Abstract
We have evaluated the possibility that the action of voluntary exercise on the regulation of brain-derived neurotrophic factor (BDNF), a molecule important for rat hippocampal learning, could involve mechanisms of epigenetic regulation. We focused the studies on the Bdnf promoter IV, as this region is highly responsive to neuronal activity. We have found that exercise stimulates DNA demethylation in Bdnf promoter IV, and elevates levels of activated methyl-CpG-binding protein 2, as well as BDNF mRNA and protein in the rat hippocampus. Chromatin immunoprecipitation assay showed that exercise increases acetylation of histone H3, and protein assessment showed that exercise elevates the ratio of acetylated : total for histone H3 but had no effects on histone H4 levels. Exercise also reduces levels of the histone deacetylase 5 mRNA and protein implicated in the regulation of the Bdnf gene [N.M. Tsankova et al. (2006) Nat. Neurosci., 9, 519–525], but did not affect histone deacetylase 9. Exercise elevated the phosphorylated forms of calcium/calmodulin-dependent protein kinase II and cAMP response element binding protein, implicated in the pathways by which neural activity influences the epigenetic regulation of gene transcription, i.e. Bdnf. These results showing the influence of exercise on the remodeling of chromatin containing the Bdnf gene emphasize the importance of exercise on the control of gene transcription in the context of brain function and plasticity. Reported information about the impact of a behavior, inherently involved in the daily human routine, on the epigenome opens exciting new directions and therapeutic opportunities in the war against neurological and psychiatric disorders.
Keywords: hippocampus, histone modification, rat, synaptic plasticity
Introduction
We now know that animals and humans contain a vast potential for genetic variability, which confers upon them the ability to adapt their phenotype according to environmental demands. Epigenetic mechanisms allow for lasting modifications in the genome, and subsequent behavioral manifestations such as learning and memory, and emotions, and may be accountable for dysfunctions observed in several brain disorders (Nestler, 2009; Sweatt, 2009). The powerful impact of exercise on the biological adaptation and lifespan of individuals suggests that exercise has the extraordinary potential to promote stable changes in gene function. The effects of exercise on preserving or enhancing brain function apply under homeostatic conditions (Vaynman et al., 2004), brain challenges (Griesbach et al., 2004) and during aging (Hillman et al., 2008). The impact of exercise on brain plasticity has been associated with the action of brain-derived neurotrophic factor (BDNF) (Vaynman et al., 2004), a molecule deeply involved with neuronal excitability, learning and memory (for review see Martinowich et al., 2007).
Epigenetic mechanisms involving post-replication modifications of DNA and nuclear proteins have been shown to modulate the Bdnf gene. There are at least four Bdnf promoters in the rat, which are differentially activated in response to various types of signaling events. Transcription involving promoter IV (formerly promoter III) is responsive to neuronal activity and can mediate synapse plasticity and learning and memory, and is subjected to epigenetic regulation (Feng et al., 2007). Promoter IV transcription is suppressed by methyl-CpG-binding protein (MeCP2), which belongs to a family of methylcytosine-binding proteins that contribute to the gene-silencing effect of DNA methylation (Chao & Zoghbi, 2009). MeCP2 occupies a site on the Bdnf promoter in the absence of stimulation, which represses the transcription of Bdnf (Martinowich et al., 2003). Neuronal depolarization dissociates MeCP2 from the Bdnf promoter resulting in demethylation within the promoter and Bdnf transcription (Chen et al., 2003). Neuronal activity is an important epigenetic regulator of the Bdnf gene, such that depression-like behavior in mice results in methylation of histone H3 and long-lasting suppression of Bdnf transcription through promoters IV and VI (Tsankova et al., 2006). In turn, exercise and BDNF have been associated with reducing depression and promoting cognitive enhancement. This implies the likely possibility that exercise can use mechanisms of epigenetic regulation to reduce depression and enhance cognitive abilities.
The influence of exercise on BDNF has been linked to hippocampal synaptic plasticity and learning and memory involving the signaling system calcium/calmodulin-dependent protein kinase II (CaMKII), and the transcription regulator cAMP response element binding protein (CREB) (Vaynman et al., 2004). In addition, the activations of CaMKII and CREB are involved in the epigenetic regulation of the Bdnf gene. We have embarked on studies to evaluate the possibility that the influence of exercise on brain plasticity may involve epigenetic modifications of the Bdnf gene in the hippocampus. These interactions could determine the capacity of exercise to promote long-lasting modifications in the brain to help cope with challenges.
Materials and methods
Exercise paradigm
Adult male Sprague-Dawley rats of approximately 3 months of age (Charles River Laboratories, Inc., Wilmington, MA, USA) were maintained under a 12/12 h light/dark cycle at 22–24 °C with food and water ad libitum. Animals were randomly divided into two groups (sedentary and exercised) and all rats were individually housed. Exercise group animals were given access to a running wheel (diameter 31.8 cm, width 10 cm) that freely rotated against a resistance of 100 g, and the running revolutions were recorded and analyzed using VitalViewer Data Acquisition System software (MiniMitter, Bend, OR, USA). The animals ran an average of 1.55 ± 0.15 km/day (± SEM). Sedentary animals were single housed in their home cages without access to a running wheel. After 7 days of exercise, all animals were killed by decapitation and their hippocampi were rapidly dissected out and stored at −70 °C. These studies were performed in accordance with the guidelines of the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by UCLA Animal Research Committees.
RNA isolation and real-time quantitative polymerase chain reaction
An RNA STAT 60 kit (Tel-Test Inc., Friendswood, TX, USA) was used for total RNA isolation and the procedure was followed by the manufacturer’s protocol. Real-time quantitative polymerase chain reaction (RT-PCR) was performed to measure the mRNA levels of BDNF, histone deacetylase (HDAC)5 and HDAC9.
Glyceraldehyde-3-phosphate dehydrogenase served as an internal control for sample normalization. Total RNA (100 ng) was used for cDNA synthesis (iScript cDNA Synthesis kit, Bio-Rad Laboratories Inc., Hercules, CA, USA). The synthesized cDNA was the template for the real-time polymerase chain reaction (PCR) amplification carried out by the CFX96 real-time PCR detection system (Bio-Rad Laboratories Inc.) using a SsoFast EvaGreen Supermix kit (Bio-Rad Laboratories Inc.) and forward/reverse primers. The sequences of primers were as follows: HDAC5 primer pair (Tsankova et al., 2006), forward: 5′-TGTCACCGCCAGATGTTTTG-3′, reverse: 5′-TGAGCAGAGCCGAGACACAG-3′; HDAC9 primer pair (Tsankova et al., 2006), forward: 5′-GCGAGACACAGATGCTCAGAC-3′, reverse: 5′-TGGGTTTTCCTTCCATTGCT-3′; Bdnf exon IV primer pair (Lubin et al., 2008), forward: 5′-TGCGAGTATTACCTCCGC CAT-3′, reverse: 5′-TCACGTGCTCAAAAGTGTCAG-3′; and glyceraldehyde-3-phosphate dehydrogenase primer pair, forward: 5′-TGC CACTCAGAAGACTGTGG-3′, reverse: 5′-TTCAGCTCTGGGATG ACCTT-3′. Glyceraldehyde-3-phosphate dehydrogenase primers were generated with primer3 software. The reverse transcription reaction steps for each cycle consisted of an initial 30 s incubation step at 95 °C for enzyme activation and were followed by 5 s at 95 °C for denaturation and then 5 s at 60 °C for annealing/extension. Three negative (no template) controls were performed to verify the sample genomic DNA contamination. Quantification of the RT-PCR results was performed using the CFX manager software version 1.6 (Bio-Rad Laboratories Inc.).
Western blot
Hippocampal tissue from one hemisphere was dissected and sonicated briefly in lysis buffer (137 mm NaCl, 20 mm Tris–HCl, pH 8.0, 1% NP-40 (Tergitol-type NP-40), 10% glycerol, 1 mm PMSF (phenylmethylsulfonyl fluoride), 10 μg/mL aprotinin, 0.1 mm benzothonium, 0.5 mm sodium vanadate). This procedure is known to yield whole cell extracts (including the nucleus), and has been used successfully to measure acetylated histone H3 (AceH3), histone H3 and other nuclear proteins (Chakrabarti et al., 2003). After centrifuging at 12 500 g for 20 min, supernatants were collected and immediately processed for total protein concentration determination according to the Micro BCA procedure (Pierce, Rockford, IL, USA), using bovine serum albumin as standard. All chemicals were obtained from Sigma (St Louis, MO, USA) unless otherwise noted.
Protein levels of BDNF, phospho-MeCP2, AceH3, histone H3, acetylated histone H4 (AceH4), histone H4, HDAC5 phospho-CaMKII, CaMKII and phospho-CREB were analyzed by western blot analysis. Actin was utilized as an internal control, and each blot was standardized to its corresponding actin value. Protein samples were separated by electrophoresis on a 10% polyacrylamide gel and electrotransferred to a PVDF (polyvinylidene fluoride) membrane. Non-specific binding sites were blocked in Tris-buffered saline with 2% bovine serum albumin and 0.1% Tween-20 for 1 h at room temperature (20 °C). Membranes were rinsed in buffer (0.1% Tween-20 in Tris-buffered saline) and incubated at 4 °C overnight, with the following primary antibodies: anti-BDNF (1 : 1000, catalog # sc-546; Santa Cruz Biotechnology Inc., CA, USA), anti-MeCP2 (1 : 1000, catalog # 07–013; Millipore, Temecula, CA, USA), anti-Ace H3 (1 : 1000, catalog # 07–353; Millipore), anti-Ace H4 (1 : 500, catalog # 06–598; Millipore), anti-histone H4 (1 : 500, catalog # 07–108; Millipore), anti-phospho-CaMKII (1 : 1000, catalog # sc-12886-R; Santa Cruz Biotechnology Inc.), anti-CaMKII (1 : 1000, catalog # sc-9035; Santa Cruz Biotechnology Inc.), anti-phospho-CREB (1 : 1000, catalog # 06–519; Millipore) and anti-CREB (1 : 1000, catalog # 06–863; Millipore) were followed by anti-rabbit IgG horseradish peroxidase conjugate (1 : 100 000; Santa Cruz Biotechnology Inc.); anti-HDAC5 (1 : 1000; Santa Cruz Biotechnology Inc.) and total anti-histone H3 (1 : 1000, catalog # 05–499; Millipore) were followed by anti-mouse IgG horseradish peroxidase conjugate (1 : 100 000; Santa Cruz Biotechnology Inc.); and anti-actin (1 : 1000, catalog # sc-1616; Santa Cruz Biotechnology Inc.) was followed by anti-goat IgG horseradish peroxidase conjugate (1 : 100 000; Santa Cruz Biotechnology Inc.). After rinsing in buffer four times for 10 min, immunocomplexes were analyzed by chemiluminescence using the ECL Plus kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA), according to the manufacturer’s instructions. The final data were expressed as the percent change from the mean sedentary values. Ratios of phospho-CREB to total CREB, phospho-CaMKII to total CaMKII, AceH3 to total histone H3 and AceH4 to total histone H4 for each rat were calculated to aid in the interpretation of the data, and group means were calculated for this measure of activation; upshift MeCP2 bands were used for phospho-MeCP2 quantification (Zhou et al., 2006; Tao et al., 2009).
DNA methylation analysis
Bisulfite genomic conversion and sequencing analysis of individual clones was performed according to published protocols (Feng et al., 2010). Briefly, DNA was extracted from hippocampal tissue, digested with Bgl II enzyme and treated with sodium bisulfite. The converted DNA was amplified with nest-PCR, which was performed with two pairs of primers designed using Methprimer software (Feng et al., 2010): outside forward: TTATTTTAAATTTTGGTTAAAGATTTAA AA, reverse: CCTTCAATAAAAAACTCCATTTAATCTAA; and inside forward: TTTATAAAGTATGTAATGTTTTGGAA, reverse: AAA ACTCCATTTAATCTAAACAAAAACTAA. The PCR products were cloned into pCR4.1 TOPO vector (Invitrogen, Carlsbad, CA, USA). Individual clones were then sequenced to detect methylation sequencing patterns, and the methylation percentage was calculated by dividing methylated clones by total clones in particular CpG sites.
Chromatin immunoprecipitation analysis
Analysis was performed using an acetyl-histone H3 chromatin immunoprecipitation (ChIP) kit (Upstate, Temecula, CA, USA). In brief, whole rat hippocampus was used to extract chromatin, and samples obtained before immunoprecipitation (Input) and after acetyl-histone H3 immunoprecipitation (AceH3) were subjected to PCR amplification. The PCR was performed using seven primers corresponding to different regions of Bdnf as previously described (Chen et al., 2003): exon I, forward: GCAGTTGGACAGTCATTGGTA ACC, reverse: ACGCAAACGCCCTCATTCTG; exon II, forward: GCAGAGTCCATTCAGCACCTTG, reverse: TGGCTTGACAGC-GAGGAAAAG; exon IV (−637 to −535 bp), forward: AACAA-GAGGCTGTGACACTATGCTC, reverse: CAGTAAGTAAAGGCT AGGGCAGGC; exon IV (−73 to +14 bp), forward: TCTATTTCGAG GCAGAGGAGGTATC, reverse: AATGGGAAAGTGGGTGGGAG; exon IV (+71 to +155 bp), forward: GCATGAAATCTCCCAGTCTC TGC, reverse: TGGAAATTGCATGGCGGAG; exon IV (+723 to +1007 bp), forward: TTGGGATGGGAAAGATGGG, reverse: CAGA GTAGGAGGGAACAAGTGTGAC; and exon VI, forward: TTTGG GGCAGACGAGAAAGC, reverse: GGCAGTGGAGTCACATTGTT GTC. Levels of histone H3 acetylation at the Bdnf promoter IV region (−73 to +14 bp) were determined by quantitative real-time PCR analysis using the iQ SYBR Green Supermix (Bio-Rad Laboratories Inc.) system. Briefly, the first threshold cycle (Ct) was normalized by subtracting the negative control value for each corresponding sample. A ΔCt representing the difference between input and immunoprecipitated (ip) samples was calculated using the formula: ΔCt = Ctip − Ctinput, and the fold difference was then determined by raising 2 to the ΔCt power.
Statistical analysis
We used eight animals per group for mRNA and protein measurements, and four animals per group were used for DNA methylation assays and ChIP assays. One-way anova (spss 16.0 software) was performed to determine differences between groups. The results were converted to percentages of sedentary controls for presentation in figures, and values represent the mean ± SEM. Statistical differences were considered significant when P < 0.05.
Results
We have previously reported that voluntary exercise elevates hippocampal BDNF levels with important effects on learning and memory performance (Vaynman et al., 2004). Here we examine the possibility that exercise can influence BDNF using mechanisms of epigenetic regulation. Given that Bdnf promoter IV (formerly promoter III) plays a key role in activity-dependent Bdnf regulation in rats (Chen et al., 2003), we focused our studies on the Bdnf promoter IV region.
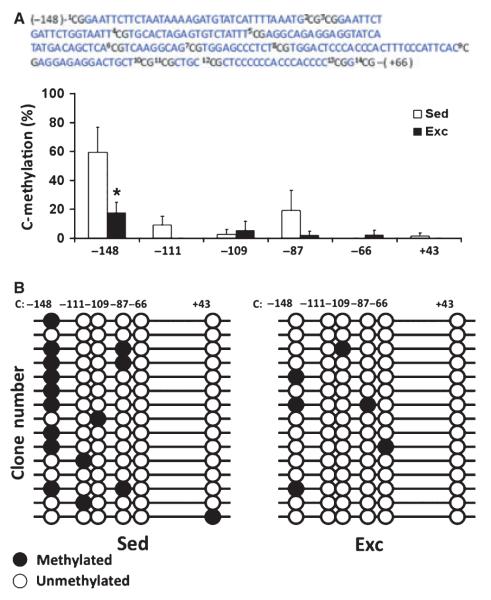
Exercise reduces methylation of CpG in Bdnf promoter IV (Fig. 1)
Fig. 1.
Exercise reduced DNA methylation of Bdnf exon IV promoter in rat hippocampus. (A) The bar graph shows the DNA methylation levels of exercised (Exc) and sedentary (Sed) control animals on six CpG sites. Bisulfite sequencing analysis showed that the DNA methylation level was less in animals exposed to exercise, the −148 CpG site showing the most dramatic DNA demethylation. (B) The number on top of the diagram labels the position of CpG sites relative to the transcription starting site (+1), and each horizontal line represents the result for one clone (open circles, unmethylated CpGs; filled circles, methylated CpGs). The DNA methylation level was calculated by the number of methylated CpGs divided by the total number of CpGs analyzed. Values represent the mean + SEM; *P < 0.05; n =4/group.
Animals were exposed to 1 week of voluntary exercise, a period that elevates BDNF mRNA levels and learning and memory performance. To determine whether exercise has an effect on the CpG methylation pattern within the Bdnf gene, we assessed the methylation status within a 230 bp region extending from −148 to +82 bp of the rat Bdnf exon IV promoter. This region, which included 14 CpG sites located immediately upstream of the transcription initiation site (base pair +1), has been shown to be crucial for Bdnf expression regulation. We performed bisulfite-sequencing analysis in six CpG sites of a total of 14 sites, and found that one CpG site (−148 bp) was significantly less methylated (unmethylated clone/total clone) in animals exposed to exercise. In particular, we found that the CpG site at −148 bp, which is associated with MeCP2 binding, showed the most frequent methylation in the sedentary rats. Interestingly, the effects of exercise were sufficient to reduce methylation of this CpG site from 59.2% (sedentary rats) to 18.4% (F1,7 = 6.409, P = 0.045, Fig. 1A and B).
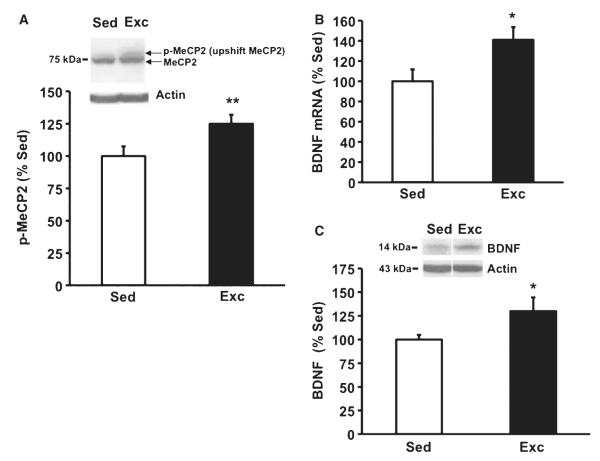
Exercise affects the methyl-CpG-binding protein level in conjunction with Bdnf (Fig. 2)
Fig. 2.
Exercise elevates (A) phospho-MeCP2 levels, (B) BDNF mRNA levels and (C) BDNF protein levels. MeCP2 is known to work with methylated chromatin to suppress promoter IV transcription and to modulate Bdnf expression and function, and according to our results these events could be influenced by exercise. Proteins were measured using western blot analysis and BDNF mRNA was measured using relative quantitative RT-polymerase chain reaction; upshift MeCP2 bands were quantified for levels of phospho-MeCP2. Values represent the mean + SEM; *P < 0.05, **P < 0.01. Sed, sedentary; Exc, exercise n =8/group.
It has been shown that MeCP2 can modulate Bdnf gene expression and function by interacting with methylated chromatin and suppressing promoter IV transcription (Martinowich et al., 2003). In the absence of stimulation, MeCP2 occupies a site on the Bdnf promoter repressing the transcription activation of Bdnf, but neuronal depolarization phosphorylates MeCP2 such that phospho-MeCP2 is dissociated from the Bdnf promoter enabling Bdnf transcription (Chen et al., 2003; Martinowich et al., 2003). Western blot analysis using the same animals as for the methylation assay showed that exercise promoted a 25% increase (F1,7 = 38.861, P = 0.001) in phospho-MeCP2 relative to sedentary rats (Fig. 2A). To verify the effects of exercise on Bdnf, we measured the levels of BDNF mRNA in exon IV and BDNF protein in the same animals used to assess the levels of phospho-MeCP2. We found that Bdnf mRNA was increased by 41% (F1,15 = 6.574, P = 0.025, Fig. 2B) and Bdnf protein was increased by 30% (F1,15 = 6.874, P = 0.024, Fig. 2C) in the exercised rats compared with sedentary rats.
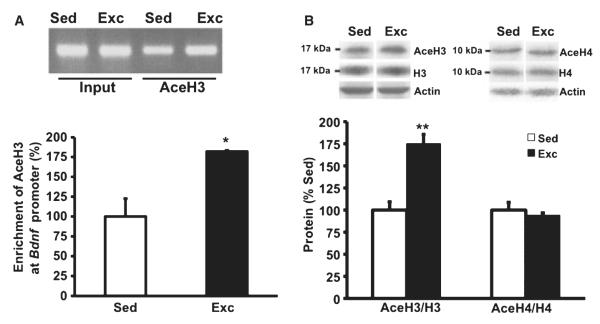
Exercise induces acetylation of histone H3 (Figs 3 and 4)
Fig. 3.
(A) Voluntary exercise increased histone H3 acetylation in hippocampi of rats assessed using the ChIP assay. Primers specific to Bdnf promoter IV were used to amplify the DNA from the AceH3 immunoprecipitates, and the relative enrichments of the Bdnf promoter IV in the AceH3 immunoprecipitates were measured using real-time PCR. Equal amounts of DNA from sedentary (Sed) or exercised (Exc) rat hippocampi were used for immunoprecipitation (Input). DNA gel shows products of real-time PCR. Data are presented as means + SEM. *P < 0.05; n =4/group. (B) We assessed levels of AceH3 and histone H3 by western blot analysis in the same hippocampal tissue used for the ChIP assay, and found a significant (**P < 0.01) increase in the ratio AceH3 : histone H3 in the exercised group compared with sedentary rats. For comparative reasons we assessed levels of AceH4 and histone H4, and did not find significant changes in the ratio AceH4 : histone H4 in the exercised rats relative to sedentary rats.
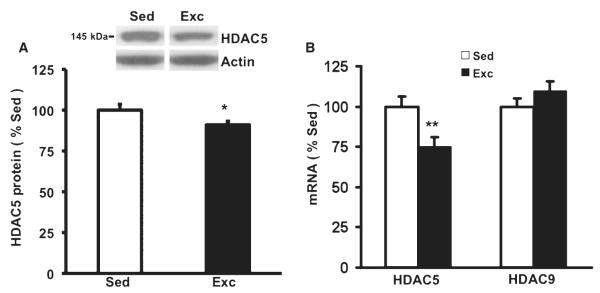
Fig. 4.
Based on indications that HDAC5 is important for the regulation of the Bdnf gene (Tsankova et al., 2006), we measured levels of HDAC5 in our paradigm. We found that exercise reduced levels of (A) HDAC5 protein (*P < 0.05) and (B) HDAC5 mRNA (**P < 0.01, n =8/group) but not HDAC9 mRNA (n =8/group). Values represent the mean + SEM. Sed, sedentary; Exc, exercise.
We used a ChIP assay to evaluate modification at histone H3 following exercise, and found that exercise increased acetylation of histone H3 in the Bdnf promoter IV region. To examine in which region AceH3 modification occurred, we assessed relative ChIP enrichments by using seven primer pairs corresponding to different Bdnf gene regions. We found that AceH3 is mostly associated with the Bdnf exon IV promoter region of base pairs −73 to +14, but not exon I, II or VI. We then performed real-time PCR to assess AceH3-bound DNA, and found 75% higher levels (F1,7 = 23.718, P = 0.003) of AceH3 binding DNA in exercised rats compared with sedentary rats (Fig. 3A).
In the hippocampal tissues for the ChIP assay, we assessed the levels of AceH3 and total histone H3 by western blot analysis. We found that the level of AceH3 was significantly increased (60%, P = 0.006) in the exercised group compared with sedentary rats, but no significant changes were observed in total histone H3. The ratio of AceH3 to total histone H3 was 75% higher in the exercised rats compared with sedentary rats (F1,7 = 90.309, P = 0.001, Fig. 3A). We measured the protein levels of AceH4 and total histone H4 in the same sample used above. No significant differences of AceH4 or its ratio (AceH4 : total histone H4) were found in the exercised group compared with the sedentary group (F1,7 = 0.171, P = 0.686, Fig. 3B). The fact that modification of acetylation was observed particularly on histone H3 indicates that histone H3, but not histone H4, is related to Bdnf epigenetic regulation after exercise. Acetylation vs. deacetylation of histones is a dynamic process controlled by specific enzymes or deacetylases (HDAC). Based on indications that HDAC5 seems to be associated with the regulation of the Bdnf gene in animal models of depression (Tsankova et al., 2006), we measured the levels of HDAC5 in our paradigm (Fig. 4). We found that exercise reduced the levels of HDAC5 mRNA by 25% (F1,15 = 9.68, P = 0.008, Fig. 4B) and its protein by 91% (F1,15 = 5.718, P = 0.038, Fig. 4A) in the hippocampal tissue used for the histone assessments. For comparative reasons, we also measured the mRNA levels of HDAC9, but no significant changes were found in the exercised and sedentary groups (F1,15 = 1.667, P = 0.218, Fig. 4B).
Exercise elevated phospho-cAMP response element binding protein and phospho-calcium/calmodulin-dependent protein kinase II levels (Fig. 5)
Fig. 5.
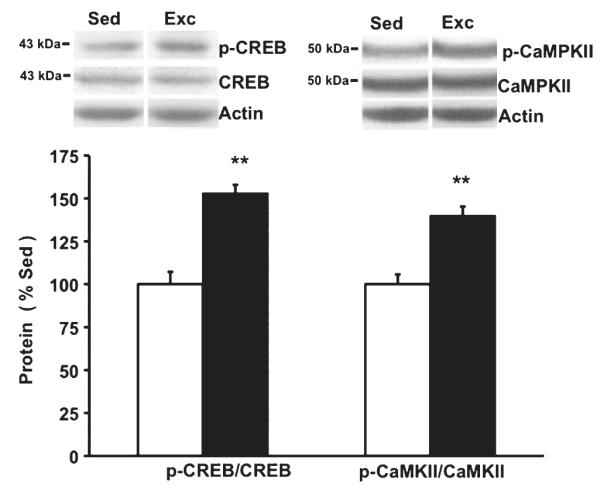
Exercise elevates levels of phospho-CREB and phospho-CaMKII. We measured levels of total and phosphorylated CaMKII and CREB based on their involvement in intracellular signaling effects on chromatin remodeling. Results of western blot analysis showed that the ratios of phospho-CREB : total CREB and phospho-CaMKII : total CaMKII were increased in exercised (Exc) rats compared with sedentary (Sed) rats. Values represent the mean + SEM. **P < 0.01; n = 8/group.
We assessed indexes of activation for CaMKII and CREB in our paradigm based on the involvement of these systems in the mechanisms by which intracellular signaling affects chromatin remodeling. CaMKII activation can lead to phosphorylation of CREB and, in turn, phospho-CREB can recruit CBS (CREB-binding protein) endowed with strong histone acetylation transferase-promoting activity. It has previously been shown that neural activity elicits the phosphorylation of MeCP2 involving CaMKII phosphorylation, thereby activating Bdnf transcription. Results of western blot analysis showed that phospho-CREB/CREB was increased by 53% (F1,15 = 9.282, P = 0.009, Fig. 5) in exercised rats compared with sedentary rats. Results also showed that exercise elevated the ratio of phospho-CaMKII : CaMKII by 40% in the exercised rats compared with sedentary rats (F1,15 = 30.049, P = 0.001, Fig. 5).
Discussion
Evidence accumulated in the last few years indicates that exercise exerts a strong influence on brain plasticity and cognition, through mechanisms centered on the action of BDNF. Our results show that an exercise regimen known for its capacity to elevate hippocampal levels of BDNF mRNA and protein, functional for exercise-induced learning and memory enhancement, promotes remodeling of chromatin containing the Bdnf gene. We have focused these studies on the promoter IV (formerly promoter III) region of the Bdnf gene as this region is highly responsive to neuronal activity and is the target of epigenetic regulation. We report that exercise affects histone acetylation and DNA methylation localized to the promoter IV region of the Bdnf gene. Exercise also affects the levels of phospho-MeCP2, a molecule important for Bdnf gene transcription regulation. The effects of exercise were also sufficient to elevate the levels of phospho-CaMKII and phospho-CREB (molecules intimately involved in the pathways by which neural activity engages mechanisms of epigenetic regulation to stimulate Bdnf transcription). The results are consistent with the notion that exercise influences epigenetic mechanisms to promote stable elevations in Bdnf gene expression, which may have important implications for the regulation of synaptic plasticity and behavior (Fig. 6).
Fig. 6.
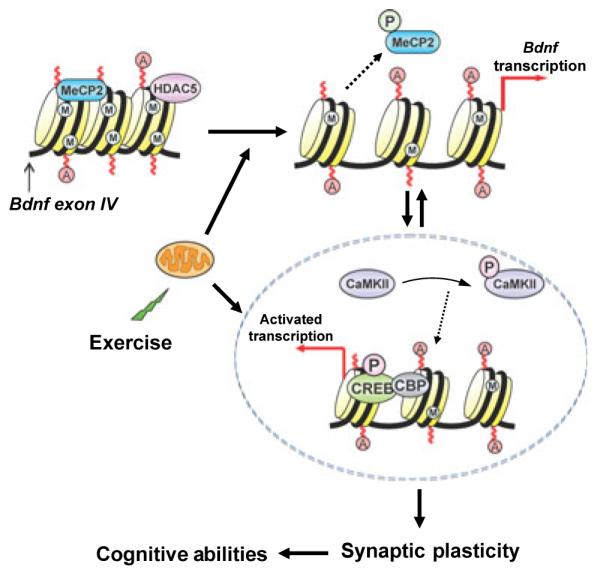
Proposed mechanism by which exercise impacts synaptic plasticity and cognitive abilities by engaging aspects of epigenetic regulation. Exercise promotes DNA demethylation in Bdnf promoter IV, involving phosphorylation of MeCP2, and acetylation of histone H3. These events may result in dissociation of MeCP2 and chromatin remodeling events leading to Bdnf gene transcription. The effects of exercise on BDNF regulation may also involve the action of HDAC5 implicated in the regulation of the Bdnf gene (Tsankova et al., 2006). Exercise elevates the activated stages of CaMKII (phospho-CaMKII) and CREB (phospho-CREB), which in turn can contribute to regulate Bdnf transcription, as well as participate in the signaling events by which BDNF influences synaptic plasticity and cognitive abilities. The impact of exercise on the remodeling of chromatin containing the Bdnf gene emphasizes the importance of exercise on the control of gene transcription in the context of brain function and plasticity. A: histone acetylation; CBP: CREB-binding protein; M: CpG site methylation; P: phosphorylation.
Exercise influences DNA methylation
Our results show that exercise increases the hypomethylated stage (decrease in CpG methylation) of the Bdnf gene promoter region. DNA methylation plays a crucial role in the remodeling of chromatin in the brain, and contributes greatly to the regulation of Bdnf as it can repress the transcription and function of Bdnf (Martinowich et al., 2003). Therefore, the fact that our results show that exercise induces hypomethylation of the Bdnf promoter IV suggests the possibility that DNA methylation may be a crucial step by which exercise regulates BDNF expression. These results are in agreement with the current and previous findings showing that exercise elevates levels of hippocampal BDNF mRNA (Vaynman et al., 2004) and protein (Ding et al., 2006).
Exercise influences intracellular signaling involved with activity-dependent regulation of Bdnf
Our results showed that exercise elevated levels of phospho-MeCP2 in the hippocampus. Although the fine points of the interaction between MeCP2 and Bdnf need to be further elucidated, there is consensus that MeCP2 is crucial for Bdnf regulation. It has been shown that MeCP2 recruited to methylated DNA contributes to repress Bdnf promoter IV (formerly promoter III) transcription (Martinowich et al., 2007; Chao & Zoghbi, 2009). Neural activity is an important activator of MeCP2, such that phosphorylation of MeCP2 by neural activity leads to the release of MeCP2 and activation of Bdnf transcription. The influence of MeCP2 on Bdnf regulation is more obvious during conditions of MeCP2 deficiency in the brain, which leads to a reduction of BDNF (Abuhatzira et al., 2007), whereas MeCP2 overexpression increases levels of BDNF (Larimore et al., 2009). Although more information is required for a full picture of the action of MeCP2 on Bdnf regulation, there are grounds to hypothesize that exercise may serve to activate MeCP2, thereby increasing BDNF mRNA and protein.
Our results showed that exercise elevated the activated stage of CaMKII and CREB in the hippocampus. The signaling modulator CaMKII and the transcription activator CREB have been implicated in the mechanisms by which intracellular signaling can affect chromatin remodeling. For example, the activation of CaMKII leads to phosphorylation of CREB and MeCP2, such that phospho-CREB can recruit CREB-binding protein with strong histone acetylation transferase-promoting activity. The CaMKII-dependent phosphorylation of MeCP2 seems to be a crucial step in the activity-dependent regulation of BDNF, as it has been shown that membrane depolarization increases Ca-dependent transcription of Bdnf exon IV. Previous studies have shown that the functions of CaMKII and CREB are necessary steps for the action of exercise on BDNF-mediated synaptic plasticity and cognition (Vaynman et al., 2003). For example, blocking the function of CaMKII in the hippocampus during exercise by using a specific inhibitor reduces BDNF mRNA and abrogates the cognitive enhancement elicited by exercise (Vaynman et al., 2007). The overall evidence seems to indicate that the involvement of phospho-CaMKII and phospho-CREB in the effects of exercise on BDNF and cognition may be associated with epigenetic mechanisms.
Exercise influences histone acetylation
It is known that the acetylation of histones associated with the DNA can promote a permissive or active chromatin that can lead to selective transcription of specific genes such as Bdnf (Tsankova et al., 2006). Interestingly, inhibiting histone deacetylation leads to an increase in BDNF mRNA levels (Tian et al., 2010). Accordingly, we directed our studies to determine the effects of exercise on histone acetylation, and we assessed histone H3 and histone H4 based on their potential involvement in experience-dependent plasticity (Sweatt, 2009). Using a ChIP assay, we found that acetylated histone 3 was mostly enriched within the Bdnf promoter IV sequence (−73 to +14 bp, Fig. 2), and that this enrichment was significantly enhanced by exercise. In addition, we used western blot analysis to determine whether these chromatin modifications could be related to changes in protein levels. The results showed that exercise increased the relative proportion of hippocampal AceH3 but did not affect histone H4 levels. These results argue in favor of the relative specificity of the action of exercise on histone H3, which could lead to a facilitation of Bdnf transcription.
The question arises how the effects of exercise can translate into synaptic plasticity engaging processes of epigenetic regulation. The physiology of exercise is directly related to adjustments in energy biogenesis, such that exercise is perceived as a viable strategy to combat obesity and mental disorders. Emerging research indicates that the regulation of energy metabolism through Bdnf is an important mechanism by which exercise can influence synaptic plasticity and cognitive function (Vaynman et al., 2006; Gomez-Pinilla et al., 2008). The results of proteomic studies have shown that most of the proteins that are upregulated by exercise are associated with energy metabolism and synaptic plasticity (Ding et al., 2006). The functions of several of these proteins on cognition and plasticity are achieved by interacting with BDNF, in which BDNF acts as a mediator between metabolism and brain plasticity (Gomez-Pinilla et al., 2008). In turn, growing evidence indicates that the main proteins that modulate bioenergetic transactions in the cell can drive modifications in the epigenome (Wallace & Fan, 2010), portraying energy metabolism as a driving force for the occurrence of epigenetic events. For example, calorie levels can determine the production of ATP, acetyl-CoA, s-adenosyl-l-methionine and NADH (nicotinamide adenine dinucleotide), which are directly implicated in the modifications of histone and chromatin. This evidence is probably the tip of the iceberg for a body of molecular events by which calorie changes driven by environmental stimulations such as exercise and diet can promote profound modifications in the genome (for review see Feinberg, 2007; Wallace & Fan, 2010). In addition, our results showing the influence of exercise on the regulation of the Bdnf gene using epigenetic mechanisms are in harmony with the described influences of other environmental factors. For example, it has recently been reported that changes in DNA methylation during thermotolerance control affect CREB binding to BDNF (Yossifoff et al., 2008), and that environmental enrichment induces epigenetic modifications of the Bdnf gene (Kuzumaki et al., 2010).
Functional implications
Although all mammalian tissues contain an immense number of genes, only a small fraction of these genes becomes functional at a given time, in a process in which epigenetic regulation plays a crucial role. The powerful influence of exercise on biological adaptation probably engages epigenetic mechanisms to control gene function. Abundant evidence exists to show that exercise contributes to promoting mental health and alleviating the effects of depression, anxiety, schizophrenia, cognitive impairment and drug addiction. Therefore, the current results may shed light on the mechanisms by which exercise contributes to alleviating psychiatric disorders. For example, the antidepressant actions of exercise and BDNF (for review see Duman et al., 2008) harmonize well with a separate set of studies showing the involvement of epigenetic regulation in the antidepressant action of BDNF (Tsankova et al., 2006). Abnormal BDNF levels have also been associated with the etiology of various cognitive disorders such as Alzheimer’s disease (Caraci et al., 2010). In turn, acetylation of histone H3 and DNA methylation in the hippocampus have been shown to play a role in Bdnf gene regulation during consolidation of fear memory (Lubin et al., 2008). Given the demonstrated action of exercise and BDNF in supporting learning and memory and synaptic plasticity (Vaynman et al., 2004), it is likely that epigenetic regulation of Bdnf by exercise can serve to modulate learning and memory events.
Our results suggest the fascinating possibility that epigenetic regulation of the Bdnf gene can be a biological mechanism by which exercise can promote mental health and resistance to neurological disorders. These results showing the influence of behavior or experience on the epigenome open new avenues and therapeutic prospects in the war against neurological and psychiatric disorders. The original concept of epigenetics implies the idea that modifications in DNA expression and function can contribute to the inheritance of information (Waddington, 1942). Although this principle has not been fully demonstrated in mammals, there is compelling evidence in the epidemiological literature suggesting that environmental factors can be inherited (Feinberg, 2007). In addition, animals studies show that environmental factors, such as toxins known for their effects on decreasing male fertility, can alter DNA methylation, and that these traits can be transmitted across generations (Anway et al., 2005). Furthermore, based on the strong adaptive force of exercise, exercise remains as a crucial candidate for promoting stable heritable biological adaptations with profound implications for public health.
Acknowledgements
This work was supported by National Institutes of Health Grants NS50465 and NS56413 (F.G.-P.) and NS51411 (G.F.).
Abbreviations
- AceH3
acetylated histone H3
- AceH4
acetylated histone H4
- BDNF
brain-derived neurotrophic factor
- CaMKII
calmodulin-dependent protein kinase II
- ChIP
chromatin immunoprecipitation
- CpG
cytosine-phosphate-guanine
- CREB
cAMP response element binding protein
- HDAC
histone deacetylase
- MeCP2
methyl-CpG-binding protein 2
- PCR
polymerase chain reaction
References
- Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics. 2007;2:214–222. doi: 10.4161/epi.2.4.5212. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraci F, Copani A, Nicoletti F, Drago F. Depression and Alzheimer’s disease: neurobiological links and common pharmacological targets. Eur. J. Pharmacol. 2010;626:64–71. doi: 10.1016/j.ejphar.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J. Biol. Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY. The yin and yang of MeCP2 phosphorylation. Proc. Natl Acad. Sci. USA. 2009;106:4577–4578. doi: 10.1073/pnas.0901518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Ding Q, Vaynman S, Souda P, Whitelegge JP, Gomez-Pinilla F. Exercise affects energy metabolism and neural plasticity-related proteins in the hippocampus as revealed by proteomic analysis. Eur. J. Neurosci. 2006;24:1265–1276. doi: 10.1111/j.1460-9568.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Duman CH, Schlesinger L, Russell DS, Duman RS. Voluntary exercise produces antidepressant and anxiolytic behavioral effects in mice. Brain Res. 2008;1199:148–158. doi: 10.1016/j.brainres.2007.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Vaynman S, Ying Z. Brain-derived neurotrophic factor functions as a metabotrophin to mediate the effects of exercise on cognition. Eur. J. Neurosci. 2008;28:2278–2287. doi: 10.1111/j.1460-9568.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbach GS, Hovda DA, Molteni R, Wu A, Gomez-Pinilla F. Voluntary exercise following traumatic brain injury: brain-derived neurotrophic factor upregulation and recovery of function. Neuroscience. 2004;125:129–139. doi: 10.1016/j.neuroscience.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Hillman CH, Erickson KI, Kramer AF. Be smart, exercise your heart: exercise effects on brain and cognition. Nat. Rev. Neurosci. 2008;9:58–65. doi: 10.1038/nrn2298. [DOI] [PubMed] [Google Scholar]
- Kuzumaki N, Ikegami D, Tamura R, Hareyama N, Imai S, Narita M, Torigoe K, Niikura K, Takeshima H, Ando T, Igarashi K, Kanno J, Ushijima T, Suzuki T. Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus. 2010 doi: 10.1002/hipo.20775. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Larimore JL, Chapleau CA, Kudo S, Theibert A, Percy AK, Pozzo-Miller L. Bdnf overexpression in hippocampal neurons prevents dendritic atrophy caused by Rett-associated MECP2 mutations. Neurobiol. Dis. 2009;34:199–211. doi: 10.1016/j.nbd.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat. Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms in psychiatry. Biol. Psychiatry. 2009;65:189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol. Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl Acad. Sci. USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Marini AM, Lipsky RH. Effects of histone deacetylase inhibitor trichostatin A on epigenetic changes and transcriptional activation of Bdnf promoter 1 by rat hippocampal neurons. Ann. NY Acad. Sci. 2010;1199:186–193. doi: 10.1111/j.1749-6632.2009.05175.x. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Interplay between BDNF and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience. 2003;122:647–657. doi: 10.1016/j.neuroscience.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur. J. Neurosci. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Wu A, Gomez-Pinilla F. Coupling energy metabolism with a mechanism to support brain-derived neurotrophic factor-mediated synaptic plasticity. Neuroscience. 2006;139:1221–1234. doi: 10.1016/j.neuroscience.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. The select action of hippocampal calcium calmodulin protein kinase II in mediating exercise-enhanced cognitive function. Neuroscience. 2007;144:825–833. doi: 10.1016/j.neuroscience.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yossifoff M, Kisliouk T, Meiri N. Dynamic changes in DNA methylation during thermal control establishment affect CREB binding to the brain-derived neurotrophic factor promoter. Eur. J. Neurosci. 2008;28:2267–2277. doi: 10.1111/j.1460-9568.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]