Abstract
Human cytomegalovirus (CMV) UL54 DNA polymerase (pol) mutants with known patterns of resistance to current antivirals ganciclovir (GCV), foscarnet (FOS), and cidofovir (CDV) were tested for cyclopropavir (CPV) susceptibility by a standardized reporter-based yield reduction assay. Exonuclease and A987G (region V) mutations at codons commonly associated with dual GCV-CDV resistance in clinical isolates paradoxically conferred increased CPV susceptibility. Various polymerase catalytic region mutations conferring FOS resistance with variable low-grade GCV and CDV cross-resistance also conferred CPV resistance, with 50% effective concentration (EC50) increases of 3- to 13-fold. CPV EC50 values against several pol mutants were increased about 2-fold by adding UL97 mutation C592G. Propagation of a CMV exonuclease mutant under CPV selected for pol mutations less often than UL97 mutations. In 21 experiments, one instance each of mutations E756D and M844V, which were shown individually to confer 3- to 4-fold increases in CPV EC50, was detected. Unlike GCV and CDV, exonuclease mutations are not a preferred mechanism of CPV resistance, but mutations in and near pol region III may confer CPV resistance by affecting its recognition as an incoming base for DNA polymerization.
INTRODUCTION
Cyclopropavir (CPV) is a methylenecyclopropane nucleoside analog (Fig. 1), currently in phase I human clinical safety studies, being developed for treatment of human cytomegalovirus (CMV) infection on the basis of favorable in vitro potency and cytotoxicity profiles (11) and efficacy in an immunodeficient mouse model (10). Similar to ganciclovir (GCV; Fig. 1), initial phosphorylation by the viral UL97 kinase is required for the anti-CMV action of CPV (8), and UL97 mutations may result in drug resistance by impairing this phosphorylation. Among the 7 canonical UL97 mutations found in >80% of GCV-resistant clinical isolates (M460V/I, H520Q, C592G, A594V, L595S, and C603W) (13), M460I and H520Q are commonly selected under CPV in vitro and confer 12- and 20-fold increased resistance to CPV, respectively, while L595S confers no CPV resistance (2, 9).
Fig 1.
Structures of cyclopropavir (CPV) and ganciclovir (GCV).
Mutations in the CMV UL54 DNA polymerase (pol) gene may confer resistance to GCV, cidofovir (CDV), and/or foscarnet (FOS). Assuming the same antiviral drug target, pol mutations are expected to confer resistance to CPV as well. Many pol mutations that confer resistance to GCV, FOS, and CDV have been characterized (13), but those that confer CPV resistance have not, nor has their relative frequency. In this study, we tested the CPV susceptibility of CMV recombinant strains containing pol mutations selected after exposure to current antivirals and examined the UL97 and pol sequences of an error-prone exonuclease mutant (3) after serial propagation under CPV to study the relative frequency and phenotype of pol mutations selected.
MATERIALS AND METHODS
Antiviral compounds.
CPV (ZSM-I-62) was synthesized at Microbiotix according to a published method (16). Purity was >98%, as determined by analytical high-pressure liquid chromatography. It was diluted into aqueous media from a 10 mM stock solution in dimethyl sulfoxide. GCV sodium salt (Cytovene; Roche), FOS (Foscavir; Astra), and CDV (Vistide; Gilead) were used as aqueous solutions from pharmaceutical materials supplied by their respective manufacturers.
Viral strains and cells.
Mutant and control recombinant CMV strains based on laboratory strain AD169 modified with a secreted alkaline phosphatase (SEAP) reporter gene cassette (CMV strain T2211) were constructed, as previously published, by homologous recombination of genomic viral DNA in fibroblast cultures (4, 5, 14) or, as described more recently, by mutagenesis of bacterial artificial chromosome (BAC) clones of T2211 (1), followed by transfection of mutant BACs into fibroblasts to reconstitute infectious CMV. Single and double mutants of interest were constructed to define the drug resistance phenotypes conferred. Most of the recombinant strains tested have been published (1, 4, 14). The mutants newly reported in this study were constructed using the same methods, i.e., by homologous recombination in fibroblasts (4) for pol M844T with or without UL97 C592G and by BAC mutagenesis (1) for pol P744T, E756D, and M844V. All recombinant strains were sequenced throughout the mutagenized gene (pol, UL97) to verify the presence of the intended mutation and absence of extraneous changes, and BAC clones were analyzed for a compatible HindIII digest pattern without detectable deletions (1). CMV strains were propagated in human foreskin fibroblast (HFF) cultures under standard conditions.
Phenotypic assays.
Drug susceptibility was assayed by the drug concentration required to reduce the accumulation of SEAP activity (chemiluminescent substrate) in HFF culture supernatants by 50% at 6 to 7 days postinoculation (50% effective concentration [EC50]), as previously standardized (1–5). Criteria for valid assays included an input multiplicity of 0.01 to 0.03, as judged by supernatant SEAP activity at 24 h, appropriate EC50 values of known drug-sensitive and -resistant control strains, and a good curve fit of SEAP values observed at various drug concentrations (2, 5). The conventional criterion for phenotypic resistance was a 2-fold elevation of EC50 values over that of a baseline control strain (13). To validate the statistical significance of differences in EC50 values, the unpaired Student t test was used to compare the observed means and standard deviations for the number of replicates performed for each mutant and its corresponding control strain. The relative growth fitness of recombinant strains over multiple cycles of infection was assessed by comparison of supernatant SEAP activities at 24 h and daily on days 4 to 8 after inoculation into 4 wells per strain of the same batch of 24-well confluent HFF monolayer cultures. An equivalent multiplicity of infection (MOI) of 0.02 for each strain was calibrated by SEAP activity measured at 24 h postinoculation, which for each strain was within 15% of the mean value of all strains being compared. This method has been used for several previous multicycle growth curve comparisons (4, 5).
Selection of mutations under CPV in vitro.
CMV exonuclease mutants (pol D413A, strains T2294 and T3360) with error-prone replication that accelerates the emergence of resistance mutations were propagated under CPV to assess the mutations selected in vitro (2, 3). Either strain was propagated in HFF cultures starting with an MOI of ∼0.1 under 0.2 μM CPV. At weekly intervals, cells were trypsinized and ∼30% were dispersed to fresh nearly confluent HFF monolayers. As viral cytopathology became less inhibited by CPV, the drug concentration was increased during propagation to a maximum of 4 μM (up to 30 μM in 3 cases). DNA was extracted from 5 to 15 aliquots of infected cell suspensions saved at various passages, PCR amplified, and sequenced using a dye terminator sequencing kit (BigDye, version 3.1; Applied Biosystems) for UL97 codons 300 to 670 and pol codons 300 to 1000, where functional kinase or conserved polymerase domains have been identified (13).
RESULTS
CPV susceptibility of pol mutants.
pol mutants were selected from available and newly constructed recombinant viruses to represent various pol functional domains and drug susceptibility phenotypes. Originally, the phenotypes conferred by most of the pol mutations were known because they had emerged in clinical isolates after exposure to currently licensed anti-CMV drugs (1, 4, 13, 14). The CPV susceptibility phenotypes are listed in Table 1. Except for P744T, all the mutants listed showed significant (P < 0.01) differences in CPV EC50 values from their wild-type control strain. For comparison, their GCV, FOS, and CDV phenotypes are listed in Table S1 in the supplemental material; these are previously published (1, 4, 13) for all strains except the mutants that were newly phenotyped and described in Table 2. Susceptibility data for E756D from a current BAC recombinant correlate well with those from an older E756D recombinant that was tested by a traditional plaque reduction assay (13). Exonuclease domain mutations (N408K, F412L, P522A, L545W) at codons commonly associated with GCV-CDV dual resistance, as well as the relatively common region V mutation A987G (13), conferred no CPV resistance. Instead, the EC50 ratios were significantly lower, implying CPV hypersensitivity. Various pol catalytic region mutations mainly linked to FOS resistance (1, 13) with variable GCV and/or CDV cross-resistance and clustered in region III (codons 805 to 845) conferred various degrees of CPV resistance: EC50 ratios, 2- to 3-fold for E756K, V781I, and A809V; 4- to 8-fold for Q578H, T813S, A834P, and M844T; and 13-fold for G841A. The addition of UL97 mutation C592G to pol mutation A809V, T813S, G841A, or M844T further increased the CPV EC50s by 1.7- to 2.3-fold, similar to the effect of UL97-pol double mutations on GCV resistance (4).
Table 1.
Genotypes and cyclopropavir susceptibility phenotypes of CMV pol mutants
| Virusa | Genotypeb |
Cyclopropavir phenotype |
||||
|---|---|---|---|---|---|---|
| UL97 | pol | EC50c (μM) | SDd | EC50 ratioe (fold change) | Nf | |
| Control strains | ||||||
| T2211 | Baseline | Baseline | 0.24 | 0.05 | 55 | |
| T3261 | Baseline | Baseline | 0.21 | 0.06 | 24 | |
| T3265 | Baseline | Baseline | 0.24 | 0.06 | 41 | |
| T3259 | C592G | 0.65 | 0.15 | 3.1 | 26 | |
| Exonuclease mutants | ||||||
| T2293 | N408K | 0.14 | 0.06 | 0.6 | 15 | |
| T3267 | F412L | 0.11 | 0.04 | 0.5 | 11 | |
| T3005 | P522A | 0.12 | 0.05 | 0.5 | 7 | |
| T3400 | L545W | 0.14 | 0.05 | 0.6 | 14 | |
| Amino-terminal catalytic domain mutants | ||||||
| T3426 | Q578H | 1.51 | 0.16 | 6.3 | 7 | |
| T3408 | D588N | 0.41 | 0.09 | 1.7 | 14 | |
| Catalytic (palm 1) domain mutants | ||||||
| T3525 | P744T | 0.23 | 0.07 | 1.0 | 11 | |
| T3658 | E756D | 0.74 | 0.11 | 3.1 | 10 | |
| T3430 | E756K | 0.66 | 0.10 | 2.8 | 8 | |
| Catalytic (finger) domain mutants (regions VI and III) | ||||||
| T3417 | V781I | 0.69 | 0.20 | 2.9 | 9 | |
| T2417 | A809V | 0.70 | 0.15 | 2.9 | 10 | |
| T2784 | C592G | A809V | 1.58 | 0.52 | 6.6 | 8 |
| T2542 | T813S | 1.89 | 0.59 | 7.9 | 9 | |
| T2798 | C592G | T813S | 3.09 | 0.55 | 13 | 9 |
| Catalytic (palm 2) domain mutants (region III) | ||||||
| T2291 | A834P | 1.35 | 0.48 | 5.6 | 16 | |
| T2311 | N408K, A834P | 0.61 | 0.25 | 2.5 | 13 | |
| T2420 | G841A | 3.16 | 0.59 | 13 | 8 | |
| T2817 | C592G | G841A | 6.07 | 1.74 | 25 | 11 |
| T2483 | M844T | 0.98 | 0.30 | 4.1 | 14 | |
| T2785 | C592G | M844T | 2.03 | 0.48 | 8.5 | 16 |
| T3652 | M844V | 1.06 | 0.15 | 4.4 | 10 | |
| Catalytic (thumb) domain mutants (region V) | ||||||
| T2222 | 981-2del | 0.33 | 0.08 | 1.4 | 8 | |
| T2261 | C592G | 981-2del | 0.65 | 0.19 | 2.7 | 16 |
| T3429 | A987G | 0.12 | 0.04 | 0.5 | 7 | |
Recombinant virus strain (for corresponding BAC clones, see Table S1 in the supplemental material).
Amino acid change in recombinant virus.
By SEAP yield reduction assay; mean value of the number of assays shown.
Standard deviation of the number of assays shown.
Ratio of mean EC50 value to that of matching control with baseline genotype. Items shown in bold have a CPV EC50 ratio of >1.9.
N, number of assays (replicates performed over at least 4 independent assay setups).
Table 2.
GCV, FOS, and CDV susceptibility phenotypes of pol mutants
| BAC | Virus | UL54 pol genotype |
Ganciclovir |
Foscarnet |
Cidofovir |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutationa | Otherb | EC50c | SDd | Ne | EC50 ratiof (fold change) | EC50 | SD | N | EC50 ratio (fold change) | EC50 | SD | N | EC50 ratio (fold change) | ||
| Controls | |||||||||||||||
| T2211 | S897L | 1.11 | 0.18 | 96 | 39 | 12 | 106 | 0.21 | 0.07 | 129 | |||||
| BA31 | T3265 | FRT, S897L | 1.08 | 0.32 | 121 | 39 | 11 | 114 | 0.20 | 0.06 | 101 | ||||
| Newly phenotyped recombinants | |||||||||||||||
| BA143 | T3525 | P744T | FRT, S897L | 0.96 | 0.24 | 10 | 0.9 | 43 | 7 | 10 | 1.1 | 0.21 | 0.06 | 7 | 1.1 |
| BA189 | T3658 | E756D | FRT, S897L | 1.20 | 0.22 | 21 | 1.1 | 107 | 14 | 9 | 2.7 | 0.22 | 0.07 | 12 | 1.1 |
| T2483 | M844T | 1.59 | 0.52 | 16 | 1.4 | 96 | 22 | 16 | 2.5 | 0.28 | 0.10 | 10 | 1.3 | ||
| T2785 | M844T | C592G (UL97) | 2.75 | 0.89 | 19 | 2.5 | 79 | 18 | 18 | 2.0 | 0.25 | 0.04 | 9 | 1.2 | |
| BA187 | T3652 | M844V | FRT, S897L | 2.50 | 0.54 | 11 | 2.3 | 84 | 27 | 12 | 2.2 | 0.32 | 0.08 | 22 | 1.6 |
Mutation transferred into baseline strain or clone.
Other pol sequence changes from strain AD169 or UL97 change in case of T2785. FRT, FLP recombinase recognition sequence used for BAC mutagenesis.
Mean drug concentration (μM) required to reduce SEAP growth by 50% at 6 to7 days postinfection. Values in bold indicate drug resistance (EC50 > 1.9 times of control value).
Standard deviation of the EC50 values.
N, number of assays (replicates performed over at least 4 independent assay setups).
Ratio of EC50 to that of matching baseline strain.
Viral mutations selected under CPV.
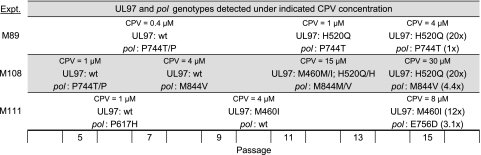
In 21 separate experiments, including 18 already published in connection with UL97 (2), serial propagation of an error-prone D413A pol exonuclease mutant consistently resulted in UL97 mutation: M460I alone (11 cases), H520Q alone (6 cases), both M460I and H520Q (3 cases), and C603R (1 case). All mutations occurred within 15 passages. UL97 mutations were typically detected after 7 to 10 passages at CPV concentrations of 2 to 4 μM. In the same 21 experiments, a pol mutation was detected by dye terminator sequencing in only 3 cases, as detailed in Fig. 2. In experiment M89, P744T appeared to be coselected with UL97 mutation H520Q, whereas in experiment M108, pol M844V appeared before UL97 mutations M460I and H520Q, ultimately selecting for an M844V-H520Q double mutation. In experiment M111, pol E756D was added to the preexisting UL97 mutation M460I. The pol sequence variants P617H and P744T were transiently detected early in the course of experiments M108 and M111 but were no longer detected as drug concentrations were escalated.
Fig 2.
Evolution of UL97 and pol mutations in strain T2294 under CPV. In each experiment (M89, M108, M111), exonuclease mutant strain T2294 was passaged under increasing concentrations of CPV and genotypes were determined periodically at the passage number shown on the bottom scale. The degree of CPV resistance (fold change in EC50 value) conferred by the mutations selected is shown in parentheses. wt, wild type.
Phenotypes conferred by mutations selected under CPV.
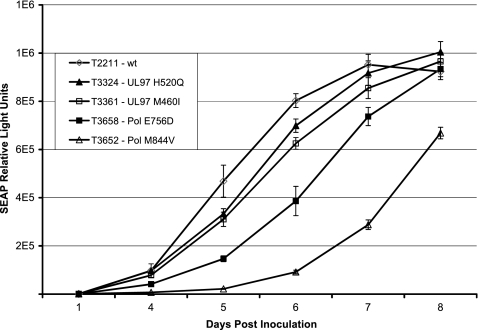
To ascertain the phenotypes conferred by the mutations selected under CPV, they were individually transferred to new recombinant BAC clones and then the clones were tested for their resistance phenotypes for CPV and current drugs (Tables 1 and 2). Mutation P744T conferred no drug resistance, while the E756D and M844V mutations conferred drug susceptibility phenotypes comparable to those conferred by E756K and M844T, with some variation in GCV susceptibility. Mutants containing E756D and M844V showed moderate growth attenuation or retardation, as observed with similar pol mutants (4), which was greater than that observed with the mutants with UL97 mutations M460I and H520Q that are more commonly selected under CPV (Fig. 3).
Fig 3.
Comparative growth curves of UL97 and pol recombinant viruses. Virus stocks were inoculated at equal MOIs of 0.02, and growth was monitored by assay of culture supernatant SEAP activities daily on days 4 to 8 postinoculation. Error bars show the standard deviation of values from 4 replicate wells per time point.
DISCUSSION
The selection of pol mutants in vitro under CPV supports a mechanism of drug action similar to that of GCV, consisting of initial phosphorylation by the UL97 kinase, followed by inhibition of viral DNA polymerase activity by the triphosphorylated compound. As with GCV, pol resistance mutations appear to be initially selected under CPV much less frequently than UL97 mutations, although mutation did occur in 1 of 21 experiments, which approximates the frequency with which pol mutations are initially selected under GCV in clinical practice (13). This probably reflects the relative growth fitness of emergent UL97 and pol mutants (Fig. 3).
As with GCV, the addition of a pol mutation to a preexisting UL97 mutation increases the overall level of CPV resistance (4), as shown by the effect of combining UL97 mutation C592G with several pol mutations in Table 1. At the highest CPV concentrations tested (up to 30 μM in experiment M108), there appeared to be an evolution of UL97 mutations conferring increasing CPV resistance (H520Q > M460I) on viral genomes with or without M844V, ultimately leading to the H520Q-M844V combination, predicted to be the most resistant combination of mutations found in that experiment, based on levels of CPV resistance conferred by the individual mutations.
A remarkable finding from Table 1 is the absence of CPV resistance based on exonuclease or region V mutations, which are frequent causes of GCV-CDV dual resistance in clinical isolates (13). Exonuclease domain mutations are thought to confer drug resistance by increasing the rate of excision of incorporated bases, including nucleoside analogs, at the cost of slowing the rate of DNA synthesis. If this model is correct, structural features of the methylenecyclopropane moiety of CPV appear to prevent its exonuclease-mediated excision, thus explaining the paradoxical CPV hypersensitivity of exonuclease mutants that have a slower overall rate of DNA synthesis.
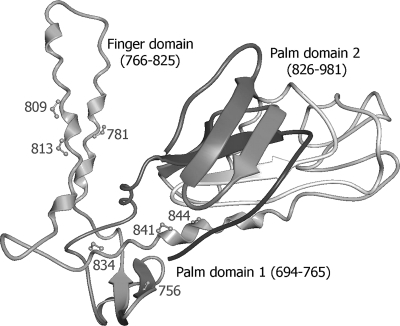
The pol mutations that confer CPV resistance map to residues that are in spatial proximity in or near the finger and palm domains of the polymerase catalytic core (Fig. 4), based on homology with the known structure of the herpes simplex virus DNA polymerase (12), and probably affect recognition of the incoming base. Mutants with such mutations generally have the phenotype of FOS resistance with borderline or low-grade GCV-CDV cross-resistance (Table 1), but the relative levels of resistance to each drug vary according to the specific mutation. The M844V mutation selected under CPV has not been reported previously in resistant CMV clinical isolates and appears to be quite attenuated in growth (Fig. 3). The other mutation, E756D, that was selected under CPV has repeatedly been found in FOS-resistant clinical isolates (13) and was twice selected under FOS in vitro as well (our unpublished data); E756K also appears to be fairly frequently encountered after FOS therapy (13). These codon 756 mutations confer relatively low-grade CPV resistance (∼3-fold increased EC50).
Fig 4.
CMV DNA polymerase catalytic region structure model. The Pol catalytic region contains palm and finger domains, as shown, and a thumb domain extending past residue 981 (data not shown). The structure is modeled on the published herpes simplex virus Pol structure (Protein Data Bank accessionr number 2gv9.pdb) using CPHmodels, version 3.0. Residues affecting CPV susceptibility are labeled; they are also involved in resistance to FOS with or without resistance to GCV/CDV. Mutation Q578H is located in a separate amino-terminal domain just left of residue 813 in this view.
Hypotheses concerning the functional consequences of exonuclease and polymerase catalytic domain mutations, as proposed above, can be tested using CPV triphosphate (CPV-TP) in biochemical assays of in vitro-expressed mutant and wild-type CMV polymerases (6, 7). Furthermore, radiolabeled CPV-TP and a suitable DNA template can be used to examine the kinetics of incorporation and excision of labeled CPV. We plan to perform such experiments in a follow-up to the present study.
Sequence variants pol P617H and P744T observed in vitro (Fig. 2) under CPV do not involve conserved residues or structural features. Their transient appearance in 2 drug selection experiments does not suggest a significant role in CPV resistance, and P744T did not confer CPV resistance when transferred to a control strain. P744T was most likely a spontaneous sequence change in an error-prone exonuclease mutant (T2294) that was coselected with H520Q in one instance, whereas in another experiment, P617H was not coselected and therefore the P617H mutant was overgrown by an emerging drug-resistant M460I mutant. Appearance of spontaneous amino acid substitutions without added drug was the original observation that identified polymerase mutants containing D413A as error prone (3) and has been used to advantage in accelerating the discovery of resistance mutations, including those later observed in clinical isolates (15).
To date, characterization of UL97 and pol CPV resistance mutations indicates a partial GCV cross-resistance based mostly on UL97 mutations (2), FOS cross-resistance based on pol mutations, and little or no CDV cross-resistance. With ongoing clinical development of methylenecyclopropane antiviral compounds with improved potency and toxicity that target the CMV DNA polymerase, understanding their individual cross-resistance profiles will help to define their potential therapeutic roles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victor Wong, Coyne Drummond, and Benjamin Houser for technical assistance.
This work was supported by NIH grant R01-AI39938 and U.S. Department of Veterans Affairs grant I01-BX00925.
Footnotes
Published ahead of print 3 October 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Chou S. 2011. Phenotypic diversity of cytomegalovirus DNA polymerase gene variants observed after antiviral therapy. J. Clin. Virol. 50:287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chou S, Bowlin TL. 2011. Cytomegalovirus UL97 mutations affecting cyclopropavir and ganciclovir susceptibility. Antimicrob. Agents Chemother. 55:382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chou S, Marousek GI. 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J. Virol. 82:246–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou S, Marousek GI, Van Wechel LC, Li S, Weinberg A. 2007. Growth and drug resistance phenotypes resulting from cytomegalovirus DNA polymerase region III mutations observed in clinical specimens. Antimicrob. Agents Chemother. 51:4160–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chou S, Van Wechel LC, Lichy HM, Marousek GI. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cihlar T, Fuller MD, Mulato AS, Cherrington JM. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382–393 [DOI] [PubMed] [Google Scholar]
- 7. Ducancelle A, et al. 2005. Phenotypic characterisation of cytomegalovirus DNA polymerase: a method to study cytomegalovirus isolates resistant to foscarnet. J. Virol. Methods 125:145–151 [DOI] [PubMed] [Google Scholar]
- 8. Gentry BG, Kamil JP, Coen DM, Zemlicka J, Drach JC. 2010. Stereoselective phosphorylation of cyclopropavir by pUL97 and competitive inhibition by maribavir. Antimicrob. Agents Chemother. 54:3093–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. James SH, et al. 2011. Cyclopropavir inhibits the normal function of the human cytomegalovirus UL97 kinase. Antimicrob. Agents Chemother. 55:4682–4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kern ER, et al. 2004. Oral activity of a methylenecyclopropane analog, cyclopropavir, in animal models for cytomegalovirus infections. Antimicrob. Agents Chemother. 48:4745–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kern ER, et al. 2005. In vitro activity and mechanism of action of methylenecyclopropane analogs of nucleosides against herpesvirus replication. Antimicrob. Agents Chemother. 49:1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu S, et al. 2006. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 281:18193–18200 [DOI] [PubMed] [Google Scholar]
- 13. Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott GM, Weinberg A, Rawlinson WD, Chou S. 2007. Multidrug resistance conferred by novel DNA polymerase mutations in human cytomegalovirus isolates. Antimicrob. Agents Chemother. 51:89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strasfeld L, Lee I, Villano S, Chou S. 2010. Virologic characterization of multi-drug-resistant cytomegalovirus infection in two transplant recipients treated with maribavir. J. Infect. Dis. 202:104–108 [DOI] [PubMed] [Google Scholar]
- 16. Zhou S, et al. 2004. Synthesis and antiviral activity of (Z)- and (E)-2,2-[bis(hydroxymethyl)cyclopropylidene]methylpurines and -pyrimidines: second-generation methylenecyclopropane analogues of nucleosides. J. Med. Chem. 47:566–575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.