Abstract
Testing P. aeruginosa efflux pump mutants showed that the LpxC inhibitor CHIR-090 is a substrate for MexAB-OprM, MexCD-OprJ, and MexEF-OprN. Utilizing P. aeruginosa PAO1 with a chromosomal mexC::luxCDABE fusion, luminescent mutants arose on medium containing 4 μg/ml CHIR-090, indicating upregulation of MexCD-OprJ. These mutants were less susceptible to CHIR-090 (MIC, 4 μg/ml) and had mutations in the mexCD-oprJ repressor gene nfxB. Nonluminescent mutants (MIC, 4 μg/ml) that had mutations in the mexAB-oprM regulator gene mexR were also observed. Plating the clinical isolate K2153 on 4 μg/ml CHIR-090 selected mutants with alterations in mexS (immediately upstream of mexT), which upregulates MexEF-OprN. A mutant altered in the putative1ribosomal binding site (RBS) upstream of lpxC and overexpressing LpxC was selected on a related LpxC inhibitor and exhibited reduced susceptibility to CHIR-090. Overexpression of LpxC from a plasmid reduced susceptibility to CHIR-090, and introduction of the altered RBS in this construct further increased expression of LpxC and decreased susceptibility to CHIR-090. Using a mutS (hypermutator) strain, a mutant with an altered lpxC target gene (LpxC L18V) was also selected. Purified LpxC L18V had activity similar to that of wild-type LpxC in an in vitro assay but had reduced inhibition by CHIR-090. Finally, an additional class of mutant, typified by an extreme growth defect, was identified. These mutants had mutations in fabG, indicating that alteration in fatty acid synthesis conferred resistance to LpxC inhibitors. Passaging experiments showed progressive decreases in susceptibility to CHIR-090. Therefore, P. aeruginosa can employ several strategies to reduce susceptibility to CHIR-090 in vitro.
INTRODUCTION
Pseudomonas aeruginosa is a serious and increasingly problematic opportunistic pathogen for which therapeutic options are limited and decreasing (21, 36). The organism is notable for its intrinsic resistance to most antibiotics and the ability to rapidly develop resistance to otherwise effective agents. These characteristics likely derive at least in part from its environmental niche, which necessitates a fundamental ability to resist killing by a wide variety of toxic agents. The combination of an impermeable outer membrane and efflux pumps, particularly those of the resistance-nodulation-cell division (RND) family (43, 45), constitutes one effective strategy for resisting toxic assault. The genome of P. aeruginosa encodes several of these RND efflux pumps; however, to date, only four have been widely investigated as important in clinical resistance to antibiotics. The MexAB-OprM pump is constitutively expressed and, as such, mediates intrinsic resistance, which can increase upon pump overexpression, resulting mainly from mutational inactivation of various regulators, including MexR, NalC, and NalD (5, 30, 51). MexXY is inducible by antibiotics targeting ribosome/protein synthesis (10, 24) or by oxidative stress (14), but constitutive overexpression is frequently encountered in clinical isolates (20, 23). MexCD-OprJ and MexEF-OprN are not appreciably expressed under typical laboratory conditions but are expressed at high levels upon mutation of regulatory or other genes, such as nfxB or mexS, respectively (29, 30, 37, 44, 53).
Efflux pumps of the RND family are arranged in a tripartite structure, with inner membrane pump components, outer membrane (OM) channel components, and so-called membrane fusion partners forming a complex spanning the inner and outer membranes. Molecules are expelled from the bacterial cell in a process driven by the inner membrane potential gradient (H+ antiporter) (4). Assembly of the complete RND pump structure itself may be dependent in part on the OM (i.e., the OM channel). Furthermore, the ability of the pump unit to effectively limit the accumulation of toxic molecules in the cell is also intimately related to the outer membrane permeability barrier, which affects the rate of compound influx (35). In organisms such as P. aeruginosa, the ability of efflux pumps to reduce intracellular accumulation is enhanced by its comparatively less permeable outer membrane. This ability of P. aeruginosa to protect itself against toxic molecules certainly partially accounts for the fact that no truly novel antibacterials effective against the pathogen have reached the market in several decades.
In addition to its role as a permeability barrier to toxins, the OM itself is essential for both viability and virulence in P. aeruginosa, and this makes OM synthesis an attractive target for the development of new therapeutic agents against the pathogen (26, 46). The outer membrane is an asymmetrical bilayer with an inner leaflet of phospholipid but defined by its outer leaflet, composed primarily of lipopolysaccharide (LPS). LPS consists of a lipid A anchor, which forms the outer leaflet, to which a nonrepeating core polysaccharide and a repeating O-antigen polysaccharide chain that extend outward from the cell surface are attached (26). The synthesis of lipid A has been the focus of much attention, since it represents the essential anchor of the outer leaflet and is relatively structurally conserved in Gram-negative bacteria (46). Indeed, target-based efforts directed toward developing small-molecule inhibitors of the LpxC protein, which encodes a metalloamidase mediating the first committed step in the synthesis of LPS, have yielded promising inhibitors with potent antimicrobial activity (8, 40). One of these, designated CHIR-090, demonstrates potency against P. aeruginosa on par with current antibiotics, such as ciprofloxacin (1, 2, 39), and as such represents one of the few reported successes in achieving excellent whole-cell antibacterial activity against this recalcitrant pathogen. The therapeutic longevity of new antimicrobials may be lessened by the emergence of resistance mechanisms, so an early understanding of the resistance potentials of novel antibiotics is critical to maximizing their development and use. Therefore, we undertook a preliminary in vitro study to gain insight into the potential mechanisms that may lead to the development of mutationally acquired resistance to CHIR-090.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli strains were routinely grown in Luria broth (LB) or LB agar. For passaging experiments, P. aeruginosa strains were grown in Mueller-Hinton II broth (Becton Dickinson). Plasmids were maintained in E. coli by supplementation as appropriate with 50 μg/ml kanamycin, 100 μg/ml ampicillin, 10 μg/ml tetracycline, or 30 μg/ml chloramphenicol unless otherwise specified. For P. aeruginosa, growth media were supplemented as necessary with 100 to 200 μg/ml carbenicillin, 100 μg/ml tetracycline, 300 μg/ml chloramphenicol, or 300 μg/ml gentamicin. For growth analysis of strain CDR0026, strains were grown overnight in 2 ml LB and then diluted the following morning to an optical density at 600 nm (OD600) of 0.02 in a total of 20 ml LB. The strains were grown at 37°C and 225 rpm, and OD600 readings were taken at 1 h and 2 h and then every 30 min over the course of 7 h. For single-step isolation of mutants with decreased susceptibility to CHIR-090, P. aeruginosa was grown to mid-log phase (OD600, approximately 0.6) in Mueller-Hinton broth, pelleted by centrifugation, and resuspended in fresh medium. Aliquots were plated on Mueller-Hinton agar containing various levels of CHIR-090 to select for resistant isolates. Serial dilutions were also plated on Mueller-Hinton agar without compound for enumeration. Resistance frequencies were calculated as the number of CFU on drug-containing plates divided by the number of CFU plated.
Table 1.
Strains and plasmids used in this study
| Strain | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| K767 | PAO1 prototroph | 38 |
| K1523 | K767 mexB in-frame deletion | K. Poole |
| K1525 | K767 mexXY in-frame deletion | K. Poole |
| K1542 | K767 mexB, mexXY in-frame deletions | K. Poole |
| K1454 | K767 mexR (MexAB-OprM upregulated) | K. Poole |
| K1536 | K767 nfxB (MexCD-OprJ upregulated) | K. Poole |
| K2153 | Clinical isolate; MexXY upregulated; active mexT | 52 |
| K2376 | K2153 mexS in-frame deletion (MexEF-OprN upregulated) | 53 |
| K2918 | K2153 mexF in-frame deletion | K. Poole |
| CDR0017 | K767 mutS (hypermutator); Gmr | This study |
| CDR0019 | K767 mexC::luxCDABE fusion placed at att site | This study |
| CDR0062 | Derivative of CDR0019 with decreased susceptibility to CHIR-090; nfxB; constitutive luminescence | This study |
| CDR0064 | Derivative of CDR0019 with decreased susceptibility to CHIR-090; mexR; nonluminescent | This study |
| CDR0063 | Derivative of K2153 with decreased susceptibility to CHIR-090; mexS | This study |
| CDR0026 | Derivative of K767 with decreased susceptibility to CHIR-090; C-A substitution at position −11 upstream of lpxC; overexpresses LpxC | This study |
| CDR0061 | Derivative of K767 with decreased susceptibility to CHIR-090; fabG (C494T) | This study |
| CDR0066 | Derivative of CDR0017 with decreased susceptibility to CHIR-090; lpxC (LpxC L18V) | This study |
| CDJ0011 | K767 with lpxC gene replaced by lpxC variant from CDR0066 encoding LpxC L18V | This study |
| PA14 | P. aeruginosa serotype 06 clinical isolate | 47 |
| CDB0011 | PA14 with lpxC gene replaced by lpxC variant from CDR0066 encoding LpxC L18V | This study |
| CDJ0012 | Derivative of CDR0017 with reduced susceptibility to CHIR-090; C-G substitution at position −11 upstream of lpxC; overexpresses LpxC | This study |
| CDJ0013 | Derivative of CDR0017 with reduced susceptibility to CHIR-090; C-G substitution at position −11 upstream of lpxC; overexpresses LpxC | This study |
| E. coli | ||
| S17 | thi pro hsdR recA Tra+; mobilizer strain | 50 |
| SM10 | thi-1 thr leu tonA lacY supE recA RP4-2-Tc::Mu; Kmr; moblizer strain | 50 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacΧ74 recA1 araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pAK1900 | E. coli-P. aeruginosa shuttle vector; Apr Cbr | A. Kropinski |
| pAK-lpxC | pAK1900 harboring lpxC and 100 bp upstream untranslated leader sequence from K767 | This study |
| pAK-lpxCRBS | pAK-lpxC with C-A nucleotide substitution 11 bp upstream of lpxC | This study |
| pMMB-10 | Gateway-adapted pMMB low-copy expression vector; Apr Cbr; IPTG inducible | 16 |
| pMMB-10-fabG | IPTG-inducible fabG expression vector | This study |
| pUCGm | Source of aacC1 Gmr insert; Apr | 49 |
| pEX18Tc | Gene replacement vector; Tcr | 22 |
| pEX18-mutS::aaCC1 | Gene replacement vector for inactivation of mutS; Gmr | This study |
| pEX18ApGW | Gateway-adapted gene replacement vector; Apr Cbr | 7 |
| pEX18ApGW-lpxC-C52G | Gene replacement vector with lpxC C52G allele | This study |
| pMini-CTX-luxCDABE | Promoterless luxCDABE fusion plasmid; Tcr | 3 |
| pFLP2 | Flip recombinase vector; Apr Cbr | 22 |
| pDONR221 | Gateway donor plasmid; Kmr | Invitrogen |
| pET-30b | Protein expression vector | Novagen |
| pET30-PalpxC | Wild-type P. aeruginosa LpxC expression vector | This study |
| pET30-PalpxC C52G | LpxC L18V variant expression vector | This study |
Apr, ampicillin resistance; Cbr, carbenicillin resistance (P. aeruginosa); Kmr, kanamycin resistance; Gmr, gentamicin resistance; Tcr, tetracycline resistance.
DNA protocols.
Standard protocols were employed for restriction endonuclease digestion, gel electrophoresis, ligations, and plasmid isolation (48). Genomic DNA was isolated from P. aeruginosa using the Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, MN) in accordance with the supplied instructions. The PCR primers used in this study are listed in Table 2. PCRs were carried out using an Accuprime GC-rich DNA polymerase kit (Invitrogen, Carlsbad, CA) or the Epicentre Failsafe PCR Kit (Epicenter Biotechnologies, Madison, WI) in accordance with the supplied instructions. PCR fragments were isolated from agarose gels by using a QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA) in accordance with the supplied instructions. DNA sequencing was done by Genewiz, Inc., and sequencing reactions were prepared according to Genewiz guidelines. PCR products were treated with ExoSap-It (USB) prior to sequencing submission.
Table 2.
Primers used in this studya
| Primer | Sequence (5′ to 3′)a | Comment |
|---|---|---|
| MexCforEcoRI | CATGAATTCTCCTCGAAGCGCTTCCGCACGACGATG | |
| MexCrevHindIII | CATAAGCTTGTGAGGACTGATCTTCCCGAGTG | |
| mutSforEcoRI | GATGAATTCGAGCTGCTGATTCCAGACGACTG | |
| mutSrevBamHI | GATGGATCCGAACGGTGTCTCCAGCACCTGC | |
| mutSfor2 | CGTGCTGGACATCACCAGCG | |
| mutSrev2 | GAAGCTGCCGATGTGCGCAAGCAGCAC | |
| lpxCfor | CATGAATTCGAGATCGTCGGCAATCCACGCCTG | |
| lpxCrev | CATGAATTCCAGGAGTAGAGATGTGATTGGTG | |
| lpxCQCfor | GATCATGGCTTTGGCCTCTTAAGCGCTGACTGCG | |
| lpxCQCrev | CGCAGTCAGCGCTTAAGAGGCCAAAGCCATGATC | |
| attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT | Gateway adapter |
| attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT | Gateway adapter |
| attB1-02 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGAGGAGGATATTC | Gateway adapter for expression constructs |
| attB2-02 | GGGGACCACTTTGTACAAGAAAGCTGGGTTTCA | Gateway adapter for expression constructs |
| lpxC 5′ | TACAAAAAAGCAGGCTCAGGAGTAGAGATGTGATTGGTG | lpxC cloning |
| lpxC 3′ | TACAAGAAAGCTGGGTGAGATCGTCGGCAATCCACGCCTG | lpxC cloning |
| lpxC US | CAACCAGTCTCACGGCAG | Amplify lpxC; 200-bp upstream region |
| 5′ fabG | TCGAGGAGGATATTCATGAGTCTGCAAGGTAAGGTCG | fabG cloning |
| 3′ fabG | CAAGAAAGCTGGGTTTCATCAGCTCATGTACATCCCACCATTG | fabG cloning |
| PaLpxCNdeIFP | GGGAATTCCATATGATCAAACAACGCACCTTGAAG | |
| PaLpxCXhoIRP | CCGCTCGAGCTACACTGCCGCC |
Restriction sites are in boldface.
Whole-genome sequencing.
Genomic DNA was isolated using the Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis MN) according to the supplied instructions. Whole-genome fragment libraries were prepared using Illumina's Multiplexing Sample Preparation Oligonucleotide Kit, multiplexing sequencing primers, a PhiX Control Kit v2, and a Paired-End Sample Preparation Kit. Briefly, 5 μg of each genomic DNA sample was fragmented using the nebulization technique for 10 min with compressed air at 32 lb/in2. The ends were repaired with the addition of a base on the 3′ end and ligation of index paired-end adapter oligonucleotide mix. After the addition of adapters, the sample was gel purified, and we selected for 300-bp fragment size to amplify by 18 cycles of PCR. After amplification, the product was purified using Agencourt's PCR Purification System. The library was quality controlled using an aliquot on a Bio-Rad Experion. The DNA sample was hybridized onto the flow cell by using a Paired-End Cluster Generation Kit v2, using a dilution of the 10 nM library. The flow cell was then transferred to the Genome Analzyer II, and at each end of DNA fragments 40 cycles of sequencing were performed using the 36 Cycle Sequencing Kit v3. Data Collection Sequence Control Software version 2.3 was used on the Genome Analzyer II. The sequencing images were analyzed using Solexa pipeline v. 1.3.2.0 software, and the resulting reads from the parental K767 strain were aligned with the P. aeruginosa PAO1 sequence using SOAP software (soap@genomics.org.cn). Nucleotide changes from the reference sequence were called where at least six separate reads called the alternate letter and where the sum of the Solexa quality scores for the alternate letter was 5-fold greater than the sum of the Solexa quality score for the reference letter. A new reference sequence was generated incorporating these changes, and the reads were then aligned with SOAP against the edited genomic sequence one more time. This putative K767 genome was used to align reads from CHIR-090-resistant mutant genomic DNA, and differences were assessed as described above. A BLASTX search of the putative mutant sequence containing the predicted mutation showed that the mutation would cause a nonsynonymous change compared to the reference protein and that the predicted amino acid sequence was not observed in any protein in NCBI's BLASTX database.
Construction of mexC::luxCDABE fusion strain CDR0019.
To construct the mexC::luxCDABE reporter fusion in vitro, a 1,098-bp PCR product encompassing the mexC promoter region and upstream nfxB repressor gene was generated from strain K767 by colony PCR using the Epicentre Failsafe PCR Kit and primers MexCforEcoRI and MexCrevHindIII. The product was digested with EcoRI and HindIII and ligated into pMini-CTX-luxCDABE. This construct was then digested with SalI and religated to remove all but 55 bp of nfxb to avoid having two copies of the repressor in the resulting reporter fusion strain. Therefore, the fusion would be controlled by the native nfxB gene, allowing the selection and characterization of nfxB mutants (see below). This final construct was used to place the fusion onto the genome of strain K767 (as described in reference 6) to generate mexC reporter fusion strain CDR0019.
Construction of mutS (hypermutator) strain CDR0017.
To inactivate mutS on the genome of P. aeruginosa strain K767, the mutS gene was PCR amplified from a resuspended colony with primers mutSforEcoRI and mutSrevBamHI using the Epicentre Failsafe PCR kit according to the manufacturer's directions. The resulting product was digested with EcoRI and BamHI and ligated to pEX18Tc, cut with the same enzymes. The aacC1 gentamicin resistance determinant was then isolated from pUCGm as an 850-bp SmaI fragment and ligated into the unique XcmI site within the mutS gene in the same orientation as the mutS gene. This construct (pEX18-mutS::aacC1) was used to introduce the inactivated gene onto the genome of strain K767 as described previously (6) to generate strain CDR0017. The position of aacC1 within mutS on the genome of CDR0017 was confirmed by PCR using primers mutSfor2 and mutSrev2, which flanked the aacC1 insertion site in mutS. This generated a larger PCR product, consistent with the insertion of aacC1. Strain CDR0017 was then compared to K767 for the frequency of resistance to rifampin, which was at least 100-fold higher for CDR0017 (data not shown), confirming the expected hypermutator phenotype.
Cloning of lpxCPa and site-directed mutagenesis of the upstream putative RBS.
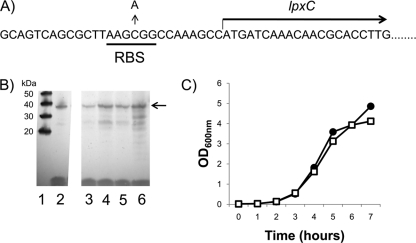
The lpxC gene and circa 100 bp upstream was PCR generated from the genome of strain K767 using the primer pair lpxCfor and lpxCrev, which introduces EcoRI sites at both ends. Plasmid pAK1900 was digested with EcoRI, releasing an approximately 200-bp fragment encompassing the lac promoter, which was then replaced with the EcoRI-digested lpxC PCR fragment to yield plasmid pAK-lpxC. Plasmid pAK-lpxC, therefore, has the P. aeruginosa lpxC gene under the control of its own promoter. This construct was then used for site-directed mutagenesis to introduce a C-to-A change into the putative ribosomal binding site (RBS) 11 bp upstream of lpxC (Fig. 1A). To introduce this change, primers lpxCQCfor and lpxCQCrev were used with the Stratagene Quickchange site-directed mutagenesis kit according to the manufacturer's protocol. The lpxC gene from representative mutated plasmids was then sequenced to confirm the expected change and to confirm that no additional mutations were introduced into lpxC. One confirmed altered plasmid, designated pAK-lpxCRBS, was selected for further study.
Fig 1.
Characterization of LpxC overexpression in P. aeruginosa. (A) Location of a C-to-A nucleotide substitution 11 bp upstream of lpxC in mutant strain CDR0026. (B) Western blot of P. aeruginosa cytoplasmic extracts using anti-LpxC polyclonal antiserum. Lane 1, Molecular size markers; lane 2, purified LpxCPa; lane 3, K767(pAK1900); lane 4, CDR0026 (RBS mutation on genome); lane 5, K767(pAK1900-lpxC) (multicopy); lane 6, K767 (pAK1900-lpxCRBS). The position of LpxC is indicated by the arrow. (C) Growth of P. aeruginosa LpxC-overexpressing strain CDR0026 (□) compared to that of K767 (●), showing no significant growth impairment.
Introduction of the lpxC C52G mutation onto the genome of P. aeruginosa.
The lpxC gene was amplified from CDR0066 with the primers lpxC 5′ and lpxC 3′ using AccuPrime GC-Rich DNA polymerase (Invitrogen). Gateway adapter sequences were added in a second round of PCR with the attB1/attB2 primer pair, and the resulting PCR fragment was cloned into pDONR221 using Gateway technology (Invitrogen). A sequence-confirmed pDONR221-lpxC-C52G clone was used to create pEX18ApGW-lpxC-C52G using Gateway technology according to the supplied instructions. Plasmid pEX18ApGW-lpxC-C52G was introduced into PAO1 strain K767 via conjugation using the mating strain SM10 as described previously (6), except that the recipient P. aeruginosa strain was grown at 37°C rather than 42°C. Merodiploids were selected on Pseudomonas Isolation Agar (Becton Dickinson) containing 200 μg/ml carbenicillin and then resolved in LB without selection. Sucrose-resistant and carbenicillin-sensitive resolvants were screened for resistance to 2 μg/ml CHIR-090, and the presence of the lpxC C52G allele in CHIR-090-resistant colonies was confirmed by sequencing. A similar approach was used to introduce the mutant allele into P. aeruginosa strain PA14 lpxC (strain CDB0011) (Table 1).
Cloning of fabG and complementation of the growth defect in strain CDR0061.
The fabG gene was cloned into the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression plasmid pMMB-10 (15, 16) as follows. fabG was PCR amplified from K767 using the 5′fabG/3′fabG primer pair (Table 2). Gateway adapters were added using primers attB1-02 and attB2-02 (Table 2) in a second round of PCR, and the resulting PCR fragment was cloned into pDONR221 using Gateway technology (Invitrogen). A sequence-confirmed pDONR221-fabG clone was used to create pMMB-10-fabG using Gateway technology. The IPTG-inducible expression plasmid pMMB-10-fabG was introduced into CDR0061 by electroporation according to the method of Enderle and Farwell (12) and selected on LB agar plates containing 150 μg/ml carbenicillin and 1 mM IPTG. K767 cells or CDR0061 cells harboring pMMB-10-fabG were grown at 37°C in 100 μl LB medium supplemented with a range of IPTG levels (0 to 0.5 mM) in a 96-well polystyrene, round-bottom microtiter plate. Growth was monitored by measurements of the optical density at 600 nm (OD600) using a Spectramax 340PC (Molecular Devices) plate reader.
Expression plasmid construction and protein overexpression for P. aeruginosa LpxC and LpxC L18V.
P. aeruginosa lpxC was amplified by PCR using P. aeruginosa K767 genomic DNA as the template and primers PaLpxCNdeIFP and PaLpxCXhoIRP. The PCR fragment was digested with NdeI and XhoI and cloned into pET-30b digested with the same enzymes to generate pET30-PalpxC. An expression vector for the lpxC C52G mutant of strain CDJ0011 (pET30-PalpxC C52G, encoding PaLpxC-L18V) was similarly constructed, except that genomic DNA from strain CDJ0011 was used as the PCR template. These constructs were transformed into E. coli One Shot BL21(DE3)pLysS competent cells for protein expression. Cells containing the expression plasmids were grown in 1 liter terrific broth (TB) containing 50 μg/ml kanamycin and 17 μg/ml chloramphenicol. The cultures were induced with 0.5 mM IPTG and 100 μM zinc sulfate at an OD600 of 0.6. After growth at 30°C for an additional 4 h, the cells were harvested, resuspended in buffer A (20 mM Na2HPO4, pH 7.0), and stored at −80°C if not used immediately.
Pseudomonas aeruginosa LpxC protein purification.
The BL21(DE3)pLysS cells containing either native PaLpxC protein or PaLpxC-L18V were lysed with a French press (∼1,100 lb/in2; 2 passages). DNA in the lysate was precipitated using 5% streptomycin sulfate (∼40% of the lysate volume) with stirring on ice for 20 min, followed by centrifugation (13,000 rpm; 50 min). Subsequently, saturated ammonium sulfate [(NH4)2SO4] solution was slowly added to the supernatant to 50% saturation, and the mixture was stirred at 4°C for an additional 30 min to precipitate the proteins. The (NH4)2SO4-protein pellets were saved after centrifugation (13,000 rpm; 20 min) and stored at −80°C if not used immediately.
The (NH4)2SO4-protein pellets were resuspended in buffer A (4 ml per liter of culture) and dialyzed against buffer A (1 liter) at 4°C overnight. The protein solution was filtered through a 0.45-μm syringe filter and injected onto the AKTA fast protein liquid chromatography (FPLC) system with a 5-ml HiTrap Q FF column (GE Healthcare) attached. The protein was eluted with a 100-ml linear gradient (total volume) of 0 to 100% buffer B (20 mM K2HPO4, pH 7.0, 500 mM NaCl). Fractions (2.5 ml each) were analyzed by SDS-PAGE. The desired fractions were combined and concentrated, followed by incubation with zinc sulfate (500 μM) at 4°C for ∼2 h. Then, the protein was further purified with a size exclusion column (HiLoad 16/60 Superdex 200 preparative-grade column; GE Healthcare) using SEC buffer {20 mM sodium phosphate, pH 7.0, 150 mM NaCl, 1 mM TCEP [tris(2-carboxyethyl)phosphine], and 10% glycerol}. The purified protein was aliquoted and stored at −80°C. The concentration of the protein was determined by Bradford assay or by A280. The presence of the desired protein was confirmed by liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS).
Determination of inhibition of LpxC activity by CHIR-090 (Kiapp determination).
K1app (apparent dissociation constant for the enzyme-inhibitor complex) was used to determine the inhibition of LpxC by CHIR-090, since CHIR-090 is a tight-binding inhibitor (2). The assay was carried out using the LC-MS/MS method as described previously (33). The LpxC substrate, UDP-3-O-R-3-hydroxydecanoyl)-N-acetylglucosamine, was synthesized by the Alberta Research Council (Alberta, Canada). Briefly, assays were performed in a 96-well plate containing PaLpxC or PaLpxC-L18V (1 nM), substrate (2 μM), and CHIR-090 (various concentrations). After incubation at room temperature for 30 min, the reaction was quenched with the stop solution (20% acetic acid). The product formation was quantified using the LC-MS/MS method. The Kiapp value was determined in GraFit (version 5.0) by fitting the data with the following equations:
where [E]T is the total enzyme concentration and [I]T is the total inhibitor concentration.
P. aeruginosa anti-LpxC antiserum production.
The purified P. aeruginosa LpxC protein (13.5 mg/ml, 400 μl, in SEC buffer) was sent to GenScript Corporation (Piscataway, NJ) for polyclonal antibody production using their Partial Polyclonal Antibody Package (mouse). A total of 2.5 ml of antisera from five mice was obtained after four immunizations per mouse. The antibody specificity was confirmed by enzyme-linked immunosorbent assay (ELISA) and Western blotting.
Gel electrophoresis and Western blot analysis for LpxC expression.
Strains K767, CDR0026 (RBS mutation on the genome), K767 containing pAK1900-lpxC, and K767 containing pAK1900-lpxCRBS (Table 1) were inoculated into 20 ml LB. The cultures were incubated overnight to stationary phase at 37°C and 220 rpm. Cells were harvested by centrifugation for 10 min at 3,000 × g, resuspended in 4 ml 30 mM Tris (pH 8.0), and lysed using a French pressure cell. Cytoplasmic extracts were isolated by centrifugation for 30 min at 40,000 × g, normalized according to protein content (A280), loaded onto a NuPage 4 to 12% Bis-Tris precast gel (Invitrogen) as indicated, and run according to the supplied protocol. Protein standards (MagicMark XP; Invitrogen) and purified LpxC (described above) were included. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes using the Invitrogen iBlot Dry blotting system, and Western blots were performed using the SNAP i.d. Protein Detection system (Millipore) according to the standard supplied protocol. The blocking solution was phosphate-buffered saline (PBS) with 0.1% Tween20, 0.25% milk. The LpxC primary mouse antibody was prepared at 1:3,300 in blocking solution. The secondary antibody (Pierce rabbit anti-mouse IgG plus IgM peroxidase conjugate) was prepared at 1:3,300 in blocking solution. The blot was developed using a TMB Substrate Kit for Peroxidase (Vector Laboratories). For mutants isolated during passaging experiments (including CDJ0012 and CDJ0013) (see Fig. 3), cells were grown overnight in LB, harvested, and normalized to an OD600 of 10 in NuPage LDS sample buffer (Invitrogen). The samples were boiled for 10 min and run on a NuPage Novex Bis-Tris Mini gel (Invitrogen). For reference, Precision Plus Protein Dual Color Standards (Bio-Rad) and purified LpxC were also run on the gel. The proteins were transferred to PVDF membranes, and the Western blot was processed as described above.
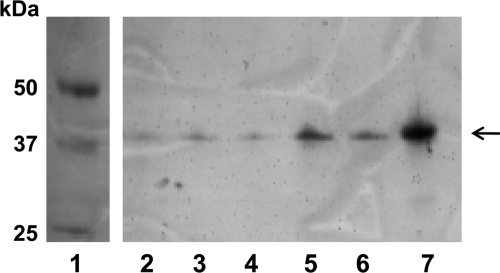
Fig 3.
Western blot of P. aeruginosa isolates from a CHIR-090 passage experiment. Samples were boiled and probed with anti-LpxC polyclonal antiserum. Lane 1, Molecular size markers; lane 2, CDR0017 colony 1; lane 3, CDR0017 colony 2; lane 4, CDR0017 colony 3; lane 5, CDR0017 colony 4 (CDJ0012); lane 6, CDR0017 colony 5 (CDJ0013); lane 7, purified LpxCPa. The position of LpxC is indicated by the arrow. A C-to-G substitution 11 bp upstream of lpxC was confirmed for the mutants in lanes 5 and 6.
Antibiotic susceptibility testing.
MIC determinations were carried out using the broth microdilution procedure in accordance with standard Clinical and Laboratory Standards Institute guidelines (9) with the following modification. The BBL Prompt kit (Difco) was used according to the manufacturer's protocol to prepare an initial suspension of 1.5 × 108 CFU/ml from stationary-phase cells, which corresponds to a McFarland turbidity standard of 0.5. The bacterial suspension was subsequently diluted in Mueller-Hinton broth (cation adjusted) to achieve a final concentration of 5 × 105 CFU/ml. Growth was assessed after approximately 18 h at 37°C. For passaging, an initial MIC determination in a 96-well plate was set up as described above, and the results were designated passage 0. One microliter of bacteria from passage 0 (from the first well below the MIC with growth similar to that in the no-compound control well) was used to inoculate passage 1. After overnight incubation of passage 1, the inoculation process was repeated and continued for a total of 10 passages.
RESULTS AND DISCUSSION
CHIR-090 is a substrate of multiple efflux pumps.
To examine the role of efflux in mediating the intrinsic susceptibility of P. aeruginosa to CHIR-090, mutants lacking MexAB-OprM or MexXY-OprM function were compared to the wild-type PAO1 parent strain K767 (Table 3). Deletion of mexB alone caused a 4-fold increase in susceptibility, indicating that CHIR-090 is a substrate of MexAB-OprM (strain K1523) (Table 3). Deletion of mexXY (strain K1525) did not change the susceptibility; however, deletion of mexB and mexXY simultaneously (strain 1542) (Table 3) caused a slightly larger shift (8-fold) than did loss of mexB alone (4-fold), suggesting that both pumps may efflux CHIR-090, although any contribution from MexXY, at least in the presence of functional MexAB-OprM, appears slight. The MexXY pump components are expressed at low levels unless induced by antibiotics targeting the ribosome or by oxidative stress (10, 14, 24). For certain MexXY-inducing antibiotics, like tetracycline, which also appear to be substrates of both MexAB-OprM and MexXY-OprM, both pumps need to be inactivated to see a substantial shift in susceptibility (10). We have observed that certain non-MexXY-inducing antibiotics, such as ciprofloxacin, are also substrates of both MexAB-OprM and MexXY-OprM and that both pumps need to be inactivated to see a full shift in susceptibility. This indicates that there is sufficient basal expression of MexXY in K767 to impact susceptibility to some noninducer pump substrate antibiotics. Consistent with CHIR-090 being a substrate of MexAB-OprM, overexpression of this pump, resulting from mutation of the mexR gene encoding a repressor of MexAB-OprM expression, led to an 8-fold decrease in susceptibility (strain K1454) (Table 3). Strain K1536, which expresses the normally silent MexCD-OprJ pump due to mutation of the nfxB repressor, was also 8-fold less susceptible to CHIR-090, indicating that CHIR-090 is also a substrate of MexCD-OprJ.
Table 3.
Role of efflux in determining susceptibilities of P. aeruginosa strains to CHIR-090 and other antibiotics
| Strain | Efflux status | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| CHIR-090 | CAR | CHL | OFX | ||
| K767 | Wild type | 0.5 | 64 | 32 | 0.5 |
| K1523 | mexB | 0.125 | 2 | 8 | 0.25 |
| K1525 | mexXY | 0.5 | 64 | 64 | 0.5 |
| K1542 | mexB-mexXY | 0.0625 | 1 | 4 | 0.125 |
| K1454 | mexR (MexAB-OprM upregulated) | 4 | >256 | 256 | 4 |
| K1536 | nfxB (MexCD-OprJ upregulated) | 4 | 64 | 32 | 4 |
| K2153 | Clinical isolate (MexXY upregulated); active mexT | 1 | 128 | 64 | 2 |
| K2376 | K2153 mexS (MexEF-OprN upregulated) | 16 | 128 | 2048 | 16 |
| K2918 | K2376 mexF | 2 | 128 | 64 | 2 |
| CDR0063 | K2153 mexS | 32 | 16 | 1,024 | 8 |
| CDR0019 | K767 mexC::luxCDABE | 1 | 64 | 64 | 1 |
| CDR0062 | CDR0019 nfxB | 4 | 8 | 128 | 8 |
| CDR0064 | CDR0019 mexR | 4 | 256 | 128 | 4 |
Strain K767, from which all of the above-described pump mutants were derived, has an inactive mexT allele, which in its active form functions as a positive activator of MexEF-OprN expression (27, 28, 37). Therefore, in order to evaluate the role of MexEF-OprN, we employed strain K2153, a clinical isolate with the active form of mexT used previously for studies of the role of mexS mutations in controlling upregulation of MexEF-OprN (53). A derivative of this strain (K2153) with mexS inactivated (K2376) (which causes strong overexpression of MexEF-OprN) was 16-fold less susceptible to CHIR-090, indicating that the compound is also a substrate for the MexEF-OprN pump. Inactivation of mexF in the mexS background (K2918) (Table 3) largely restored susceptibility, confirming that the MexEF-OprN pump itself is the resistance determinant. It should be noted that strain K2153 is a pan-aminoglycoside-resistant clinical isolate that constitutively overexpresses MexXY (52). The isolate is not substantially less susceptible to CHIR-090 than other P. aeruginosa strains, such as K767 or PA14, suggesting that overexpression of MexXY does not mediate a meaningful decrease in susceptibility. Consistent with this, deletion of mexXY in strain K2153 did not increase susceptibility (data not shown).
CHIR-090 selects for efflux pump-overexpressing mutants.
Since CHIR-090 was shown to be a substrate for several efflux pumps, we predicted that the inhibitor would directly select for pump-overexpressing mutants. To allow rapid identification of nfxB mutants overexpressing MexCD-OprJ from among the mutant population, the K767 derivative CDR0019 containing a mexC::luxCDABE fusion was employed for these studies. Since CDR0019 lacks a functional MexT, it was expected that MexEF-OprN-overexpressing mutants would be exceptionally rare. Therefore, luminescence from mutant colonies would indicate MexCD-OprJ expression while nonluminescent colonies would be good candidates for overexpression of MexAB-OprM or other nonefflux mechanisms. As predicted, plating strain CDR0019 on 4 μg/ml CHIR-090 selected both luminescent and nonluminescent colonies. An example of a luminescent colony that was 4-fold less susceptible to CHIR-090 (CDR0062) (Table 3) contained a C deletion at position 211 in the MexCD-OprJ repressor gene nfxB. As expected for an nfxB mutant, CDR0062 was also less susceptible to the MexCD-OprJ substrate antibiotics chloramphenicol (CHL) and ofloxacin (OFX), but not the MexAB-OprM-specific substrate carbenicillin (CAR) (Table 3). Indeed, susceptibility to CAR increased somewhat, possibly reflecting downregulation of MexAB-OprM when MexCD-OprJ is overexpressed (19). Two additional luminescent mutants were examined, revealing a G insertion in nfxB at position 475 and a C-to-T substitution at position 154, respectively. None of these nfxB mutants had mutations in lpxC or mexR, the repressor of MexAB-OprM expression. An example of a nonluminescent colony that was also 4-fold less susceptible to CHIR-090 (strain CDR0064) (Table 3) had a G-to-T substitution (introducing a stop codon) at position 80 in mexR, encoding the repressor of MexAB-OprM. Consistent with overexpression of MexAB-OprM in this mutant, susceptibility to the MexAB-OprM substrate antibiotic CAR (as well as CHL and OFX) also decreased (Table 3). An additional nonluminescent mutant was examined, and it had a G deletion at position 307 in mexR.
Strain K2153 was used to test for direct selection of MexEF-OprN-overexpressing mutants. Plating K2153 on 4 μg/ml CHIR-090 selected mutants at a frequency of approximately 1 × 10−7, and of 4 mutants examined, all had mutations in mexS, which has been shown to result in hyperexpression of MexEF-OprN (53). One of these, strain CDR0063, had a CCCGG repeat at position 69 in mexS and was 32-fold less susceptible to CHIR-090 than the parent strain, K2153 (Table 3). Characteristic of MexEF-OprN expression, susceptibility to CHL was dramatically reduced (Table 3). Three additional mutants were examined for changes in mexS, revealing C764A and T725C substitutions and a single-base-pair deletion at position 74, respectively. Therefore, exposure to CHIR-090 in vitro selects for efflux pump-overexpressing mutants, and corresponding multidrug resistance, at compound levels where overexpressed efflux pumps can mediate survival.
Amino acid substitution in LpxC can reduce susceptibility to CHIR-090.
To increase our chances of finding lpxC target mutations decreasing susceptibility to CHIR-090 that might occur at lower frequencies, we conducted plating experiments using a hypermutator strain, CDR0017 (K767 mutS). This strain was plated on a larger amount of CHIR-090 (6 to 8 μg/ml) to minimize the survival of efflux-overexpressing mutants. Several mutants arising on these plates were examined for mutations in lpxC, and one example contained a mutation (C52G) encoding an LpxC L18V substitution. To confirm that this was specifically responsible for decreasing susceptibility to CHIR-090, the chromosomal lpxC was replaced with the mutant allele in P. aeruginosa PAO1 (K767 non-mutS) and in strain PA14. Both of the engineered strains (CDJ0011 and CDB0011) were 16-fold less susceptible to CHIR-090 (Table 4), confirming that alteration in LpxC specifically mediated a decrease in susceptibility. No changes in susceptibility to the efflux pump substrate antibiotics CAR, CHL, and OFX were observed, indicating that efflux overexpression was not selected during strain construction. Mapping the position of the L18V substitution onto the previously published structure of Aquifex aeolicus LpxC bound to CHIR-090 (1) indicates that the L18 residue is located in the CHIR-090 binding pocket within 4 Å of the bound CHIR-090. A recent analysis of E. coli LpxC and P. aeruginosa LpxC structures complexed with a CHIR-090 analog also revealed that the proximal phenyl ring of the inhibitor faces the methyl groups of L18, indicating there is a hydrophobic interaction between the L18 residue and the inhibitor at the entrance of the substrate-binding passage (34). Therefore, amino acid substitutions at this position would be expected to alter compound binding and the corresponding antibacterial activity. Indeed, Kiapp determination showed circa 57-fold (57 ± 11) reduction in LpxC L18V inhibition by CHIR-090 (Kiapp, 9.56 ± 1.76 nM compared to 0.17 ± 0.01 nM for the native protein). Furthermore, this L18V variant protein shows activity comparable to that of the native LpxC protein, consistent with the lack of an obvious growth defect for strain CDJ0011 (data not shown).
Table 4.
Roles of nonefflux mechanisms in determining susceptibility to CHIR-090 in P. aeruginosa
| Strain | Efflux status | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| CHIR-090 | CAR | CHL | OFX | ||
| K767 | Wild type | 0.5 | 64 | 32 | 0.5 |
| CDJ0011 | K767 LpxC L18V | 8 | 64 | 32 | 0.5 |
| PA14 | Clinical isolate | 0.5 | NDb | ND | ND |
| CDB0011 | PA14 LpxC L18V | 8 | 128 | 32 | 0.5 |
| CDR0026 | K767 LpxC upregulated | 16 | 64 | 32 | 0.5 |
| K767(pAK1900) | 0.5 | >256 | 32 | 0.5 | |
| K767(pAK-lpxC) | LpxC overexpressed | 4 | >256 | 32 | 0.5 |
| K767(pAK-lpxCRBS) | LpxC overexpressed | 32 | >256 | 32 | 0.5 |
| CDR0061 | K767 fabG (C494T) | 4 | 32 | 32 | 0.5 |
| CDR0061(pMMB-10-fabG)a | 1 | >256 | 16 | 0.5 | |
With IPTG induction. Strains CDJ0011, CDR0026, and CDR0061 did not change in susceptibility to tobramycin, polymyxin or ciprofloxacin relative to the wild-type K767 parent.
ND, not done.
LpxC overexpression can reduce susceptibility to CHIR-090.
During a plating experiment with a compound structurally related to CHIR-090 (and with similar antibacterial activity) at a level of compound higher than that which would allow the survival of efflux pump-overexpressing mutants, we encountered several slow-growing mutants (described below), but also one normally growing mutant (CDR0026). Isolate CDR0026 was 32-fold less susceptible to CHIR-090 than its parent, K767, and no change in susceptibility to the pump substrate antibiotics CAR, CHL, and OFX was seen, indicating that efflux was unlikely to be involved (Table 4). Sequencing of lpxC and its upstream region revealed a C-to-A mutation located 11 bp upstream of lpxC in a putative ribosomal binding site (Fig. 1A). Western blotting using anti-LpxC antiserum revealed that CDR0026 overexpressed LpxC (Fig. 1B), presumably mediating the decrease in susceptibility to CHIR-090. Transcriptional profiling of CDR0026 did not show an increased lpxC transcript titer (data not shown), suggesting that this mutation caused an increase in LpxC translation. To investigate this further, the wild-type lpxC gene was amplified, along with circa 100 bp of upstream sequence, and cloned into plasmid pAK1900, from which the lac promoter at the multicloning site had been removed, to generate pAK-lpxC. When introduced into a K767 background, multicopy expression from this vector resulted in increased LpxC levels compared to K767(pAK1900), as judged by Western blot analysis [Fig. 1B, compare K767(pAK-lpxC) to K767]. This also resulted in an 8-fold decrease in susceptibility to CHIR-090 [Table 4, K767(pAK-lpxC)], confirming that target overexpression was a resistance determinant for this inhibitor. Site-directed mutagenesis was then employed to introduce the RBS mutation identified in strain CDR0026 into pAK-lpxC, and this new construct mediated an even higher level of LpxC expression [Fig. 1B, K767(pAK-lpxCRBS)], and correspondingly, a 64-fold decrease in susceptibility to CHIR-090 (Table 4). This strongly supports the notion that the upstream putative RBS mutation specifically mediates an increase in LpxC protein expression. It has been reported that LpxC levels in E. coli are tightly regulated via proteolytic cleavage by FtsH in order to prevent a toxic disruption of the balance of LPS and phospholipid production (17, 18, 25, 41). This implied that resistance to LpxC inhibitors caused by a sustained increase in LpxC levels might be unlikely. However, a recent report demonstrated that the FtsH-mediated proteolytic control of LpxC levels described for E. coli does not extend to P. aeruginosa, and overexpression of LpxC from a regulated promoter construct did not impair growth of P. aeruginosa (32). Consistent with this, strain CDR0026 did not show any discernible growth defect (Fig. 1C), and therefore, overexpression of LpxC at levels sufficient to substantially alter susceptibility to CHIR-090 appears to be well tolerated in P. aeruginosa.
Mutation of fabG reduces susceptibility to CHIR-090.
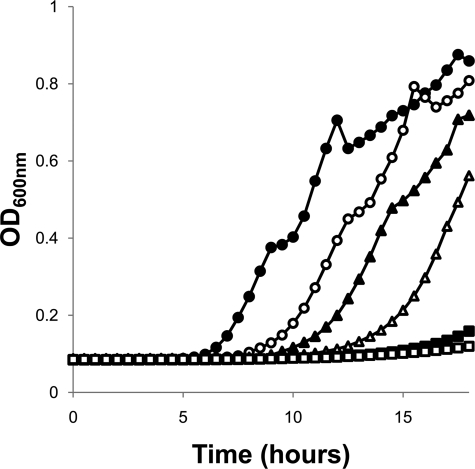
As mentioned above, plating P. aeruginosa PAO1 on an LpxC inhibitor related to CHIR-090 gave rise to mutants taking 24 to 48 h to appear on plates, indicating a significant growth defect. Selection experiments done directly with CHIR-090 (8 μg/ml) also yielded several slow-growth mutants. Whole-genome sequencing of one slow-growth mutant revealed a change in the fatty acid biosynthetic pathway gene fabG, and follow-up PCR and sequencing of fabG from a selection of these mutants confirmed fabG mutations in 5 example mutants analyzed. Example mutant CDR0061 had a C494T substitution encoding FabG T165I and was 8-fold less susceptible to CHIR-090, with no significant change in susceptibility to the pump substrate antibiotics CAR, CHL, and OFX (Table 4). Additional mutations identified were C476A, G3A (mutating the ATG start codon), and a CA deletion (underlined) at the start codon (CATG). This suggests that compromised FabG function causes both a severe growth defect and decreased susceptibility to LpxC inhibitors in P. aeruginosa. Confirming this, induction of FabG expression from pMMB-10-fabG restored wild-type growth and CHIR-090 susceptibility to mutant strain CDR0061 (Fig. 2 and Table 4). It should be noted that CDR0061 was unstable, presumably due to the severe growth defect. Indeed, faster-growing putative suppressor/revertant mutants arose readily from strain CDR0061 plated on drug-containing or non-drug-containing medium, and the behavior of the pMMB-10-fabG plasmid itself was highly unstable (losing IPTG regulation, etc.), hampering complementation studies and MIC determinations. To ensure our complementation resulted from fabG in trans, we used PCR primers designed to specifically amplify the genomic copy of fabG from the complemented strain to confirm the presence of the original genomic C494T mutation (data not shown). Our identification here of several mutations in fabG causing growth defects, including examples changing the ATG start codon, shows that there are several mutations that can disrupt FabG expression/stability or function. A previous report of an inability to genetically inactivate fabG (31) implies that FabG function is reduced rather than eliminated in the mutants identified here, although this remains to be proven. A similar relationship between disruption of the fatty acid biosynthetic pathway and decreased susceptibility to LpxC inhibitors was reported from studies with E. coli, specifically due to mutation of fabZ (8). Reduced FabZ function was proposed to result in an accumulation of the substrate for LpxA (the first step of the lipid A biosynthesis pathway upstream of LpxC), resulting in increased lipid A synthesis and a corresponding necessity for stronger inhibition by LpxC inhibitors to block LPS synthesis and kill cells. It is interesting that in E. coli only fabZ mutations were identified, whereas in our studies with P. aeruginosa, only fabG mutations have been identified to date. Further studies are necessary to determine why this is the case; however, one possibility is that, since the acyl chain lengths on lipid A differ between E. coli (14 carbons) and P. aeruginosa (10 carbons), the substrates for the respective LpxA enzymes correspondingly differ (11), requiring blocking of the fatty acid biosynthesis cycle at a different step in order to accumulate the correct substrate acyl chain length for the P. aeruginosa LpxA.
Fig 2.
Growth defect of fabG C494T mutant strain CDR0061. Complementation of a growth defect by IPTG-inducible expression of wild-type FabG from plasmid pMMB-10-fabG induced with IPTG (0 mM, □; 0.03125 mM, ■; 0.0625 mM, ▵; 0.125 mM, ▴; 0.5 mM, ○). Growth of K767 is shown (●).
Decreasing susceptibility upon repeated passaging in CHIR-090.
Given the identification of several mechanisms that can reduce susceptibility to CHIR-090 in P. aeruginosa and the likelihood that other as-yet-unidentified mechanisms exist, it was of interest to perform a simple passaging experiment to evaluate the decrease in susceptibility over time and to measure the possible cumulative effect of several resistance mechanisms operating together. As mentioned above, MexEF-OprN overexpressors are not readily selected from strain K767 due to its inactive mexT gene, whereas they are present in populations of K2153 (with an active mexT), since any loss-of-function mutation in mexS can cause overexpression of MexEF-OprN in this background. The single-step mexS mutants described above were fairly nonsusceptible (CDR0063 [Table 3]; MIC, 32 μg/ml), suggesting that K2153 might show a faster decrease in susceptibility than K767 during passaging due to initial emergence of MexEF-OprN expression. Strains K767 and K2153, however, showed a similar progressive decrease in susceptibility in this experiment, culminating in a 128-fold change at passage 10 (Table 5) (susceptibility of the population, 128 μg/ml). The mechanisms emerging over time within the populations of mutants at each stage remain to be characterized, but nonetheless, the passaging results show that the cumulative effect of multiple mechanisms can substantially and fairly rapidly reduce susceptibility to CHIR-090, and presumably other LpxC inhibitors, in P. aeruginosa. The rate of mutation, and the corresponding resistance development, varies among different isolates of P. aeruginosa from different infections, and therefore, these two strains provide only a snapshot of resistance potential. Indeed, in the case of chronic P. aeruginosa infection of the cystic fibrosis (CF) lung, it has been reported that a large percentage of isolates are hypermutators, which may increase resistance development (13, 20, 42). Therefore, it would be prudent to examine resistance in hypermutators to better understand resistance potential in chronic CF infection. Thus, a passaging experiment was done with a mutS derivative of strain K767 (CDR0017). As expected, the mutS derivative became less susceptible to CHIR-090 much more quickly than did the K767 parent strain, with a decrease of at least 256-fold (susceptibility of the population > 128 μg/ml) at passage number 4. Aside from providing some perspective on the in vitro resistance potential of hypermutators, the mutS strain can also be helpful for isolation, identification, and characterization of rare mutations. The ribosomal binding site mutation mediating LpxC overexpression in mutant strain CDR0026 is likely a relatively rare mutation, since only one such mutant was confirmed throughout all of the plating experiments done during this work. To determine if this mechanism emerged during passaging, we isolated 5 single colonies from the fifth passage of the K767 mutS strain. We surmised that examining the hypermutator strain would improve the chances of finding an LpxC overexpressor. Examination of LpxC protein levels using Western immunoblotting showed that 2 of the 5 isolated colonies did produce comparatively elevated levels of LpxC (strains CDJ0012 and CDJ0013) (Fig. 3), suggesting that this mechanism would likely emerge over time. The lpxC gene (including ∼200 bp of the upstream region) was amplified from these mutants using the lpxC US/lpxC 3′ primer pair (Table 2) and sequenced. Consistent with the overexpression of LpxC in these mutants, each had an alteration 11 bp upstream of the lpxC structural gene, in the same position as the mutation in the originally identified LpxC overexpressor CDR0026. However, in both CDJ0012 and CDJ0013, the alteration was a C-to-G change, as opposed to the C-to-A change identified in strain CDR0026. Thus, either of these substitutions can apparently alter the translation of LpxC. One of the isolates not showing elevated LpxC levels (colony 3 [Fig. 3, lane 4]) contained no mutation in the upstream region.
Table 5.
Decrease in susceptibility of P. aeruginosa strains to CHIR-090 upon passaging in CHIR-090
| Passage | CHIR-090 susceptibilitya (μg/ml) |
||
|---|---|---|---|
| K767 | K2153 | CDR0017 | |
| 0 | 1 | 1 | 1 |
| 1 | 4 | 4 | 8 |
| 2 | 4 | 4 | 16 |
| 3 | 8 | 16 | 64 |
| 4 | 8 | 16 | >128 |
| 5 | 16 | 32 | >128b |
| 6 | 32 | 32 | ND |
| 7 | 32 | 64 | ND |
| 8 | 64 | 64 | ND |
| 9 | 64 | 64 | ND |
| 10 | 128 | 128 | ND |
Indicated as susceptibility rather than MIC. This is a measure of the susceptibility of the mixed population determined using a nonstandardized inoculum from the previous passage as described in Materials and Methods. Examples of isolated colonies from each step had susceptibilities within one dilution step of that reported in the table, determined with standard MIC testing. ND, not done.
Colonies were isolated from this well for Western blot analysis (Fig. 3).
Conclusions.
In this study, we demonstrated that P. aeruginosa can reduce susceptibility to the LpxC inhibitor CHIR-090 in vitro through a variety of mechanisms, including efflux pump upregulation, target overexpression, target mutation, and interference with fatty acid biosynthesis. The focus in this study was a survey of potential mechanisms rather than an accurate estimation of the frequency of each of them. In general, selection of efflux mutants at drug levels well within what pump overexpression could accommodate was relatively frequent (e.g., circa 1 × 10−7 for MexEF-OprN-expressing mutants), which is expected, since they require simple loss-of-function mutations in genes controlling expression. Target mutations and mutations mediating target overexpression are likely to be relatively rare, but in the latter case, they will likely impact all LpxC inhibitors regardless of structure. For mechanisms such as this, the starting potency of any LpxC inhibitors will be an important factor determining the ultimate level of nonsusceptibility achieved. Similarly, fabG mutations could be expected to affect susceptibility to all LpxC inhibitors, but unlike the other mechanisms described here, they incur a severe fitness cost. Reflecting the diversity of mechanisms identified (and any additional unidentified mechanisms), passaging studies revealed a progressive and substantial decrease in susceptibility, which was exacerbated in a hypermutator strain. Finally, this study again highlights the notion that increasing the starting cellular potency and circumventing recognition by efflux pumps might improve the potential therapeutic longevity of LpxC inhibitors. The contributions of these mechanisms to decreasing susceptibility in vivo remain to be examined.
ACKNOWLEDGMENTS
We thank Keith Poole and Andrew Kropinski (Queens University), Herbert Schweizer (Colorado State University), Fred Ausubel (Massachusetts General Hospital), and Steve Lory (Harvard University) for strains and plasmids. We also thank Srijan Ranjitkar and Jade Bojkovic (Novartis Infectious Diseases) for mass spectrometry assistance and Sovanda Som for assistance with the manuscript. We thank Simon Bushell and Ruben Tommasi (Novartis, Global Discovery Chemistry) for helpful discussions.
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Barb AW, Jiang L, Raetz CR, Zhou P. 2007. Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: time-dependent inhibition and specificity in ligand binding. Proc. Natl. Acad. Sci. U. S. A. 104:18433–18438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barb AW, et al. 2007. Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46:3793–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952 [DOI] [PubMed] [Google Scholar]
- 4. Blair JM, Piddock LJ. 2009. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 12:512–519 [DOI] [PubMed] [Google Scholar]
- 5. Cao L, Srikumar R, Poole K. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423–1436 [DOI] [PubMed] [Google Scholar]
- 6. Caughlan RE, et al. 2009. Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob. Agents Chemother. 53:5015–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clements JM, et al. 2002. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob. Agents Chemother. 46:1793–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CLSI 2006. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically (M7-A7). CLSI, Wayne, PA [Google Scholar]
- 10. Dean CR, Visalli MA, Projan SJ, Sum PE, Bradford PA. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dotson GD, Kaltashov IA, Cotter RJ, Raetz CR. 1998. Expression cloning of a Pseudomonas gene encoding a hydroxydecanoyl-acyl carrier protein-dependent UDP-GlcNAc acyltransferase. J. Bacteriol. 180:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enderle PJ, Farwell MA. 1998. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques 25:954–956, 958 [DOI] [PubMed] [Google Scholar]
- 13. Feliziani S, et al. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fraud S, Poole K. 2011. Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuchs EL, et al. 2010. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J. Bacteriol. 192:3553–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuchs EL, et al. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J. Bacteriol. 192:2779–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhrer F, Langklotz S, Narberhaus F. 2006. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 59:1025–1036 [DOI] [PubMed] [Google Scholar]
- 18. Fuhrer F, et al. 2007. Sequence and length recognition of the C-terminal turnover element of LpxC, a soluble substrate of the membrane-bound FtsH protease. J. Mol. Biol. 372:485–496 [DOI] [PubMed] [Google Scholar]
- 19. Gotoh N, et al. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ho J, Tambyah PA, Paterson DL. 2010. Multiresistant Gram-negative infections: a global perspective. Curr. Opin. Infect. Dis. 23:546–553 [DOI] [PubMed] [Google Scholar]
- 22. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 23. Hocquet D, et al. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51:3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeannot K, Sobel ML, El Garch F, Poole K, Plesiat P. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katz C, Ron EZ. 2008. Dual role of FtsH in regulating lipopolysaccharide biosynthesis in Escherichia coli. J. Bacteriol. 190:7117–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King JD, Kocincova D, Westman EL, Lam JS. 2009. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15:261–312 [DOI] [PubMed] [Google Scholar]
- 27. Kohler T, Epp SF, Curty LK, Pechere JC. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohler T, et al. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345–354 [DOI] [PubMed] [Google Scholar]
- 29. Kohler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar A, Schweizer HP. 2005. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Deliv. Rev. 57:1486–1513 [DOI] [PubMed] [Google Scholar]
- 31. Kutchma AJ, Hoang TT, Schweizer HP. 1999. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J. Bacteriol. 181:5498–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langklotz S, Schakermann M, Narberhaus F. 2011. Control of LPS biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all Gram-negative bacteria. J. Bacteriol. 193:1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langsdorf EF, et al. 2010. Screening for antibacterial inhibitors of the UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC) using a high-throughput mass spectrometry assay. J. Biomol. Screen. 15:52–61 [DOI] [PubMed] [Google Scholar]
- 34. Lee CJ, et al. 2011. Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design. Chem. Biol. 18:38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li XZ, Zhang L, Poole K. 2000. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:433–436 [DOI] [PubMed] [Google Scholar]
- 36. Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22:582–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maseda H, Saito K, Nakajima A, Nakae T. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107–112 [DOI] [PubMed] [Google Scholar]
- 38. Masuda N, Ohya S. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McClerren AL, et al. 2005. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry 44:16574–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mdluli KE, et al. 2006. Molecular validation of LpxC as an antibacterial drug target in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:2178–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogura T, et al. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31:833–844 [DOI] [PubMed] [Google Scholar]
- 42. Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254 [DOI] [PubMed] [Google Scholar]
- 43. Poole K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12–26 [DOI] [PubMed] [Google Scholar]
- 44. Poole K, et al. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713–724 [DOI] [PubMed] [Google Scholar]
- 45. Poole K, Srikumar R. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59–71 [DOI] [PubMed] [Google Scholar]
- 46. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 48. Sambrook R. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Schweizer HD. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834 [PubMed] [Google Scholar]
- 50. Simon R, O'Connell M, Labes M, Puhler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640–659 [DOI] [PubMed] [Google Scholar]
- 51. Sobel ML, Hocquet D, Cao L, Plesiat P, Poole K. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sobel ML, McKay GA, Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sobel ML, Neshat S, Poole K. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187:1246–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]