close to their origins (Fig. 2 B, bold segments denote PCMs). Although observed step sizes in our data did not change markedly during a PCM (Fig. 2 C, blue), these two modes of motion could be clearly distinguished by tracking the distance traveled by a patch from its initial position over time (Fig. 2 C, red plot): periods in which the displacement shows uniform and concerted changes over several time points stand out among the small fluctuations in the otherwise horizontal plot. To automatically identify these PCMs, we searched for segments in which the cumulative displacement over three consecutive time intervals (1.2 s) was in the top 15th percentile of three-step displacements (Fig. 2, B [bold regions] and C [shaded regions]). Modifying these criteria by including a range of two to five consecutive steps or a displacement percentile range of 5–25% did not markedly affect our results (unpublished data). Using these criteria, we identified 91 individual PCMs from 36 trajectories in five datasets.
The measured velocities during PCMs conformed to a tight normal distribution, with a mean speed of 0.19 µm/s (SD = 0.07 µm/s; Fig. 2 D). The distributions of the duration of PCMs and total distance traveled during each instance exhibited exponential decay, characteristic of processive, motor-driven motions measured both in vivo and in vitro (e.g., Vale et al., 1996 [kinesin]; Veigel et al., 1999 [myosin I]; Sakamoto et al., 2000 [myosin V]; Reck-Peterson et al., 2006 [dynein]). Calculation of the decay coefficient allowed us to estimate a mean PCM run length of 1.9 s (95% confidence interval, 1.6–2.4 s) and mean translocation in a single PCM of 0.49 µm (95% confidence interval, 0.39–0.63 µm), with some motions of >2 µm—traversing almost one fifth of the circumference of a nucleus (∼11 µm; Fig. 2, E and F).
To facilitate interpretation of our 3D data recorded at 2-s intervals, we computationally undersampled our higher speed 2D data by considering only every fifth time point. When we overlaid the displacement plot using all frames at 400-ms intervals (Fig. 2 C, red) on a plot of the steps observed between every fifth frame (2 s; Fig. 2 C, green), it was clear that PCMs corresponded well with the intermittent large step sizes observed at lower temporal resolution (e.g., Fig. 2 A). Indeed, all of the steps of >0.4 µm in 2 s corresponded to a PCM, and thus, the fraction of these large steps in our 3D datasets provides a good measure of the frequency and prevalence of PCMs.
During periods that did not meet the criteria for PCMs, sequential steps exhibited constant changes in direction and limited displacement (e.g., Fig. 2 C, 24–38 s) and were therefore analyzed in a different way, by considering their cumulative effects over larger time intervals. Plots of the root mean square displacement (MSD; RMSD) provide a direction-independent measure of the distance traveled over all time intervals, minimizing the effects of stochastic changes in direction. The RMSD plot for the PCMs has a steep slope, reflecting the large distances traveled in short periods. Notably, RMSD plots of the non-PCM segments of patch trajectories have a lower slope (Fig. 2 G). These differences underscore the major contribution made by PCMs to the mobility of patches and to the rate at which they explore the nuclear surface, even though patches are engaged in PCMs only ∼15% of the time. Plotting the MSD over time (Fig. 2 H) highlights the mode of movement: objects that move by free diffusion generate linear MSD plots, whereas those undergoing active motion or biased diffusion result in MSD plots with upward curvature (Saxton, 1993). We detected upward curvature in the MSD plot of PCMs alone, as expected for coordinated motion. However, MSD plots corresponding to all patch motions or non-PCM segments do not show upward curvature, consistent with diffusion being the prevailing mode of motion.
In summary, rapid 2D imaging indicates that the saltatory large steps in our 3D data reflect periods in which chromosome sites associated with the NE translocate processively along linear paths, consistent with motor-driven motion along rigid cytoskeletal elements. Such motions occupy individual patches only a minority of the time but contribute markedly to overall mobility. Analysis of images acquired at high frame rates provides a physical explanation for the variable step sizes seen in our lower resolution data (Fig. 2 A and Discussion) and also observed by Baudrimont et al. (2010).
Chromosomes undergo PC-driven motion
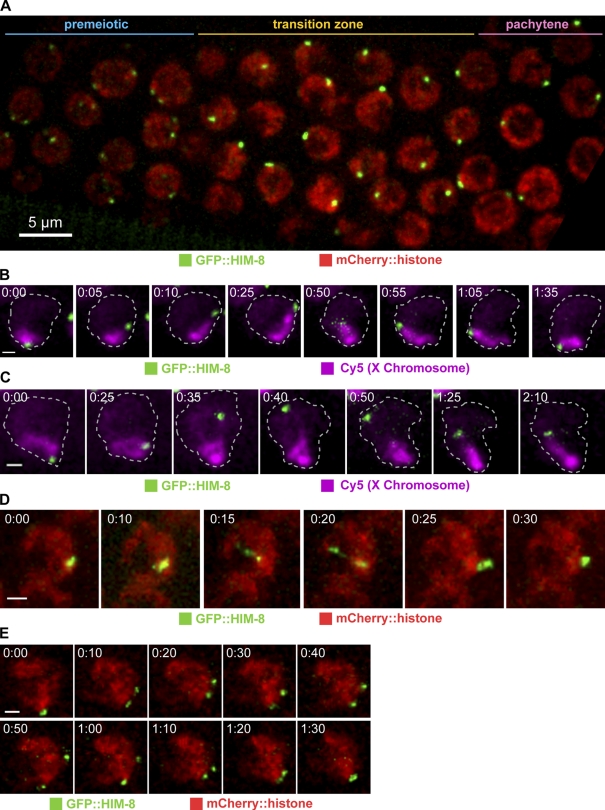
To enable analysis of the motion of specific chromosomes, we generated strains expressing HIM-8 fused to GFP (see Materials and methods; hereafter GFP::HIM-8). The fusion protein recapitulated the localization of endogenous HIM-8 (Phillips et al., 2005), associating with the X chromosome PCs (X PCs) in both premeiotic and meiotic prophase nuclei in the germline (Fig. 3 A). GFP::HIM-8 was crossed to a strain expressing mCherry::histone H2B (McNally et al., 2006), which allowed us to assign GFP::HIM-8 foci to specific nuclei, to differentiate premeiotic, TZ, and pachytene nuclei based on chromosome morphology, and to computationally align adjacent time points in time-lapse data (see Materials and methods). In most TZ nuclei, GFP::HIM-8 localized to a single focus, consistent with prior evidence that X chromosome PCs pair rapidly after the onset of meiosis (Figs. 3 A and S2; Phillips et al., 2005).
Figure 3.
GFP::HIM-8 reveals X PC dynamics. (A) Projection image of 12 optical sections from a single time point showing a field of nuclei expressing GFP::HIM-8 and mCherry::histone. Meiosis progresses from left to right. At the left edge of the field, nuclei have not yet entered meiosis. Pachytene nuclei lacking a crescent-shaped chromosome mass are seen toward the right. (B and C) Projections of selected time points showing a single nucleus expressing GFP::HIM-8 in which the X chromosome is visualized by Cy5-dUTP incorporation. Nuclear outlines are based on the mCherry::histone signal (not depicted). The dotted lines demarcate the nuclear outline, as observed by imaging of mCherry::histone. (D and E) Projections of selected time points showing GFP::HIM-8 (red) and mCherry::histone. These examples show nuclei in which X PCs (green) stretch (D) or dissociate temporarily (E) after colocalizing. Times are given in minutes and seconds. Bars: (A) 5 µm; (B–E) 1 µm.
To follow X PCs over time, we collected two-color stacks of optical sections using spinning-disk confocal microscopy. The requirement for multiwavelength 3D imaging reduced our collection rate to 5-s intervals. As elaborated in the next section, these imaging conditions confirmed that X PCs in TZ nuclei undergo PCMs, as expected based on the ZYG-12::GFP imaging (Video 4). In some experiments, we specifically labeled the X chromosomes in a subset of germline nuclei by injecting fluorescent deoxynucleotides (Jaramillo-Lambert et al., 2007).
In TZ nuclei, the PC end of the X chromosome was highly mobile, whereas the distal end of the chromosome remained virtually static throughout data collection (Fig. 3 B and Video 5). Chromosomes exhibited remarkable elasticity: in some nuclei, we observed a transient separation of >2 µm between the bulk of the labeled X chromosome and the focus of GFP::HIM-8 (Fig. 3 C and Video 6). The elasticity of the PC end of the chromosome was also indicated by transient stretching of the GFP::HIM-8 signal to far beyond its normal size. In dramatic examples of this behavior, a bright GFP::HIM-8 focus split into two major foci connected by fainter fluorescence intensity, remaining apart for up to two time points (5 s; Fig. 3 D and Video 7). We found that paired GFP::HIM-8 signals, which are normally 0.4–0.8 µm wide, stretched to >1.0 µm in 28% of TZ nuclei within 5 min, indicating that this is a frequent occurrence. We also observed several instances in which paired X PCs fully separated for longer periods of time and then reassociated to form a single focus (Fig. 3 E and Video 8). This suggests that pairing may be stabilized at loci outside of the PC, allowing the PCs to separate without dissociation of the homologues.
These data indicate that chromosomes undergo rapid motions led by their PCs. The viscoelastic properties of meiotic chromosomes apparently allow them to absorb forces along their length such that regions distant from PCs undergo substantially less motion than PCs, even during large PCMs.
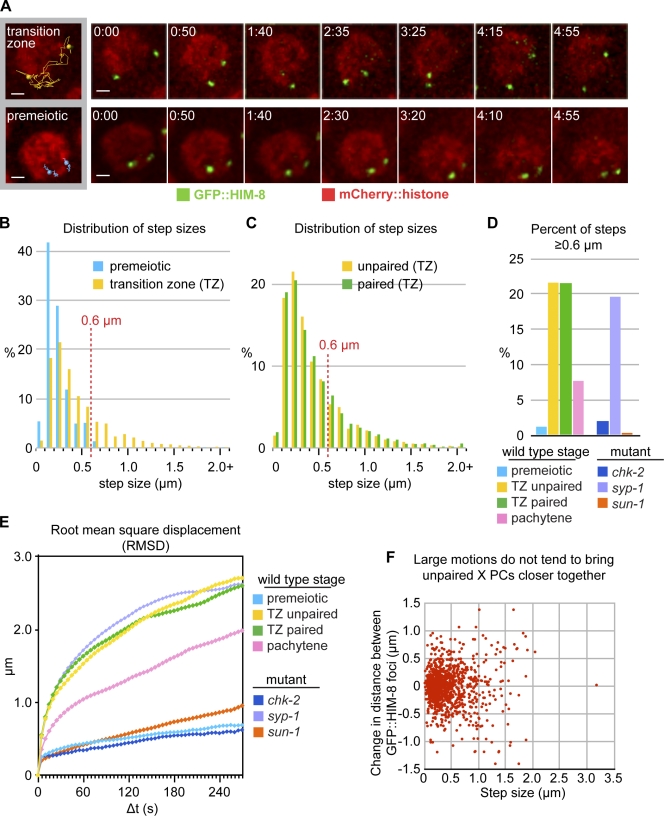
PCMs are regulated by meiotic progression but not by pairing
Like endogenous HIM-8 protein, the GFP::HIM-8 reporter localized to X PCs both before and throughout meiotic prophase, enabling us to evaluate PC dynamics as a function of meiotic entry and progression (Video 4 and Fig. 4 A). We first compared the dynamics of GFP::HIM-8 foci in TZ nuclei to measurements of ZYG-12::GFP patches at the same stage. Tracking and quantification of X PC motion in TZ nuclei yielded a mean step size of 0.412 µm, corresponding to a mean apparent speed of 0.082 µm/s. Based on the aforementioned analysis for ZYG-12::GFP motion, we defined PCMs as steps of >0.6 µm between 3D images acquired at 5-s intervals. 21.5% of observed steps met this criterion, consistent with the proportion of PCMs observed for ZYG-12::GFP (Figs. 4 B and S3). The duration and velocity of these steps also agrees well with our measurements of ZYG-12 motion, adjusted for the different sampling rate (Fig. S4). We also acquired single-plane images of GFP::HIM-8 motion at 400-ms intervals to confirm that large step sizes observed at 5-s intervals correspond to PCMs (Fig. S3). Although we note that the mean velocity of X PC PCMs is somewhat higher than the velocity measured for all PCMs using ZYG-12::GFP, the similarity between the data collected for these two reporters indicated that PCMs of X PCs are a good proxy for all PCMs (compare Figs. 2 and S3).
Figure 4.
PCMs are regulated by meiotic progression but not by pairing. (A) Projections of selected time points showing GFP::HIM-8 and mCherry::histone from single nuclei in the TZ or the premeiotic zone. The left images show complete X PC trajectories overlaid on a projection of the initial time point. Times are given in minutes and seconds. Bars, 1 µm. (B) Step-size distributions of X PCs in premeiotic zone nuclei and TZ nuclei with unpaired X PCs (premeiotic: 2,248 steps, 38 trajectories, and 3 datasets; TZ unpaired: 1,443 steps, 26 trajectories, and 3 datasets). Mann–Whitney U test, P = 10−212. (C) Step-size distributions of unpaired and paired X PCs in TZ nuclei (TZ paired: 3,779 steps, 65 trajectories, and 6 datasets). Mann–Whitney U test, P = 0.85. (D) Fraction of steps >0.6 µm in different meiotic stages and in syp-1(me17), chk-2(me64) and sun-1(jf18) mutant animals (syp-1(me17): 2,698 steps, 45 trajectories, and 3 datasets; chk-2(me64): 1,620 steps, 30 trajectories, and 2 datasets; sun-1(jf18): 2,650 steps, 45 trajectories, and 3 datasets). (E) RMSD plots for all trajectories. The plateau at ∼2.5 µm reflects the distance limit for travel on the surface of a sphere ∼3.5 µm in diameter. (F) The change in distance between unpaired GFP::HIM-8 foci in TZ nuclei (vertical axis) is plotted as a function of the size of individual steps (horizontal axis). Longer steps, corresponding to PCMs, do not tend to bring X PCs closer together.
Analysis of GFP::HIM-8 dynamics in premeiotic nuclei, which occupy the region of the gonad distal to the TZ, indicated that PCMs were absent. This was evident from both 3D data acquired at 5-s intervals (Fig. 4, A and B; and Video 9) and 2D images recorded at 400-ms intervals (Figs. S3 and S5). In premeiotic nuclei, the mean observed step size was 0.195 µm, and only 1.2% of steps traversed >0.6 µm—far below the proportion seen in TZ nuclei (Fig. 4, B and D).
Notably, PCs in premeiotic nuclei showed relatively restricted motion within the NE: in TZ nuclei, the mean displacement of PCs over 20 s was 1.2 µm, a magnitude that was never observed over the entire 5-min time course in premeiotic nuclei, which showed a mean displacement of 0.7 µm after 5 min (Fig. 4 E). This mean displacement of 0.7 µm corresponds to a search area of 1.54 µm2 on the NE, only ∼4% of the total area.
The absence of PCMs in the premeiotic zone indicates that PCMs are a regulated aspect of the meiotic program. To further investigate how chromosome dynamics are regulated at meiotic entry, we analyzed X PC motion in the gonads of chk-2(me64) mutant animals, which fail to initiate many events of the meiotic program: the appearance of patches of SUN-1 and ZYG-12 in the TZ (Penkner et al., 2009; Sato et al., 2009), pairing and synapsis, and initiation of recombination (MacQueen and Villeneuve, 2001). GFP::HIM-8 foci in the TZ region of chk-2 mutants showed similar mobility to those in the premeiotic region of wild-type (WT) animals with respect to step size, displacement, and absence of PCMs (Figs. 4, D and E; and S4). PCMs were also absent in gonads of sun-1(jf18) mutant animals, which have small NE patches and defective pairing and synapsis (Figs. 4, D and E; and S4; Penkner et al., 2007; Sato et al., 2009). These findings reinforce the idea that aggregation of SUN-1 and ZYG-12 into patches is required for normal early prophase dynamics because HIM-8 is associated with the NE (but not with patches) in WT premeiotic nuclei and during early meiosis in sun-1(jf18) and chk-2(me64) hermaphrodites (Phillips et al., 2005; Penkner et al., 2007, 2009; Sato et al., 2009).
We compared the dynamics of unpaired HIM-8 foci, which were occasionally observed in early TZ nuclei, to paired foci, and found that the step-size distributions and RMSD plots were indistinguishable (Fig. 4, C and D). The fraction of steps of >0.6 µm were 21.5 and 21.6% for unpaired and paired foci, respectively. This result indicates that X chromosome dynamics remain constant throughout the TZ, before and after pairing and synapsis. Moreover, it implies that the forces exerted on PCs are sufficiently strong, or scale proportionately, to be insensitive to the increased drag imposed by association with another chromosome.
Analysis of TZ nuclei with unpaired GFP::HIM-8 signals also allowed us to test whether PCMs are biased in direction such that they directly promote pairing by bringing PCs closer together. Specifically, we tested whether the speed of X PC motion was correlated with the change in distance between homologous foci during the same interval (Fig. 4 F). This analysis revealed that both smaller fluctuations and PCMs move X PCs away from each other as often as they bring them closer together, consistent with pairing being a consequence of random collisions (see Discussion). This reinforced our conclusion from the ZYG-12::GFP data that individual PCs move independently and nondirectionally during early prophase. It also corroborated our finding that the observed motion is dominated by PC movements along the nuclear surface and is not a consequence of nuclear drift or rotation (Fig. S1).
Upon completion of synapsis, nuclei exit the TZ stage and enter pachytene, which is marked by the dispersal of SUN-1 and ZYG-12 throughout the NE. However, the GFP::HIM-8 reporter remains associated with synapsed X chromosomes and with a single remaining focus of ZYG-12 and SUN-1 (Sato et al., 2009). Compared with TZ nuclei, X PCs in pachytene nuclei showed markedly reduced motion. This was apparent as a much smaller proportion of steps >0.6 µm and a lower slope of the RMSD plot (Figs. 4, D and E; and S4). We also measured the motion of X PCs in syp-1(me17) mutant hermaphrodites, in which the absence of chromosome synapsis results in an elongated region of leptotene/zygotene stage nuclei, often referred to as an extended TZ (MacQueen et al., 2002). PC motion in the TZ region of syp-1 mutants was similar to that in WT TZ nuclei (Figs. 4, D and E; and S4), consistent with our conclusion that synapsis does not markedly alter chromosome motions. These motions continued with indistinguishable velocities, step sizes, and displacements throughout the extended TZ region (unpublished data). In contrast to a recent study (Baudrimont et al., 2010), we did not find a significant difference between step sizes in syp-1 mutants compared with WT (P = 0.5, Mann–Whitney U test). This reinforces the idea that motion is regulated by the meiotic stage of each nucleus and not by extrinsic signals such as its position in the gonad.
PCMs depend on MTs and dynein but not on actin
As described in the aforementioned results and in Figs. 2 and 4, PCMs in TZ nuclei show velocities and a distribution of durations consistent with motor-driven motion along linear cytoskeletal elements. Prior work has shown that disruption of MTs abrogates pairing and synapsis (Sato et al., 2009). Consistent with these findings, we observed that MT destabilization through injection of 0.1 M colchicine into the gonad eliminated all PCMs, as indicated by a distribution of step sizes similar to premeiotic nuclei (Fig. 5, A and B; compare Fig. 5 B with Fig. 4 B) and elimination of most steps >0.6 µm (1.9 vs. 22.1% for buffer-injected control animals; compare Fig. 5 C with Fig. 4 D). In contrast, PCMs were not dependent on actin: animals microinjected with 10 µM latrunculin A exhibited PC motion indistinguishable from control animals, as judged by the step-size distributions, the fraction of steps >0.6 µm (26.2%), and RMSD plots (Fig. 5, A–D). Thus, PCMs are completely dependent on MTs but insensitive to destabilization of actin filaments.
Figure 5.
PCMs are dependent on MTs but not on actin. (A) Projections of selected time points showing TZ nuclei expressing GFP::HIM-8 and mCherry::histone. X chromosomes are selectively labeled by incorporation of Cy5-dUTP. Animals were injected with colchicine, latrunculin A, or buffer. Bars, 1 µm. Leftmost images show the complete GFP::HIM-8 trajectory for each 5-min series overlaid on a projection of the initial z stack. Times are given in minutes and seconds. (B) Distributions of step sizes from worms injected with colchicine (left) or latrunculin A (right) compared with controls (colchicine: 2,583 steps, 46 trajectories, and 2 datasets; latrunculin A: 1,363 steps, 24 trajectories, and 1 dataset; buffer injected: 1,855 steps, 32 trajectories, and 1 dataset; bins = 0.1 µm). (C) Fraction of steps >0.6 µm observed for each condition. (D) RMSD plots for all trajectories analyzed in B and C.
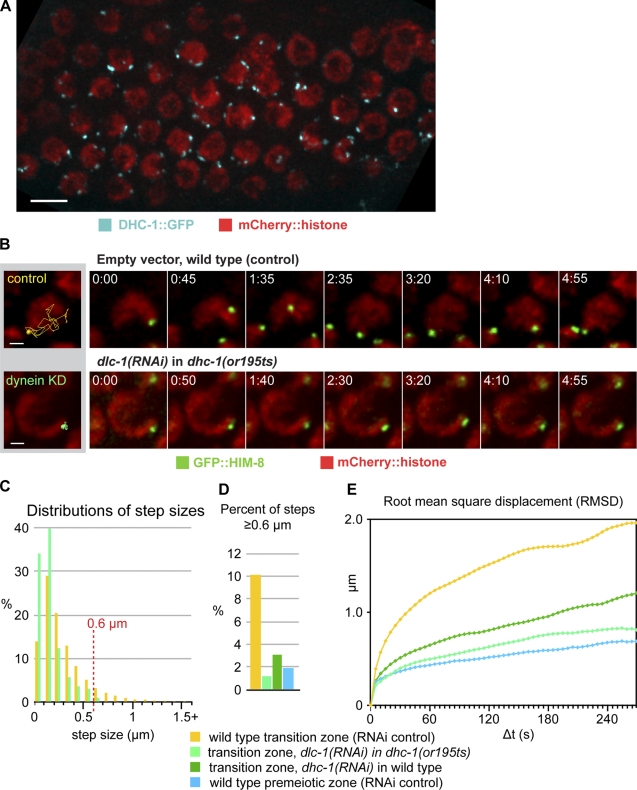
The activity of cytoplasmic dynein enhances the rate of homologue pairing and is required for synapsis (Sato et al., 2009). Fluorescently tagged dynein (DHC-1::GFP; Gassmann et al., 2008) localized to bright, dynamic foci associated with the NE of TZ nuclei (Fig. 6 A), consistent with immunolocalization of dynein in fixed gonads (Sato et al., 2009). Importantly, dynein foci were not restricted to patches undergoing PCMs but rather showed similar fluorescence intensity at all patches (Video 10), indicating that dynein recruitment is not sufficient for PCMs.
Figure 6.
Dynein activity is required for PCMs. (A) Maximum intensity projection image from a recording of a hermaphrodite expressing DHC-1::GFP and mCherry::histone. Meiotic progression is from left to right; TZ nuclei show prominent dynein foci at the nuclear surface. Bar, 5 µm. (B) Selected projection images from a time series showing GFP::HIM-8 and mCherry::histone from single nuclei in the TZ of dynein knockdown (KD; dlc-1 RNAi in dhc-1(or195ts)) or WT control animals. (left) Tracks represent all steps over a 5-min data collection superimposed on a projection image from a single frame. Bars, 1 µm. (C) Distributions of step sizes in TZ nuclei from dynein knockdown (dlc-1 RNAi in dhc-1(or195ts)) animals compared with TZ nuclei of RNAi control animals (dynein knockdown [dlc-1 RNAi in dhc-1(or195ts)]: 2,599 steps, 45 trajectories, and 3 datasets; control RNAi: 2,588 steps, 45 trajectories, and 3 datasets). Bins = 0.1 µm. (D) Proportion of steps >0.6 µm under the indicated conditions; data from WT premeiotic nuclei are shown for comparison (dynein knockdown [dhc-1 RNAi 30 h]: 2,396 steps, 40 trajectories, and 3 datasets). (E) RMSD plots for all trajectories. Colors correspond to those in D.
We used several approaches to knock down dynein function in the germline. RNAi targeting the dynein light chain gene dlc-1 in animals carrying a temperature-sensitive allele of dynein heavy chain dhc-1(or195) at a restrictive temperature is an effective way to inhibit dynein activity during meiosis, with only limited mitotic defects (O’Rourke et al., 2007; Sato et al., 2009). We found that this double knockdown approach eliminated PCMs (Fig. 6, B–E; and Video 10). The step-size distribution in dynein knockdown animals showed a downward shift (Fig. 6 C), and the fraction of steps >0.6 µm was reduced to 1.2% compared with 10.1% in control knockdowns (Fig. 6 D); note that PCMs in control animals were also somewhat reduced, likely because of the increased age and/or elevated growth temperature required for this dynein knockdown protocol. High-speed imaging of both GFP::HIM-8 and ZYG-12::GFP confirmed an absence of PCMs (Fig. S5). Similar results were obtained when dhc-1 was knocked down by RNAi in WT animals at 20°C (Fig. S5). Collectively, these data demonstrate that dynein is essential for PCMs, which likely reflect movements along cytoplasmic MTs.
Increased PC mobility in meiotic prophase results from a combination of PCMs and reduced constraints to diffusion
Although PCMs first appear at meiotic entry, concomitant with homology search and the initiation of synapsis, homologue pairing does not require dynein activity (this study; Sato et al., 2009). We therefore investigated whether other features of early prophase dynamics might contribute to pairing. By comparing plots of RMSD, we asked how extensively PCs explore the nuclear periphery under various conditions. We found that even after dynein knockdown, PCs explore a larger region of the NE in early meiosis than they do in premeiotic nuclei (Fig. S4). This difference can be visualized more directly by overlaying multiple observed trajectories set to initiate at the same point on a sphere (Fig. 7 A). Trajectories from dynein knockdown recordings display a search radius intermediate between premeiotic and control TZ nuclei. This indicates that the motion of PCs is constrained in premeiotic nuclei and that this constraint is relaxed upon meiotic entry.
Figure 7.
Dynein-dependent and -independent chromosome mobility contributes to homologue pairing in early meiotic nuclei. (A) To visualize the search radius of PCs under different conditions, 40 GFP::HIM-8 tracks from WT TZ, WT premeiotic, or TZ in dynein knockdown (dlc-1 RNAi in dhc-1(or195ts)) nuclei were rotated about a 3.5-µm-diameter sphere such that all tracks initiate at the same point. (B) RMSD plots for all GFP::HIM-8 trajectories in WT TZ nuclei, the same data with PCMs computationally removed, in TZ nuclei from dynein knockdown animals, and in premeiotic nuclei. (C) Example of a simulated collision between two objects traveling in random walks on the surface of a sphere. The star indicates the point at which the objects come within 200 nm at the same time point. (D) Comparison of time required to achieve collision in 95% of simulations based on motion parameters measured for the indicated conditions. Collision times were calculated from ≥10 independent sets of 100 simulations; error bars indicate the SD for all sets. See Materials and methods for details.
The increased mobility of PCs upon meiotic entry required both MTs and the activity of the CHK-2 kinase (Figs. 4, 5, and S4). Prior work has shown that CHK-2 activity is required for SUN-1 phosphorylation and patch formation upon meiotic entry (Penkner et al., 2009). Interestingly, we found that GFP::HIM-8 foci did show increased mobility upon meiotic entry in animals homozygous for the sun-1(jf18) allele (Fig. S4), although they did not undergo PCMs. This suggests that the jf18 missense mutation impairs the engagement of PCs with dynein but does not affect the elevated mobility that occurs upon meiotic entry, consistent with the idea that this is promoted by SUN-1 phosphorylation. However, sun-1(jf18) impairs homologue pairing to a greater degree than dynein knockdown (Penkner et al., 2007; Sato et al., 2009), indicating that another activity required for pairing—perhaps the ability of SUN-1 to aggregate—is disrupted in jf18 mutants.
To understand how these differences in overall mobility might affect PC pairing, we simulated trajectories representing a random walk along the surface of a sphere of 3.5-µm diameter using the step sizes and mean displacements measured for PCs in premeiotic and TZ nuclei (see Materials and methods for details; Fig. 7, B–D). We asked how long it would take for two objects, each 200 nm in diameter, moving along independent trajectories on the same sphere from initial positions one quadrant apart to come into contact (collide; Fig. 7 C). Not surprisingly, we found that the time to collision was much lower using the parameters measured for TZ PCs than using premeiotic values: ∼6 h was required for 95% of premeiotic simulations to produce a collision versus 18.5 min (0.3 h) for the same fraction of TZ foci to collide (Fig. 7 D). Elimination of PCMs computationally or experimentally by dynein knockdown resulted in estimates of 43 or 93 min to achieve 95% collision, respectively. Interestingly, the 1.2-h delay in simulated collision time using the dynein knockdown data agrees well with the delay in X chromosome pairing that was estimated based on images of fixed samples (Sato et al., 2009). Thus, although PCMs facilitate timely pairing and play an essential role in allowing paired chromosomes to synapse (see Discussion), the release of constraints to diffusive chromosome motion strongly contributes to promote homologue pairing during meiotic prophase.
Discussion
In this work, we have developed a quantitative analysis of meiotic chromosome movements in a living animal, based on imaging at temporal and spatial resolution surpassing previous studies in any organism. We have shown that the onset of meiotic prophase in C. elegans is accompanied by the appearance of PCMs, which greatly enhance the mobility of the PC regions of chromosomes. PCMs reach speeds >0.4 µm/s, can last for several seconds and dramatically stretch chromosomes, and require dynein and the MT cytoskeleton. These chromosome dynamics are regulated by meiotic progression but are insensitive to the pairing or synapsis status of individual chromosomes. Homologue pairing is achieved in the absence of PCMs but is delayed. This analysis has also revealed that PC motion in premeiotic nuclei is constrained relative to early meiotic prophase. The release of these constraints is likely a major factor that promotes chromosome interactions to facilitate homologue pairing.
Characterizing chromosome motion in meiotic nuclei
Direct analysis of chromosome motion during meiosis poses several technical challenges. In animals, meiosis occurs within reproductive organs, which are often not amenable to in vivo imaging. C. elegans is small, optically transparent, and resilient to being immobilized under a coverslip. However, efforts to track meiotic chromosome dynamics in vivo have been thwarted by robust germline silencing mechanisms that have made it difficult to express appropriate reporters. In this work, we used a broadly expressed ZYG-12 reporter and took advantage of recent advances in C. elegans transgene technology (Frøkjaer-Jensen et al., 2008) to generate a fluorescent HIM-8 reporter.
In any time-lapse imaging, only continuous motions lasting for at least two to three times the image acquisition interval can be tracked and measured accurately (Meijering et al., 2008). Indeed, it became evident during the course of this study that the image acquisition rate was a crucial experimental parameter. To clearly differentiate PCMs from diffusive motion required multiple images per second. A recent study in C. elegans used coarser (1 µm) optical sectioning at 5-s intervals and 2D analysis to describe SUN–KASH-dependent differential step-size distributions upon meiotic entry (Baudrimont et al., 2010). Here, we have combined high-resolution 3D imaging at a limiting speed with more rapid 2D datasets to characterize PCMs and distinguish them from the remaining uncoordinated motions. This automatic classification of PCMs allowed us to measure their speed and duration and identify their signature in the slower 3D datasets, which were essential to analyze their directionality and demonstrate their independent behavior.
The horsetail motion of Schizosaccharomyces pombe nuclei during meiotic prophase is slower and more processive and has therefore been successfully characterized at lower temporal resolution (Ding et al., 1998, 2004; Meijering et al., 2008; Vogel et al., 2009). Studies in budding yeast have been primarily based on single images per time point, either single optical sections or through-focus projection images in which information along the optical axis is sacrificed (Conrad et al., 2008; Koszul et al., 2008; Wanat et al., 2008). Because yeast nuclei are very small, a reasonable fraction of their volume can be captured in an optical section. However, in animals and plants, which have much larger meiotic nuclei, rapid 3D image acquisition will likely be crucial to capture and interpret chromosome dynamics. Even in Saccharomyces cerevisiae, in which early prophase chromosome dynamics have been extensively studied at imaging rates as fast as one frame per second (fps), higher temporal resolution will likely refine our understanding.
Imaging chromosome dynamics in living worms involves at least some specimen motion because of pharyngeal pumping and other muscle contraction. In this study, we used two different methods for alignment, based on total fluorescence or on a chromatin marker (see Materials and methods). Our statistical analysis, as well as the good agreement between the different methods, suggests that computational image alignment successfully eliminated most artifactual motions and did not introduce significant biases (Figs. S1 and S4).
Biophysical properties of chromosomes and chromosome motion
We have found that dynein-driven PCMs in C. elegans often last for >4 s and tend to be unidirectional. Although in vitro dynein stepping behavior is more variable than that of kinesin motors, in vivo, it is predominantly unidirectional toward MT minus ends (Kardon and Vale, 2009). Thus, PCMs are likely to be the result of translocation along a single MT or a bundle of parallel MTs. Our measurement of PCMs >2 µm is consistent with the processivity of dynein on MTs, which produces mean run lengths of >1 µm for purified dynein in vitro (Reck-Peterson et al., 2006; Ross et al., 2006) and similar distances in vivo (e.g., Mallik et al., 2005; Kim et al., 2007). It is possible that the run lengths in our system are determined in part by the arc lengths over which MTs are tangentially associated with the NE.
Our analysis illuminates both the elastic properties of meiotic chromosomes and the forces required to generate PCMs. We observed dramatic stretching events that transiently extended the X PC, which spans ∼1 Mb (Phillips et al., 2009), from <1 to >2 µm (Fig. 3). Measurements in grasshopper spermatocytes indicated that forces on the order of 320 pN were required to stretch individual chromatids to double their original lengths at metaphase or anaphase of meiosis I (Nicklas, 1983). For chromatids ∼1 µm in diameter, this yielded an estimated elastic constant of 4.3 × 103 dyne/cm2 (430 Pa or pN/µm2), in good agreement with independent measurements of 102–103 Pa for mitotic prophase newt and Xenopus laevis chromosomes using other techniques (Poirier et al., 2000). Prophase chromosomes in C. elegans have a diameter of ∼0.5 µm or a cross section about fourfold smaller than the anaphase chromosomes in the classical Nicklas experiments. Although we do not know how meiotic chromosome architecture or NE attachment might affect their elastic behavior, a conservative estimate is that tens, if not hundreds, of piconewtons are required to generate the stretches we observed, along with corresponding deflections of the NE.
Chromosomes in the TZ switch between two distinct modes of motion: dynein-driven PCMs and nonprocessive, apparently diffusive, motion. PCMs showed a tight normal distribution of velocities, which were not sensitive to either chromosome stretching (Fig. 3) or homologue pairing (Fig. 4), each of which would likely increase the load. Moreover, the observation that the number of ZYG-12 patches is often less than half the number of PCs indicates that three or more chromosomes are often moved together, but we observed no indication of variability in patch motion as a function of patch size. These observations suggest that the force available at patches exceeds any resistance from drag or chromosome stretching and that it is likely the engagement of motors with MTs, rather than recruitment of dynein, that allows PCM commencement.
In single-molecule experiments, individual dynein motors stall under forces in the range of 1–7 pN (Kardon and Vale, 2009), which suggests that many cooperating dynein molecules are required to produce the observed motion. The bright dynein foci that we observed at NE patches (Fig. 6; Sato et al., 2009) are consistent with the idea that a large ensemble of motors is concentrated at each patch. The fact that chromosomes appear to undergo PCMs only when they are in contact with a patch and not merely associated with the NE suggests that patch formation reflects the recruitment of sufficient numbers of motors to drive a high load process. Furthermore, LIS-1, which has been shown to mediate the concerted action of multiple dynein motors (McKenney et al., 2010), is enriched at these patches (Sato et al., 2009). Interestingly, it has been hypothesized that Drosophila melanogaster Klarsicht, a KASH domain protein like ZYG-12, facilitates concerted action of multiple motors to drive transport (Welte et al., 1998), suggesting that NE patch components may regulate force generation in addition to their role in dynein recruitment.
Other motors, such as kinesins, may contribute to the mobility of meiotic chromosomes in C. elegans, but analysis of candidate proteins has not yet been fruitful. Some of the motion could reflect passive sliding along MTs, an activity that has been attributed to Hook proteins, such as ZYG-12 (Walenta et al., 2001). It also remains possible that in our dynein knockdown experiments, residual dynein activity contributed to this enhanced mobility, although any such activity was too low to produce PCMs. The mechanism driving this apparently diffusive motion remains to be characterized.
It is remarkable that such rapid and large-scale chromosome motions can occur along the NE in the presence of an intact nuclear lamina. In other cells, the lamina is thought to be a stable structure (Moir et al., 2000), so it seems likely that its properties are modified during meiosis to increase its fluidity. Phosphorylation of SUN-1 upon meiotic entry (Penkner et al., 2009) is essential for patch formation, and it seems likely that other NE proteins also undergo meiosis-specific modification. At least two kinases, CHK-2 and PLK-2, are required to promote homologue pairing and synapsis, and future identification of their targets will likely illuminate how changes that allow chromosome mobility are implemented at meiotic entry.
Conservation of the meiotic bouquet and meiotic prophase motions
Abundant evidence indicates that the attachment of PCs to the NE and the resulting motion are analogous to roles played by telomeres during meiosis in other organisms. In many organisms, telomere attachment to the NE during meiotic prophase leads to their concentration within a confined region to form the meiotic “bouquet” (Scherthan, 2001). Although a recent study suggested that NE patches in C. elegans are limited to one hemisphere of the NE (Baudrimont et al., 2010), our observation of motions that traverse the entire surface of the nucleus does not support this idea (Fig. 1 H). In other species, the degree of telomere clustering varies markedly, suggesting that clustering per se may not be functionally important. If minus end–directed motion along cytoplasmic MTs is a widespread feature of meiotic chromosomes, it is likely that the classical bouquet configuration is a byproduct of cytoplasmic MT-organizing centers because these sites are the most concentrated location of minus ends. Consistent with this, in organisms that show tight telomere bouquets, these clusters are invariably associated with MT-organizing centers. In contrast, in C. elegans prophase, nuclei are embedded in a meshwork of MTs that appear to be primarily nucleated at the plasma membrane and project inward toward the nuclear periphery (Zhou et al., 2009). This cytoskeletal organization may explain the absence of pronounced clustering of PCs that we have observed in both fixed tissue and living animals.
An early hypothesis was that tethering of chromosomes to the nuclear surface might facilitate pairing by reducing homology search from a 3D to a 2D problem, and concentration of the telomeres within a limited region of the NE might further enhance the search (Zickler and Kleckner, 1998). Such a mechanism would depend on the ability of attachment sites to diffuse readily within the NE. Here, we have provided evidence that the onset of meiosis is accompanied by a marked increase in the mobility of chromosome attachment sites, even in the absence of PCMs, and that PCs consequently explore larger regions of the NE during prophase than in premeiotic nuclei (Fig. 7). In cases in which motion in prophase is reduced to levels seen in premeiotic nuclei, such as in animals injected with colchicine or in chk-2 mutants (Figs. 4, 5, and S4), homologues do not pair.
Although simply reducing the homology search volume may indeed be a way that chromosome attachment to the NE facilitates pairing, it now seems likely that its primary purpose is to connect chromosomes to force-generating machinery. Live-cell imaging in single-celled fungi and maize anthers has uncovered meiotic prophase chromosome dynamics that share features with the motions we have described. Specifically, chromosomes in budding yeast and maize undergo rapid motions that seem to reflect independent behavior of individual chromosomes or groups of chromosomes (Conrad et al., 2008; Koszul et al., 2008; Sheehan and Pawlowski, 2009). These rapid chromosome movements coincide with the bouquet stage in maize and persist throughout pachytene in S. cerevisiae, which has only a transient bouquet stage. Our evidence from C. elegans supports the idea that rapid, independent chromosome end–directed motion may be a widely conserved feature of meiotic prophase.
Perhaps surprisingly, there is little consensus in the field about the function of meiotic chromosome motion. Arguments have been made that telomere-mediated motion may facilitate local homologue pairing or global alignment, disrupt nonhomologous interactions, promote crossover recombination, and/or resolve chromosome interlocks (Zickler and Kleckner, 1998; Wanat et al., 2008; Koszul and Kleckner, 2009; Ronceret and Pawlowski, 2010). Unlike telomeres, there is only a single PC region per chromosome, resulting in highly asymmetric motility of the two ends of C. elegans chromosomes during early prophase (Fig. 3). This unusual arrangement may facilitate pairing and synapsis even more effectively than the classical bouquet—e.g., it may be simpler to avoid or resolve interlocks between chromosomes attached to the NE via a single region than with both ends attached.
We have previously argued that in C. elegans, dynein-mediated PC motion is specifically required to license synaptonemal complex formation at or near PCs (Sato et al., 2009). The evidence presented here is consistent with this view and may hint at a mechanism for licensing: we observed paired PCs undergoing dramatic stretches (Fig. 3). The ability to withstand such forces without dissociating may be unique to homologously paired chromosomes. We speculate that the stretching of the chromosome or associated chromosomal or NE components may induce a structural transition to allow synapsis. We have also found that dynein greatly enhances the rate at which chromosomes explore the nuclear surface and that this likely facilitates the process of homologue pairing.
C. elegans has proved to be an advantageous model system in which to investigate the molecular mechanisms of homologue pairing and synapsis. The ability to carry out live imaging, combined with quantitative image analysis, offers an exciting new tool to understand the regulation of chromosome dynamics and their contribution to faithful transmission of genetic information.
Materials and methods
Transgenic reporters
The ZYG-12::GFP construct used in this study (Malone et al., 2003) is robustly expressed throughout the germline, recapitulates endogenous protein function, and is well suited to live imaging because of its brightness. The GFP::HIM-8 construct generated in this study included the htp-3 promoter (488 bp, ending with the first 13 bases of the first exon of htp-3), an N-terminal tag of the S65C variant of GFP containing three synthetic introns (from Fire laboratory vectors via the Seydoux laboratory vector kit; Merritt and Seydoux, 2010), and the him-8 genomic sequence, including 1,208 bp downstream of the final exon. Fragments were combined into a single vector containing the unc-119+ gene from Caenorhabditis briggsae (unc-119+) using the Multisite Gateway system (Invitrogen). This left two attB recombination sequences at the junctions that are translated in the protein product. This construct was integrated into the ttTi5605 site on chromosome II using the MosSCI (Mos1-mediated single copy gene insertion) transposon-mediated insertion system (Frøkjaer-Jensen et al., 2008). Although the GFP::HIM-8 transgene did not complement mutations in him-8, the reporter did not interfere with normal pairing and segregation of X chromosomes (Fig. S2).
Worm strains
Worm strains used in this study were as follows: CA261 (ojIs9[Ppie-1::ZYG-12::GFP] IV), CA493 (dhc-1(or195) I; ojIs9[Ppie-1::zyg-12::gfp] unc-24 IV), CA756 (ieSi1[Phtp-3::gfp::him-8 unc119+] II; itIs37[Ppie-1::mCherry-TEV-Spep::his-58 unc119+] IV), CA777 (ieSi1[Phtp-3:gfp::him-8 unc119+] II; unc-119(ed3) III; itIs37[Ppie-1::mCherry- TEV-Spep::his-58 unc119+] him-8(tm611) IV), CA778 (ieSi1[Phtp-3::gfp::him8 unc-119+] II; unc119(ed3) III; itIs37[Ppie-1::mCherry-TEV-Spep::his-58 unc-119+] IV; chk-2(me64) rol-9(sc148)/unc-51(e369) rol-9(sc148) V), CA786 (ieSi1[Phtp-3::gfp::him-8 unc-119+] II; itIs37[Ppie-1::mCherry-TEV-Spep::his-58 unc-119+]/nT1 IV; syp-1(me17)/nT1(myo-2::gfp and Unc-?) V), CA805 (dhc-1(or195ts) I; ieSi1[Phtp-3::gfp::him-8 unc-119+] II; itIs37[Ppie-1:mCherry- TEV-Spep::his-58 unc-119+] IV), and CA823 (ieSi1[Phtp-3::gfp::him-8 unc-119+] II; itIs37[Ppie-1:mCherry-TEV-Spep::his-58 unc119+] IV; sun-1(jf18)/nT1 (IV;V)).
Time-lapse and 3D fluorescence microscopy
For time-lapse imaging of meiosis, young adult worms (16–20 h after L4) were immobilized on freshly made 3% agarose pads in a drop of M9 containing 0.4 mM (0.05%) tetramisole and 3.8 mM (0.5%) tricaine. A 0.17-mm coverslip was applied without sealing 2 min after immersion in anesthetic, and stacks of images spanning the monolayer of meiotic nuclei closest to the coverslip were collected at 20°C or room temperature 2–15 min after immersion. On the OMX microscope system, excitation light was provided by a 100-mW 488-nm laser (Sapphire; Coherent, Inc.) attenuated to 10% transmission with a neutral density filter. A piezoelectric stage motor (piezosystem jena, Inc.) provided rapid z-axis stage movement, and an EM charge-coupled device (CCD) camera (iXon; Andor) was used to record images with 30-ms exposure times at a rate of 30 Hz. Stacks of 25–33 optical sections with 0.25-µm spacing were acquired every 2 s for 2 min using a 60×, 1.2 NA U Plan Apochromat objective (Olympus). The entire OMX system, including camera acquisition and stage control, is run using custom Python software. A more extensive description of the system can be found in Carlton et al. (2010). Confocal microscopy was performed using a spinning-disk confocal digital microscopy workstation (Marianas; Intelligent Imaging Innovations) equipped with a spinning disk (CSU-X1; Yokogawa), EM CCD camera (Evolve; Photometrics), 63×, 1.4 NA Plan Apochromat objective (Carl Zeiss), and a spherical aberration correction module, and images were acquired used SlideBook software (Intelligent Imaging Innovations). For 3D confocal imaging, stacks of 12 optical sections with 0.5-µm spacing were acquired every 5 s for 5 min, cycling through 75-ms exposures for each channel at each focal plane. For 2D confocal imaging, 50–100-ms exposures in each channel were acquired every 400 ms for ≤80 s at a focal plane near the apical surface of many nuclei. 3D images of fixed samples were acquired using a microscope system (DeltaVision RT; Applied Precision) equipped with a 60×, 1.2 NA U Plan Apochromat or 100×, 1.4 NA U Plan S Apochromat objective (Olympus) and a CCD camera (CoolSNAP HQ; Photometrics). Acquisition and constrained iterative deconvolution of data from the DeltaVision were performed using softWoRx software (Applied Precision).
Image processing, segmentation, and 4D tracking
All wide-field datasets were processed by constrained, iterative 3D deconvolution using a measured point spread function (Agard et al., 1989). Time points were aligned by cross-correlation based on total fluorescence using PRIISM software (Chen et al., 1992) before segmentation. Confocal data were segmented without deconvolution; however, constrained iterative deconvolution was performed using SlideBook software before generating projections for visualization.
Alignment of confocal data was performed using Imaris software (Bitplane). The positions of 15–30 nuclei were used to align the entire dataset. Nuclei were segmented based on the mCherry::histone fluorescence, using the spots function with an expected diameter of 3.5 µm. Alignment was then performed using the correct drift function by selecting the well-segmented nuclei (those for which the spots function determined a position in all time points) and applying a translational shift to each frame to minimize the movement of those nuclei. After alignment, the GFP::HIM-8 signal was segmented and tracked using the spots function with an expected diameter of 0.4–0.5 µm. Automatically generated tracks were then edited manually to eliminate inappropriate connections, including connections between foci in different nuclei or between foci of different size or intensity when more likely assignments were apparent or multiple spots assigned to the same focus and to include foci that were missed by the automatic segmentation.
For ZYG-12::GFP data, a custom Python script was used to measure the cross-correlation between maximum intensity z projections of each optical stack, and a simplex algorithm was applied to minimize the amount of rotation and translation between consecutive images. Because the identity of a patch cannot be followed unambiguously during and after merges with other patches, we generated trajectories according to the following three rules during the manual editing of tracks: First, multiple patches were assigned to a single ZYG-12 focus if they had been distinct foci at an earlier time point or split into multiple foci in a subsequent time point. Second, patches were only tracked if they persisted for 10 time points (≥20 s). Third, when the connectivity between patches was ambiguous from one time point to the next (typically because two patches were assigned to a single focus or two patches of similar size and brightness remained close together in successive time points), we assigned connections that resulted in the least apparent motion.
Data analysis
Analysis of positional data generated with Imaris was performed in MATLAB (version 7.10; MathWorks) using customized code.
Translational and rotational correlation.
Potential translational correlation in displacement among ZYG-12 patches was tested by calculating the mean of all 3D angles between all simultaneous motions in the same nucleus. Correlation was defined as any difference of the cosine of the resulting angle from zero (cos(90°) = 0). For the rotational correlation analysis, tracks were first aligned by rotation around the best-fitting center of a sphere with a radius of 1.75 µm.
PCM detection.
PCMs were defined by measuring the distance a patch moved over 1.2 s (three time points) and selecting those displacements in the top 15th percentile. Noise reduction included (a) removing stand-alone large displacements and (b) merging stretches of fast motion separated by no more than one missing large displacement.
Curve fitting.
Exponential decay curves were fitted onto the PCM duration and distance histograms, assuming that we did not detect most PCMs shorter than four time points or 0.4 µm. Curve fitting was performed using the MATLAB functions expfit and normfilt for exponential and normal curves, respectively.
Random walks and collision simulations.
Random walks on a sphere with a radius of 1.75 µm were generated to match the step-size distribution of the measured datasets. Angles between each successive step were either assigned randomly (for the TZ tracks) or were restricted to restorative (backward turning) to a degree that provided the best fit to observed RMSD plots (for the premeiotic and dynein knockdown tracks). Collisions were simulated by performing two simultaneous random walks, starting one quadrant apart. Collision time was determined as the point when the two tracks were <200 nm apart. Modifying this collision distance between 100 and 400 nm did not have a qualitative effect on the results. In total, >1,000 simulations were run for each condition.
Microinjections and drug treatments
25 nM Cy5–deoxy-UTP (dUTP; GE Healthcare) in injection buffer (200 mM potassium phosphate, pH 7.5, 30 mM potassium citrate, pH 7.5, 20% polyethylene glycol [molecular weight of 6,000–8,000], and 2.5% DMSO) with or without 100 mM colchicine or 10 µM latrunculin A was microinjected into the distal gonads of young adult worms (16–20 h after L4). Imaging was performed 5–6 h after drug injection.
Dynein knockdown
RNAi knockdown of dhc-1 or dlc-1 was performed by feeding RNAi (Timmons and Fire, 1998). Bacterial strains (Fraser et al., 2000) were cultured overnight in 20 ml Luria broth + appropriate antibiotics, spun down, and resuspended in 1 ml Luria broth + antibiotics. 70 µl RNAi bacteria was spread onto 60-mm nematode growth medium plates containing 1 mM IPTG + antibiotics, and double-stranded RNAs were induced overnight at 37°C. Shortly before RNAi bacteria were spread on plates, additional antibiotics (50 µl of 50-mg/ml stock solution per plate) and IPTG (50 µl of 1-M stock solution per plate) were top spread to enhance double-stranded RNA expression. For double knockdown of dlc-1 and dhc-1, dhc1(or195) animals were placed on freshly prepared dlc-1 RNAi plates as L4 larvae, incubated for 24 h at 15°C, and shifted to 25°C for 24 h before imaging. In controls for dynein knockdown, WT L4 larvae were placed on plates with empty vector bacteria and subjected to the identical temperature shifts used in dynein knockdown.
Immunofluorescence and pairing analysis
For immunostaining, dissected gonads were fixed in 2% formaldehyde in egg buffer containing 0.1% Tween 20 for 5 min, freeze cracked into cold methanol, and transferred to PBS + 0.1% Tween 20 at room temperature. Primary antiserum was used without affinity purification, and secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. or Invitrogen. Quantification of pairing was performed by dividing the TZ region, as defined by the presence of ZIM-2 foci (Phillips and Dernburg, 2006), into four zones of equal length. Foci were considered unpaired if they were separated by >0.5 µm.
Online supplemental material
Fig. S1 presents a computational assessment of the correlations in our motion data. Fig. S2 confirms that the GFP::HIM-8 reporter recapitulates the localization of the endogenous HIM-8 protein and does not confer dominant-negative effects. Fig. S3 presents step-size and PCM analysis for X PCs using the GFP::HIM-8 reporter as was performed in Fig. 2 for all PCs using the ZYG-12::GFP reporter. Fig. S4 provides additional comparisons of step-size distributions and RMSD plots; these illustrate the similarity between data generated using the HIM-8 and ZYG-12 reporters, provide all step-size histograms for data shown in Fig. 4, and highlight the differences between datasets in which PCMs are lost. Fig. S5 presents further analysis of the effect of different dynein knockdown strategies on nuclear morphology and position in the gonad. Video 1 shows that 3D time-lapse imaging reveals ZYG-12::GFP patch motions along the NE. Video 2 shows that ZYG-12::GFP patches exhibit heterogeneous, independent trajectories. Video 3 shows PC motion at higher temporal resolution using ZYG-12::GFP. Video 4 shows that imaging of GFP::HIM-8 reveals rapid motion of the X PCs. Video 5 shows that GFP::HIM-8 and X chromosome labeling reveal that chromosome motion is mediated by the PC. Video 6 shows that chromosome elasticity is revealed by imaging of GFP::HIM-8 and X chromosome labeling shows chromosome elasticity. Video 7 shows that during rapid motions, PCs undergo extensive stretching. Video 8 shows that paired X PCs can separate for up to several seconds before reassociating. Video 9 shows X PC motion in representative nuclei from the TZ or premeiotic regions. Video 10 shows the motion of dynein at NE patches using GFP::DHC-1 and the reduction in X PC motion after inhibition of dynein by RNAi. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106022/DC1.
Acknowledgments
We are grateful to John Sedat for the use of the OMX microscope. We thank Keith Cheveralls, Ahmet Yildiz, and members of the Dernburg laboratory for helpful discussions and Yuval Mazor and Julie Cooper for critical reading of the manuscript.
This work was supported by a National Science Foundation graduate fellowship to D.J. Wynne, a European Molecular Biology Organization Long-Term Fellowship (ALTF 564-2010) to O. Rog, and support to A.F. Dernburg from the American Cancer Society (RSG-07-187-01-GMC), the National Institutes of Health (R01 GM065591), and the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- dUTP
- deoxy-UTP
- fps
- frame per second
- MSD
- mean square displacement
- MT
- microtubule
- NE
- nuclear envelope
- PC
- pairing center
- PCM
- processive chromosome motion
- RMSD
- root MSD
- TZ
- transition zone
- WT
- wild type
References
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Bass H.W., Riera-Lizarazu O., Ananiev E.V., Bordoli S.J., Rines H.W., Phillips R.L., Sedat J.W., Agard D.A., Cande W.Z. 2000. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113:1033–1042 [DOI] [PubMed] [Google Scholar]
- Baudrimont A., Penkner A., Woglar A., Machacek T., Wegrostek C., Gloggnitzer J., Fridkin A., Klein F., Gruenbaum Y., Pasierbek P., Jantsch V. 2010. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 6:e1001219 10.1371/journal.pgen.1001219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton P.M., Boulanger J., Kervrann C., Sibarita J.B., Salamero J., Gordon-Messer S., Bressan D., Haber J.E., Haase S., Shao L., et al. 2010. Fast live simultaneous multiwavelength four-dimensional optical microscopy. Proc. Natl. Acad. Sci. USA. 107:16016–16022 10.1073/pnas.1004037107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., W.K. Clyborne, J.W. Sedat, and D.A. Agard. 1992. PRIISM: an integrated system for display and analysis of 3-D microscope images. In Biomedical Image Processing and Three-Dimensional Microscopy. Vol. 784. R.S. Acharya, C.J. Cogswell, and D.B. Goldgof, editors. SPIE, Bellingham, WA. 10.1117/12.59604.
- Conrad M.N., Lee C.Y., Chao G., Shinohara M., Kosaka H., Shinohara A., Conchello J.A., Dresser M.E. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 133:1175–1187 10.1016/j.cell.2008.04.047 [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Chikashige Y., Haraguchi T., Hiraoka Y. 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111:701–712 [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Yamamoto A., Haraguchi T., Hiraoka Y. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 6:329–341 10.1016/S1534-5807(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper N.C., Rillo R., Jover-Gil S., Assaf Z.J., Bhalla N., Dernburg A.F. 2011. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell. 21:934–947 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A.M., Engebrecht J. 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308:206–221 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Kardon J.R., Vale R.D. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., Gelfand V.I. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kleckner N. 2009. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 19:716–724 10.1016/j.tcb.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 133:1188–1201 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Villeneuve A.M. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15:1674–1687 10.1101/gad.902601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiácovo M.P., McDonald K., Villeneuve A.M. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16:2428–2442 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips C.M., Bhalla N., Weiser P., Villeneuve A.M., Dernburg A.F. 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 123:1037–1050 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik R., Petrov D., Lex S.A., King S.J., Gross S.P. 2005. Building complexity: an in vitro study of cytoplasmic dynein with in vivo implications. Curr. Biol. 15:2075–2085 10.1016/j.cub.2005.10.039 [DOI] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- McKenney R.J., Vershinin M., Kunwar A., Vallee R.B., Gross S.P. 2010. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 141:304–314 10.1016/j.cell.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F.J. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering, E., I. Smal, O. Dzyubachyk, and J. OlivoMarin. 2008. Time-lapse imaging. In Microscope Image Processing. Q. Wu, F.A. Merchant, and K.R. Castleman, editors. Elsevier/Academic Press, Amsterdam/Boston. 401–440.
- Merritt C., Seydoux G. 2010. Transgenic solutions for the germline. WormBook. 8:1–21 10.1895/wormbook.1.148.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., Goldman R.D. 2000. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168 10.1083/jcb.151.6.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. 1983. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 97:542–548 10.1083/jcb.97.2.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Dorfman M.D., Carter J.C., Bowerman B. 2007. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3:e128 10.1371/journal.pgen.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., Jantsch V. 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 12:873–885 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., Jantsch V. 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 139:920–933 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Dernburg A.F. 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 11:817–829 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., Dernburg A.F. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 123:1051–1063 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C.M., Meng X., Zhang L., Chretien J.H., Urnov F.D., Dernburg A.F. 2009. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 11:934–942 10.1038/ncb1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier M., Eroglu S., Chatenay D., Marko J.F. 2000. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell. 11:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Yildiz A., Carter A.P., Gennerich A., Zhang N., Vale R.D. 2006. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 126:335–348 10.1016/j.cell.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Pawlowski W.P. 2010. Chromosome dynamics in meiotic prophase I in plants. Cytogenet. Genome Res. 129:173–183 10.1159/000313656 [DOI] [PubMed] [Google Scholar]
- Ross J.L., Wallace K., Shuman H., Goldman Y.E., Holzbaur E.L. 2006. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 8:562–570 10.1038/ncb1421 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Amitani I., Yokota E., Ando T. 2000. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 272:586–590 10.1006/bbrc.2000.2819 [DOI] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J. 1993. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys. J. 64:1766–1780 10.1016/S0006-3495(93)81548-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621–627 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Scherthan H., Weich S., Schwegler H., Heyting C., Härle M., Cremer T. 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134:1109–1125 10.1083/jcb.134.5.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M.J., Pawlowski W.P. 2009. Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc. Natl. Acad. Sci. USA. 106:20989–20994 10.1073/pnas.0906498106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Vale R.D., Funatsu T., Pierce D.W., Romberg L., Harada Y., Yanagida T. 1996. Direct observation of single kinesin molecules moving along microtubules. Nature. 380:451–453 10.1038/380451a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Belmont A.S., Sedat J.W. 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12:1473–1483 10.1016/S0960-9822(02)01090-4 [DOI] [PubMed] [Google Scholar]
- Veigel C., Coluccio L.M., Jontes J.D., Sparrow J.C., Milligan R.A., Molloy J.E. 1999. The motor protein myosin-I produces its working stroke in two steps. Nature. 398:530–533 10.1038/19104 [DOI] [PubMed] [Google Scholar]
- Vogel S.K., Pavin N., Maghelli N., Jülicher F., Tolić-Nørrelykke I.M. 2009. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 7:e1000087 10.1371/journal.pbio.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta J.H., Didier A.J., Liu X., Krämer H. 2001. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152:923–934 10.1083/jcb.152.5.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanat J.J., Kim K.P., Koszul R., Zanders S., Weiner B., Kleckner N., Alani E. 2008. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4:e1000188 10.1371/journal.pgen.1000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A., Gross S.P., Postner M., Block S.M., Wieschaus E.F. 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 92:547–557 10.1016/S0092-8674(00)80947-2 [DOI] [PubMed] [Google Scholar]
- Zhou K., Rolls M.M., Hall D.H., Malone C.J., Hanna-Rose W. 2009. A ZYG-12–dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186:229–241 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619–697 10.1146/annurev.genet.32.1.619 [DOI] [PubMed] [Google Scholar]