Abstract
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strain MW2 harbors a plethora of toxins to mediate its virulence. However, toxin expression and regulation with simulated clinical antimicrobial exposures are unclear. This study evaluated these relationships using an in vitro pharmacodynamic hollow-fiber infection model. Clinical doses of clindamycin, linezolid, minocycline, trimethoprim-sulfamethoxazole (SXT), and vancomycin were simulated over 72 h against MW2 in the hollow fiber model. Expression levels of lukSF-PV and enterotoxin genes sec4, sek, seq, and sel2 were quantified by real-time PCR. Panton-Valentine leukocidin (PVL) was quantified by enzyme-linked immunosorbent assay (ELISA), and cytotoxicity was determined on polymorphonuclear cells (PMNs). Vancomycin produced the maximum MW2 killing (2.53 log10 CFU/ml) after the first dose, but the greatest sustained killing over 72 h occurred with linezolid and clindamycin. Vancomycin and minocycline induced gene upregulation from 0 to 8 h, followed by downregulation for the remaining simulation period. Clindamycin decreased gene expression in the first 24 h, followed by moderate increases (2.5-fold) thereafter. Linezolid increased gene expression 11.4- to 200.4-fold but inhibited PVL production (0.6 ± 0.3 versus 5.9 ± 0.2 μg/ml, linezolid versus control at 72 h; P < 0.05). Similar effects on PVL production occurred with clindamycin and minocycline. SXT increased PVL production at 48 h (2.8-fold) and 72 h (4.9-fold) of treatment (P < 0.05), resulting in increased PVL cytotoxicity on PMNs. Linezolid, clindamycin, and minocycline were the most effective agents on decreasing the virulence potential in CA-MRSA, notably after 8 h of treatment. SXT had minimal effects on toxin gene regulation, but it increased production and cytotoxicity of PVL toxin in the model and may enhance virulence when it is used to treat severe infections.
INTRODUCTION
Over the last 15 years, community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains have emerged as a principal cause of skin and soft tissue infections along with contributing to invasive disease (24, 31). The pathogenic capacity and epidemiological success of CA-MRSA are believed to result from diverse antibiotic resistance mechanisms (16) added to the production of a myriad of cell wall-associated adhesion and virulence factors that alter immune system defenses and function (8, 23).
Exotoxins and superantigen toxins in S. aureus are characterized by their shared capacities to induce high fever and cause serious life-threatening toxic shock syndromes and related illnesses (42). Although multiple toxins have been identified in S. aureus, controversy remains regarding their exact function in disease severity among infection types. The presence of genes encoding Panton-Valentine leukocidin ([PVL] lukSF-PV) toxin is an epidemiologic marker for CA-MRSA isolates. Although it is clear that PVL is cytotoxic and epidemiologically associated with staphylococcal necrotizing pneumonia (25), severe bone and joint infections (27), deep-seated abscesses, and complicated skin and soft tissue infections (43, 46), its role in disease was controversial due to inconclusive animal studies (9, 10, 33, 44, 55). This controversy has recently been resolved by evidence that the effects of PVL are species specific. PVL is toxic for human and rabbit neutrophils but not in murine or simian cells (39); studies in rabbit models have confirmed that lukSF-PV expression in the S. aureus USA300 strain is associated with more severe lesions than infection with non-lukSF-PV-expressing strains (14, 19).
In addition, sec4, sel2, sek, and seq genes encode enterotoxins that have been associated with direct tissue damage because of overstimulation or dysregulation of innate inflammatory mediators and cytokine production (42). sek and seq occur in tandem, with ∼55% nucleotide sequence identity. sec4 and sel2 are located on the genomic island Sa3 in the MW2 strain and are the new allelic forms of corresponding staphylococcal enterotoxin genes (2, 52). The diversity of virulence factors present in CA-MRSA suggests that cumulative effects of these virulent toxin genes, rather than a single identified locus, contribute to its high pathogenicity (37, 52).
As described by Stevens et al., antimicrobials can affect S. aureus toxin transcription and translation (53). Antibiotics that inhibit protein synthesis reduce toxin production in vitro and have correlated to improved outcomes in animal models of infection (1, 57). However, recent findings with CA-MRSA clonal strain USA300 propose that protein synthesis inhibitors may exacerbate the progression of disease by altering quorum-sensing gene expression (30). These in vitro studies utilized subinhibitory antibiotic concentrations (21, 30, 53), which are difficult to extrapolate to concentrations achieved with human doses. In addition, these studies are limited to end-of-treatment response evaluations. Virulence modification with antimicrobials throughout the course of therapy may differ, and beneficial effects may be linked to duration of antimicrobial exposure. The effect of simulated antibiotic exposures on CA-MRSA toxin regulation and production has not been investigated. In this study, we employ an in vitro hollow fiber infection model to analyze antibiotic pharmacodynamic activity, toxin gene regulation, and toxin production with resulting cytotoxicity in the well-described CA-MRSA virulent clone of USA400.
(Portions of this work were presented at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, 12 to 15 September 2010.)
MATERIALS AND METHODS
Bacteria.
The isolate used in this investigation was CA-MRSA MW2, a USA400 clonal type. This isolate was obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) Program supported under NIAID/NIH contract number HHSN272200700055C. This widespread strain contains a number of virulence genes, including lukSF-PV, sek, sel2, sec4, and seq, which are predicted to enhance its virulence (2).
Medium.
Todd Hewitt broth ([THB] BD Diagnostic System) medium was used for all susceptibility testing and antibiotic concentration profile simulations in the pharmacodynamic model. This medium optimizes toxin gene expression and production in vitro (51). Bacterial quantification was determined on tryptic soy agar.
Antibiotics.
Six antibiotics with activity against CA-MRSA were evaluated. Clindamycin, trimethoprim-sulfamethoxazole (SXT), vancomycin, and minocycline analytical powders were commercially purchased (RPI Corp., Mt. Prospect, IL). Linezolid (Pfizer, New York, NY) and daptomycin (Cubist, Lexington, MA) analytical powder were provided by the manufacturer. All antibiotic solutions were made fresh prior to each experiment and stored at 4°C throughout the simulated model.
Susceptibility testing.
Antibiotic MICs were determined by broth microdilution in THB. To verify antibiotic activity in THB, MICs were compared to those in Mueller-Hinton broth supplemented with 25 μg/ml calcium (50 μg/ml calcium with daptomycin) and 12.5 μg/ml magnesium (CAMHB) according to the Clinical and Laboratory Standards Institute guidelines (13). Antibiotics with greater than 1 dilution difference in the MICs between the medium types were excluded from further analysis.
Pharmacodynamic model.
A hollow-fiber pharmacodynamic infection model was used to study the effect of antibiotics on S. aureus toxin gene expression and production under conditions of simulated human pharmacokinetics. FiberCell hollow-fiber C2011 cartridges were obtained from FiberCell Systems, Inc. (Frederick, MD). The two-compartment hollow-fiber in vitro model is a well-described pharmacokinetic/pharmacodynamic (PK/PD) model that optimizes drug delivery and allows for simulation of sequestered infections (20). Initial bacterial inocula of 106 CFU/ml were achieved by injecting the extracapillary space of a hollow-fiber cartridge with 0.25 ml of a 0.5 McFarland organism suspension using a DEN-1 densitometer for each experiment. After 30 min, the antibiotic was administered into the central reservoir during the start of the organism's growth phase.
The following clinical antibiotic regimens were simulated with target pharmacodynamic indices for unbound free (f) concentrations derived from clinical pharmacokinetic studies: clindamycin at 600 mg every 8 h (47), linezolid at 600 mg every 12 h (17), minocycline at 100 mg every 12 h (56), SXT at 160 mg trimethoprim/800 mg sulfamethoxazole every 12 h (54), and vancomycin at 1,000 mg every 12 h (49). The antibiotic regimens were simulated to target pharmacokinetic parameters as described in Table 1. Simulated human pharmacokinetic profiles of the antibiotics were achieved by instilling and eliminating growth medium into the central reservoir at a flow rate set for the model volume to target the antibiotic elimination half-life. Samples were taken before each antibiotic administration from the extracapillary space at 0, 1, 2, 4, 8, 24, 32, 48, 56, and 72 h for PK/PD analysis and at 0, 4, 8, 24, 48, and 72 h for mRNA relative quantification and toxin production. Samples for bacterial colony enumeration were serially diluted and quantified by spot plating on drug-free agar. All samples were drawn in duplicate to account for variability in measurements. The change in CFU count (CFU/ml) over time was evaluated during antibiotic exposure (48). This method results in a lower limit of detection of 2.0 log10 CFU/ml. The total reduction in log10 CFU/ml was determined by plotting time-kill curves based on the number of remaining organisms over the 72-h duration. Bactericidal and bacteriostatic activities were defined as a reduction in colony count from the initial inoculum of ≥3 log10 CFU/ml and <3 log10 CFU/ml, respectively. Maximum kill (reduction in log10 CFU/ml compared to starting inoculum) and regrowth (log10 CFU/ml increase from maximal kill until end of the exposure) were also used to assess pharmacodynamic activity (48).
Table 1.
Simulated dosing regimens and targeted and observed pharmacokinetic parameters of the antibiotics in the in vitro hollow-fiber model
| Antibiotic (reference) | Simulated dosing regimen |
t1/2 (h) |
Cmax (μg/ml)a |
||
|---|---|---|---|---|---|
| Predicted | Observedb | Predicted | Observedb | ||
| Clindamycin (47) | 600 mg every 8h | 2.4 | 2.5 ± 0.7 | 2.8 | 2.6 ± 0.2 |
| Minocycline (56) | 100 mg every 12h | 13.6 | 12.6 ± 5.8 | 0.6 | 0.6 ± 0.05 |
| Linezolid (17) | 600 mg every 12h | 7 | 10.1 ± 0.8 | 17.1 | 13.2 ± 0.3 |
| SXTc (54) | 160/800 mg every 12h | 11/11 | 9.9 ± 0.06/11.4 ± 1.4 | 0.8/27 | 0.9 ± 0.02/25 ± 0.7 |
| Vancomycin (49) | 1,000 mg every 12h | 6 | 5.2 ± 0.9 | 17 | 23.7 ± 1.9 |
Cmax, maximum concentration.
Values are ± standard error.
Values represent trimethoprim/sulfamethoxazole amounts, respectively.
Antibiotic concentration and t1/2 estimation.
Samples were withdrawn from the central reservoir for determination of antibiotic concentrations during the first dose of simulation for verification of targeted PK parameters and used for regression analysis. The pharmacokinetic parameters were calculated using WinNonLin pharmacokinetic modeling software (release 5.2; WNL; Pharsight Software, Inc.) Elimination half-life (t1/2) was estimated by linear regression from four data points.
Clindamycin, minocycline, and vancomycin concentrations were evaluated by microbioassay using S. aureus 6539p as the assay organism (34). Blank 0.25-mm disks were spotted with 20 μl of the standards or samples. Each standard was tested in duplicate in agar preswabbed with a 0.5 McFarland suspension of the test organism. Plates were incubated for 18 to 24 h at 37°C, and the resulting zone sizes were measured. The interday coefficient of variation of the microbioassays was <3.6% for each drug. Linezolid and trimethoprim concentrations were determined using high-performance liquid chromatography (HPLC) on a reverse-phase XBridge BEH300 C18 column and XTerra C18 column, respectively. The mobile phase consisted of acetonitrile-ammonium formate-formic acid (40/60/0.1) for linezolid and of formic acid-acetonitrile-formic acid (75/25/0.1) for trimethoprim. Detection was accomplished using an LCQ mass spectrometer (MS) (Micromass; Quattro Micro API), which was programmed in turbo-spray-positive tandem MS (MS-MS) mode to permit measurement of the fragment ions of linezolid and trimethoprim of m/z 338/296 and 291.2/261, respectively. The limit of quantification was 0.1 μg/ml for linezolid and 0.01 μg/ml for trimethoprim. Sulfamethoxazole concentrations were determined using HPLC with fluorescence detection using an XTerra C18 column (excitation wavelength [λexc] of 267 nm; emission wavelength [λem] of 342 nm) with a 1.25 μg/ml limit of quantification.
mRNA quantification.
We determined the relative quantification of seq, sek, sec4, sel2, and lukSF-PV transcripts against an endogenous control gene, gyr, from the MW2 strain, first without antibiotics and then during the simulated exposures for each sample. The mRNA extraction, quantification, and gene expression protocols have been described in Pantrangi et al. (45). The PCR primers and probes are listed in Table 2.
Table 2.
Primers and probes used in this study
| Gene | Primer direction | Primer sequence (5′ → 3′) | Probe sequence (5′ FAM → 3′ NFQ)a | Reference |
|---|---|---|---|---|
| gyr | Forward | CAAATGATCACAGCATTTGGTACAG | AATCGGTGGCGACTTTGATCTAGCGAAAG | 55 |
| Reverse | CGGCATCAGTCATAATGACGAT | |||
| lukS-PV | Forward | AATAACGTATGGCAGAAATATGGATGT | ACTCATGCTACTAGAAGAACAACACACTATGG | 40 |
| Reverse | CAAATGCGTTGTGTATTCTAGATCCT | |||
| sec4 | Forward | ACGGCAATACTTTTTGGTATGA | TGATGCCTGCACCAGGCGAT | This study |
| Reverse | TCAACCGTTTTATTGTCGTTG | |||
| sek | Forward | TGATTTATGACCGTATTCTTCTCC | TGCCGTTATGTCCATAAATGTTG | 50 |
| Reverse | TTGTTACCGCTCAAGAGATTG | |||
| sel2 | Forward | CATCTTGCAAGTATTTTCTTAGTTTGA | CATCTATTTCTTGTGCGGTAACCATTT | This study |
| Reverse | GGCAAGCATCAAACAGTTACA | |||
| seq | Forward | TCAATCTCTTGAGCAGTTACTTCTTT | TGCTTACCATTGACCCAGAGATT | 50 |
| Reverse | GGAATTACGTTGGCAAATCAA |
FAM, 6-carboxyfluorescein; NFQ, nonfluorescent quencher.
PVL protein quantification.
PVL quantification was performed in duplicate by an antibody sandwich enzyme-linked immunosorbent assay ([ELISA] (R&D bioMérieux, Marcy l'Etoile, France) targeting LukS-PV, as described by Badiou et al. (3). The wells of microtiter plates (Sigma, Saint Quentin Fallavier, France) were coated with anti-LukS-PV monoclonal antibody in phosphate-buffered saline (PBS) overnight at room temperature. Unbound monoclonal antibody was washed out twice with a solution of PBS-Tween (0.05%) (PBST), and nonspecific binding sites were blocked with a PBST-milk (5%) solution for 30 min at 37°C. The plates were further washed with PBST before standard recombinant LukS-PV dilutions or samples were added to duplicate wells for 1.5 h at 37°C. Horseradish peroxidase-conjugated rabbit polyclonal F(ab′)2 anti-LukS-PV was then added. Subsequently, the microplates were incubated for 1.5 h at 37°C and washed with PBST before proteins were revealed by the addition of the enzyme substrate (tetramethylbenzidine). After a 30-min incubation in the dark at room temperature, the reaction was stopped by the addition of H2SO4. The plates were read at 450 nm in a model 680 microplate reader (Bio-Rad). PVL concentrations were compared as unadjusted concentrations (μg/ml) and corrected for organism inoculum at each time point for every regimen (μg/ml to log10 CFU/ml).
PVL cytotoxicity in human PMNs.
Human polymorphonuclear cells (PMNs) collected from the blood of healthy donors were isolated by density gradient centrifugation on Polymorphprep (Nycomed Pharma, Zurich, Switzerland). PMNs were washed twice in Hanks balanced salt solution (Sigma-Aldrich) and immediately used in experiments. Viable PMNs were determined by trypan blue exclusion and ranged from 98 to 99% viability. After incubation with culture supernatants from the model samples, pore formation in the cytoplasmic membrane of PMNs was assessed by the uptake of propidium iodide (PI). At 10 min, PMNs were analyzed by flow cytometry, as previously described (4).
Statistical evaluation.
All statistical analyses were performed using GraphPad Prism software (version 5.0a; GraphPad Software, Inc.). Antimicrobial activity (maximum kill, time taken for the inoculum to fall to 99.9% of its value at time zero [T99.9], and regrowth) in all regimens was compared by analysis of variance with Tukey's posthoc test for multiple comparisons. Toxin transcription (quantitative reverse transcription-PCR [qRT-PCR]) and PVL production for each antibiotic were compared to those of the controls (no antibiotic) at the same sample time points throughout the model using a Student's t test. A P value of ≤0.05 was considered statistically significant
RESULTS
Antimicrobial pharmacokinetics/pharmacodynamics.
The MW2 strain was susceptible to all antibiotics evaluated. The MICs of all tested antimicrobials were as follows: clindamycin, 0.12 μg/ml; linezolid, 2 μg/ml; minocycline, 0.5 μg/ml; SXT, 0.13/2.4 μg/ml; and vancomycin, 1 μg/ml. MICs of these antibiotics were stable up to 72 h at 37°C in THB. Although daptomycin is a viable treatment option for CA-MRSA infections, the susceptibility in Todd Hewitt broth was at least 2- to 4-fold higher than that in CAMHB adjusted with 50 μg/ml calcium as recommended. Therefore, the antimicrobial activity of daptomycin in this model with THB could not be accurately determined and was not evaluated. The calculated pharmacokinetic parameters of the antibiotics from measured concentrations are listed in Table 1.
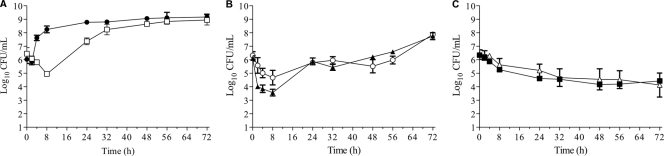
In our in vitro pharmacodynamic model, no antibiotic produced a ≥3-log bacterial reduction from the initial inoculum over the 72-h simulated exposure and therefore produced a bacteriostatic effect (Fig. 1A to C). SXT was the least active antibiotic in the model, with only a 1.5-log10 CFU/ml inoculum reduction after the first dose; this was followed by regrowth after 8 h of treatment and was equivalent to control levels thereafter. Vancomycin resulted in the highest overall bacterial kill of a 2.53-log10 CFU/ml reduction in bacterial count compared to the initial inoculum. This occurred after the first dose and was followed by consistent regrowth with continued dosing from 24 to 72 h. Minocycline followed the same profile but with a lower reduction from the starting inoculum (1.64 log10 CFU/ml). Linezolid and clindamycin had the greatest sustained reduction in bacterial load over the 72-h simulated exposure, with a maximum decrease in overall bacterial load of 2.46 log10 CFU/ml (P = 0.021) and 2.16 log10 CFU/ml (P = 0.037), respectively, occurring at 72 h.
Fig 1.
Activity of simulated concentration exposures against MW2 in the hollow-fiber infection model. (A) Circles, growth control; squares, SXT at 160 mg/800 mg every 12 h. (B) Triangles, vancomycin at 1 g every 12 h; circles, minocycline at 100 mg every 12 h. (C) Squares, clindamycin at 600 mg every 8 h; triangles, linezolid at 600 mg every 12 h.
Relative quantification of toxin gene expression in MW2.
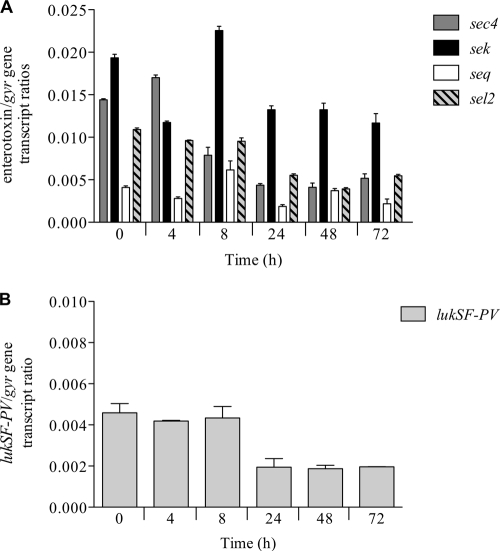
First, we evaluated the expression of lukSF-PV and the four staphylococcal enterotoxin genes, sec4, sek, seq, and sel2, in culture without antibiotic exposure (Fig. 2A and B). In this evaluation, the MW2 strain displayed a consistent downregulation from initial toxin gene expression over the 72-h model duration. As expected, different genes had different relative expression levels over the 72 h. Overall, the average relative expression of these genes was higher during the period from 0 to 8 h than from 24 to 72 h. At the 106 CFU/ml inoculum at 0 h, the expression of sek was the highest, followed by that of sec4, sel2, and seq (Fig. 2A). The lowest gene expression occurred at 24 h, with a reduction of 3.3-fold for sec4, 1.5-fold for sek, 2.2-fold for seq, and 2.0-fold for sel2 compared to the 0-h expression level. This diurnal-like gene expression was more apparent with lukSF-PV, where expression was 2-fold higher during the period from 0 to 8 h than from 24 to 72 h (Fig. 2B).
Fig 2.
Toxin gene expression without antibiotic exposure in the hollow-fiber model of enterotoxin genes sec4, sek, seq, and sel2 (A) and the PVL gene lukSF-PV (B).
Relative quantification of lukSF-PV expression with simulated antibiotic exposures.
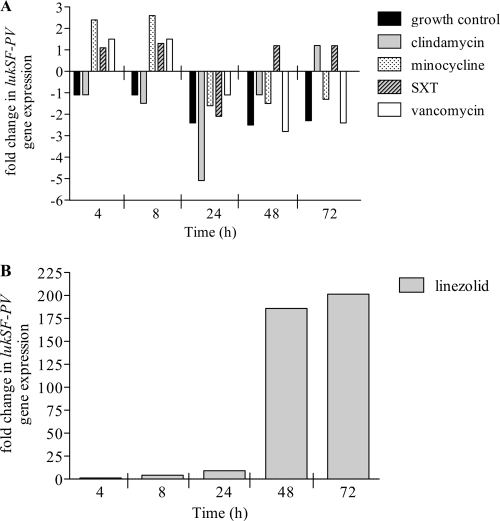
Next, we determined if the simulated dose exposures of antibiotics modulate the expression of lukSF-PV (Fig. 3). Antimicrobials that inhibit protein synthesis (clindamycin, linezolid and minocycline) displayed specific effects on lukSF-PV of MW2. Minocycline stimulated the highest expression of the lukSF-PV at 4 and 8 h before downregulating expression at 24 to 72 h compared to control levels. Clindamycin significantly downregulated lukSF-PV expression by 5.1-fold at 24 h. (P < 0.05). Similar effects were noted with vancomycin. Linezolid was effective in suppressing the expression of lukSF-PV during the first 24 h, with minimal changes observed during this phase of linezolid exposure. However, at 24 h and continuing throughout the model exposure, a consistent trend toward increased gene expression was observed (Fig. 3B). At 72 h, lukSF-PV levels were significantly upregulated (201.4-fold increase; P < 0.05). SXT exposure overall did not significantly change lukSF-PV expression at any point during the model simulation.
Fig 3.
lukSF-PV gene expression in MW2 from the hollow-fiber model with antibiotic simulations. Data represent changes in gene expression (fold increase or decrease) during antibiotic exposures compared to the initial expression level at time zero for each treatment with the following antibiotics: clindamycin, minocycline, SXT, and vancomycin (A) and linezolid (B).
Relative quantification of expression of enterotoxin genes sec4, sek, seq, and sel2 with simulated antibiotic exposures.
Substantial changes also occurred in the expression of enterotoxin genes upon exposure to the antibiotics, but their effects varied significantly relative to each other (Table 3). Minocycline exposure stimulated increased expression at 4 h and 8 h, but at 48 to 72 h of exposure gene expression was either unchanged or downregulated. Clindamycin reduced enterotoxin gene expression in the first 8 h, with seq and sel2 expression most affected (6.9-fold and 3.2-fold reduction, respectively; P < 0.05). At 72 h of clindamycin exposure, all genes except sel2 were upregulated compared to the expression at 0 h. Linezolid increased gene expression of all enterotoxins (≤8.3-fold) within the first 8 h of linezolid exposure. However, at 24 h and continuing until 72 h of exposure, extraordinarily high levels of gene expression were observed. At 72 h, there was a 161.5-fold increase in sec4 and 23- to 37-fold increases in sek, seq, and sel2 (P < 0.05) expression levels.
Table 3.
Enterotoxin gene expression in MW2 from the hollow-fiber model during growth and antibiotic exposure simulations
| Drug | Time (h) | Fold change in expression of:a |
|||
|---|---|---|---|---|---|
| sec4 | sek | seq | sel2 | ||
| Control | 4 | NC | −1.6 | −1.5 | NC |
| 8 | −1.8 | NC | 1.5 | NC | |
| 24 | −3.3 | −1.5 | −2.2 | −2.0 | |
| 48 | −3.5 | −1.5 | NC | −2.8 | |
| 72 | −2.8 | −1.7 | −1.9 | −2.0 | |
| Clindamycin | 4 | NC | NC | NC | NC |
| 8 | −1.5 | NC | −6.9 | −3.2 | |
| 24 | 3.2 | NC | −1.5 | −4.1 | |
| 48 | 1.9 | NC | NC | NC | |
| 72 | 2.5 | NC | 1.7 | −1.5 | |
| Linezolid | 4 | 2.0 | 1.8 | 2.3 | NC |
| 8 | 6.3 | 4.1 | 8.3 | 3.4 | |
| 24 | 38.4 | 7.7 | 2.1 | 4.9 | |
| 48 | 124.1 | 16.9 | 24.4 | 24.0 | |
| 72 | 161.5 | 23.3 | 37.3 | 34.5 | |
| Minocycline | 4 | −1.6 | 3.0 | 4.2 | 2.1 |
| 8 | NC | 2.9 | 3.9 | 2.2 | |
| 24 | −5.1 | NC | NC | −1.7 | |
| 48 | −6.4 | NC | NC | NC | |
| 72 | −4.6 | NC | NC | −1.7 | |
| SXT | 4 | NC | NC | −1.6 | −1.5 |
| 8 | 2.6 | 1.5 | 2.5 | 2.0 | |
| 24 | NC | NC | NC | NC | |
| 48 | NC | NC | 1.5 | NC | |
| 72 | NC | NC | NC | NC | |
| Vancomycin | 4 | −1.6 | 2.5 | 2.6 | 9.8 |
| 8 | NC | 3.3 | 2.8 | 17.5 | |
| 24 | −3.1 | NC | −2.0 | NC | |
| 48 | −9.1 | −2.3 | −2.0 | −2.5 | |
| 72 | −23.5 | −1.8 | −1.8 | −2.4 | |
Data represent change in gene expression (fold increase/decrease) compared to initial expression at time zero for each treatment. NC, no change (defined as <1.5-fold).
Vancomycin exposure upregulated toxin gene expression in the first 8 h, with the highest level observed for sel2 (17.5-fold increase; P ≤ 0.05). From 24 to 72 h of exposure, enterotoxin gene expression levels were reduced, with, most prominently, a 23.5-fold reduction in sec4 expression (P < 0.05). SXT exposure only moderately increased enterotoxin gene expression at 8 h (2.6-fold increase in sec4, 2.5-fold increase in seq, 2.0-fold increase in sel2, and 1.5-fold increase in sek), but minimal changes were observed from 24 to 72 h.
PVL protein production.
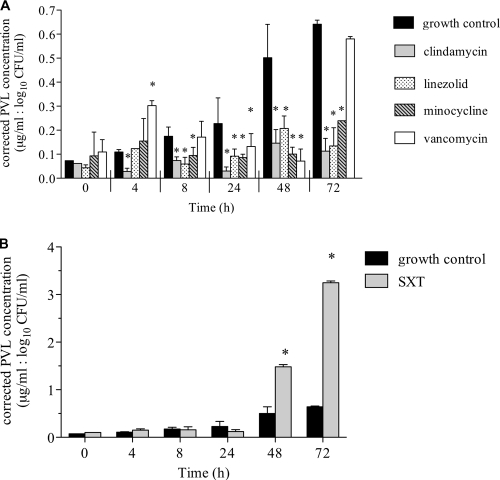
PVL concentrations were determined with and without the antibiotic-simulated exposures from 0 to 72 h. At 0 h, PVL production ranged from 0.27 to 1.22 μg/ml, and levels were similar between control and antibiotic experiments. In the absence of antibiotics, PVL production increased in the model to a maximum of 5.9 ± 0.2 μg/ml at 72 h, a 13.3-fold increase from the initial PVL concentration. Clindamycin, linezolid, and minocycline reduced PVL production compared to the control at all time points. After 72 h of exposure, PVL toxin production was inhibited most effectively with clindamycin (0.5 ± 0.23 μg/ml; P < 0.001) and linezolid (0.55 ± 0.31 μg/ml; P < 0.001). This trend was similar for the other time points studied with the hollow-fiber model. Minocycline also inhibited PVL production from 8 to 72 h compared to the control (1.88 ± 0.01 μg/ml at 72 h; P < 0.05). The PVL concentrations were also assessed after correction for log10 CFU/ml at each time point to account for differences in bacterial burdens among the treatment regimens (Fig. 4). This analysis demonstrated similar PVL protein profiles to the unadjusted data with the exception of an increase in PVL during early vancomycin therapy at 4 h (P < 0.05). All other time points with vancomycin had similar or lower adjusted PVL concentrations compared to control levels.
Fig 4.
PVL protein production (μg/ml) in MW2 during antibiotic exposure corrected for organism burden (log10 CFU/ml) per time point in the hollow-fiber PK/PD model. (A) Clindamycin, linezolid, minocycline, and vancomycin versus control. (B) SXT versus control. *, P < 0.05 for the difference in PVL concentration with antibiotic versus the control at each time point.
SXT was the only antibiotic evaluated that significantly increased PVL toxin production in both unadjusted and corrected analyses (Fig. 4B). At 48 and 72 h of SXT exposure, unadjusted PVL concentrations were 12.8 ± 0.4 and 29.1 ± 0.3 μg/ml, respectively (P < 0.05). These values represent 19.3- and 43.7-fold increases in PVL production compared to initial concentrations.
PVL cytotoxicity.
We determined the cytotoxicity of the quantified PVL protein on polymorphonuclear cells for the antibiotic regimens that resulted in significantly increased PVL production compared to the control. Therefore, only the SXT-simulated regimen was used for this analysis. The formation of membrane pores in PMNs increased as a function of SXT exposure in the model. After a 20-min incubation, mean propidium iodide (PI) uptake was 7%, 35% (P = 0.01), and 39% (P < 0.001) after 24, 48, and 72 h, respectively, of SXT exposure in the hollow-fiber model. This indicates that the accumulated toxin was active and increasingly cytotoxic as PVL toxin production increased with SXT exposure.
DISCUSSION
The pathogenicity of a CA-MRSA strain belonging to the clone USA400 has been largely attributed to a diversity of virulence factors often not present in other S. aureus strains (2, 52). It is predicted that the presence of additional superantigen-like genes enhances the virulence potential of this clone, increases disease severity, and alters host inflammation (2). Effective management of these infections includes early and appropriate antimicrobial treatment, which could also impact the production of virulent toxins. Antibiotics are known to modulate immune functions during inflammation or serious infections (29), and different classes of antimicrobials have various effects (32). Antibiotics that inhibit protein synthesis may reduce toxin production while potent cell wall inhibitors stimulate an increase in toxin release; however, these observations in vitro are limited to subinhibitory concentrations (21, 53). In the present study, we examined the impact of simulated antibiotic exposures on expression of five important toxin genes in MW2 as well as on PVL production in a hollow-fiber infection model. This model has been a valuable tool to clinically optimize antibiotic effectiveness, and we demonstrate a novel function for this system to understand antibiotic influence on bacterial virulence during therapeutic treatment.
The pharmacodynamic activities of the antibiotics used in the study are consistent with previously reported effects in two-compartment models (7). The agents with the most rapid antimicrobial activity on first dose (vancomycin, minocycline, and SXT) had limited further killing effects in the model after continuous dosing. Clindamycin and linezolid, although less active after the first dose, displayed improved and sustained activity over 72 h. All of these antibiotics demonstrate good outcomes in the treatment of complicated skin and skin structure infections, the predominant infection type in CA-MRSA (38). Interestingly, linezolid and clindamycin, both antibiotics with high tissue penetration in patients with clinical doses (15, 17), were the most effective in reducing inoculum burden at the end of exposure in this study.
A comparison of antibiotic pharmacodynamics and corresponding toxin gene expression indicated that antibiotics with the highest initial activity (vancomycin and minocycline) resulted in upregulation of lukSF-PV expression in the first 8 h of exposure. This upregulation corresponded to enhanced production of PVL with vancomycin when production was corrected for bacterial burden but no change in corrected PVL production with minocycline, confirming that the 30S ribosomal-based inhibitors at clinical concentrations inhibit the production of PVL. Linezolid and clindamycin, the two 50S ribosomal subunit inhibitors, also induced an upregulation of the enterotoxin genes at later time points where the killing activity was maximized. Animal models with these antibiotics appear to link our findings to improved outcomes. Azeh et al. observed decreased morbidity and mortality due to sepsis caused by Gram-positive bacteria with clindamycin treatment compared to bactericidal β-lactam antibiotics in a mouse model (1). More recently, Yanagihara et al. reported that linezolid reduces bacterial burden, cytokine production, and inflammation compared to vancomycin and therefore improves survival in a lukSF-PV-positive S. aureus pulmonary mouse model (57). Our study adds to these findings by demonstrating a point during therapy, in this case after 8 h, when significant reductions in PVL toxin production begin to occur and beneficial clinical effects may emerge.
Linezolid induced the highest lukSF-PV gene expression but lowest PVL production at 72 h. This can be explained by the fact that 50S ribosomal-based inhibitors block peptidyltransferase activity as well as inhibiting protease and RNase (41). Linezolid inhibits translation, and therefore tRNA levels increase from reduced consumption due to blocked translation (41). Inhibition of translation can also expose mRNA to degradation, resulting in polarity and the noncoordinate synthesis of ribosomal proteins from longer operons, further exacerbating the shortage of ribosomal proteins (18). Since linezolid also increased the expression of enterotoxin genes at 72 h, it is likely to have a similar effect on blocking the production of enterotoxins at the ribosomal level. Based on this mechanism, we believe that linezolid at clinical exposures inhibits multiple toxins produced by CA-MRSA in addition to PVL toxin as demonstrated in our study, despite an upregulation in these toxin gene transcripts.
In our model with minocycline exposure, toxin genes were upregulated at 0 and 8 h (during maximum antibacterial effect), but mRNA levels were not altered or upregulated at 24 h and onwards. A number of factors may be involved with this association, including bacterial stress or dysregulation of quorum-sensing system (12, 30, 41). Tetracyclines inhibit bacterial protein synthesis by binding to the 30S subunit and preventing the association of aminoacyl-tRNA with the bacterial ribosome (5). Later in the course of therapy beyond the 3 days studied in this model, minocycline may be more efficient in reducing gene expression and ultimately toxin production since 30S inhibitors have been shown to slow the assembly of both the 30S and 50S ribosomal subunits (41). Minocycline was effective in suppressing PVL toxin production throughout the model, but overall it had lower antibacterial activity and therefore was potentially less effective in treatment of CA-MRSA infections than other protein synthesis inhibitors with initial activity at the 50S subunit.
The only antibiotic in our study that increased PVL toxin production was SXT, despite the relatively moderate change in gene expression with this antibiotic. Previous studies of the effects of antibiotics on S. aureus virulence did not evaluate SXT (21, 53), and animal models with SXT are difficult due to the high content of thymidine in animal sera (34). We did not anticipate the notable increase in toxin production observed with SXT. The involvement of the SOS response in S. aureus may partially explain our findings (35). The SOS response is a process in which DNA repair and mutagenesis are induced as a result of DNA damage (22). This process also stimulates the replication of phages in the bacterial host. Trimethoprim, a known inducer of the SOS response, prevents bacterial DNA synthesis by inhibiting dihydrofolate reductase (DHF) and limits incorporation of thymine into bacterial DNA (35). At subinhibitory concentrations, trimethoprim has been linked to induction of phage-encoded virulence determinants such as staphylokinase (26). A number of bacteriophages encoding PVL have been identified in CA-MRSA, including in the MW2 clonal strain (6). We propose that SXT, primarily the trimethoprim component, activates genes in the PVL phage and contributes to our observations of increased PVL production with limited changes in lukSF-PV gene transcripts. Given these findings, studies of antibiotic exposures should consider the impact of modifications in phage transcription on toxin production.
The mechanism of S. aureus virulence and toxin gene expression is a complicated process that likely involves multiple regulatory pathways (11). Numerous global regulators such as agr, sarA, and mgrA, among others, independently control virulence genes and protein production, and each may be affected differently by outside influences such as host defenses and antibiotics (40). Our findings in the hollow-fiber model substantiate the complex nature of these relationships among the enterotoxin genes. Expression levels of these genes in the exponential and stationary growth phases were highly variable relative to each other. When MW2 was tested against antibiotics in the model, antibiotic killing corresponded to upregulation in gene expression regardless of the antibiotic mechanism. In addition, increased lukSF-PV expression did not necessarily result in increased PVL toxin production. The regulation of these virulence factors in human infection and treatment is even more complex (11), but this model provides some insight into the relationships between pathogen and treatment. It should be noted that this in vitro model does not simulate clearance or degradation of toxins in vivo, which is also important in the duration of the inflammatory response. Further, we used common clinical antibiotic doses in the model, and clinicians may prefer different dosing regimens of these antibiotics. It is of interest whether these effects vary with antibiotic doses or intervals and are driven by certain PK parameters such as minimum or maximum concentration (Cmin or Cmax, respectively) or area under the concentration-time curve (AUC). Comprehensive temporal gene expression studies using simulated antibiotic exposures in the hollow-fiber model will be helpful in understanding virulence control in vitro and in vivo
In conclusion, CA-MRSA MW2 expresses a variety of virulence factors in vitro. The hollow-fiber infection model that simulates clinical antibiotic pharmacodynamics is a useful method to evaluate the toxin-modulating effects of antimicrobials throughout dosing intervals and the duration of therapy. In this model, the protein synthesis inhibitors that target the 50S ribosome, clindamycin and linezolid, had the greatest sustained antibacterial activity along with the most potent inhibition of PVL production over a 3-day simulation of standard doses. These effects demonstrate a potential advantage after 8 h of treatment for using these antibiotics in severe CA-MRSA infections to reduce pathogen virulence where toxin production may contribute to disease severity. Although multiple toxin genes were evaluated in this study, only PVL protein production was determined. Our study evaluated a single CA-MRSA clonal strain, USA400, and other investigators have described marked strain-to-strain variation in virulence factors such as toxin quantities produced by PVL-positive MRSA strains in vitro (28) and differential virulence in vivo among global CA-MRSA strains (36). Analysis of these relationships with respect to other S. aureus strains, relevant toxins, and virulence factors using in vitro models, in conjunction with in vivo studies, may advance dosing strategies and optimize the length of antimicrobial therapy used to modulate pathogen virulence and improve patient outcome.
ACKNOWLEDGMENTS
This work was supported by a grant from the Alternatives Research and Development Foundation. G.L. and C.B. were supported by a grant from the European Community (EC 222718). We thank Marshfield Research Clinic Foundation for financial support.
We thank Oana Dumitrescu for scientific guidance.
Footnotes
Published ahead of print 7 November 2011
REFERENCES
- 1. Azeh I, et al. 2002. Protein synthesis inhibiting clindamycin improves outcome in a mouse model of Staphylococcus aureus sepsis compared with the cell wall active ceftriaxone. Crit. Care Med. 30:1560–1564 [DOI] [PubMed] [Google Scholar]
- 2. Baba T, et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 3. Badiou C, et al. 2010. Rapid detection of Staphylococcus aureus Panton-Valentine leukocidin in clinical specimens by enzyme-linked immunosorbent assay and immunochromatographic tests. J. Clin. Microbiol. 48:1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besseyre des Horts T, et al. 2010. A histidine-to-arginine substitution in Panton-Valentine leukocidin from USA300 community-acquired methicillin-resistant Staphylococcus aureus does not impair its leukotoxicity. Infect. Immun. 78:260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bishburg E, Bishburg K. 2009. Minocycline—an old drug for a new century: emphasis on methicillin-resistant Staphylococcus aureus (MRSA) and Acinetobacter baumannii. Int. J. Antimicrob. Agents. 34:395–401 [DOI] [PubMed] [Google Scholar]
- 6. Boakes E, et al. 2011. Distinct bacteriophages encoding Panton-Valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J. Clin. Microbiol. 49:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowker KE, Noel AR, Macgowan AP. 2008. Pharmacodynamics of minocycline against Staphylococcus aureus in an in vitro pharmacokinetic model. Antimicrob. Agents Chemother. 52:4370–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bronner S, Monteil H, Prevost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183–200 [DOI] [PubMed] [Google Scholar]
- 9. Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405–1406 [DOI] [PubMed] [Google Scholar]
- 10. Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 198:1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chambers HF. 2009. Pathogenesis of staphylococcal infection: a manner of expression. J. Infect. Dis. 199:291–293 [DOI] [PubMed] [Google Scholar]
- 12. Clements MO, Foster SJ. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458–462 [DOI] [PubMed] [Google Scholar]
- 13. Clinical Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Cremieux AC, et al. 2009. Panton-valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. PLoS One 4:e7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darley ES, MacGowan AP. 2004. Antibiotic treatment of gram-positive bone and joint infections. J. Antimicrob. Chemother. 53:928–935 [DOI] [PubMed] [Google Scholar]
- 16. Daum RS, et al. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344–1347 [DOI] [PubMed] [Google Scholar]
- 17. Dehghanyar P, et al. 2005. Penetration of linezolid into soft tissues of healthy volunteers after single and multiple doses. Antimicrob. Agents Chemother. 49:2367–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dennis PP. 1976. Effects of chloramphenicol on the transcriptional activities of ribosomal RNA and ribosomal protein genes in Escherichia coli. J. Mol. Biol. 108:535–546 [DOI] [PubMed] [Google Scholar]
- 19. Diep BA, et al. 2010. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. U. S. A. 107:5587–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drusano GL, Liu W, Brown DL, Rice LB, Louie A. 2009. Impact of short-course quinolone therapy on susceptible and resistant populations of Staphylococcus aureus. J. Infect. Dis. 199:219–226 [DOI] [PubMed] [Google Scholar]
- 21. Dumitrescu O, et al. 2007. Effect of antibiotics on Staphylococcus aureus producing Panton-Valentine leukocidin. Antimicrob. Agents Chemother. 51:1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erill I, Campoy S, Barbe J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637–656 [DOI] [PubMed] [Google Scholar]
- 23. Foster TJ, Hook M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484–488 [DOI] [PubMed] [Google Scholar]
- 24. Fridkin SK, et al. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436–1444 [DOI] [PubMed] [Google Scholar]
- 25. Gillet Y, et al. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759 [DOI] [PubMed] [Google Scholar]
- 26. Goerke C, Koller J, Wolz C. 2006. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonzalez BE, et al. 2005. Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 115:642–648 [DOI] [PubMed] [Google Scholar]
- 28. Hamilton SM, et al. 2007. In vitro production of Panton-Valentine leukocidin among strains of methicillin-resistant Staphylococcus aureus causing diverse infections. Clin. Infect. Dis. 45:1550–1558 [DOI] [PubMed] [Google Scholar]
- 29. Hirata N, et al. 2001. Pretreatment of mice with clindamycin improves survival of endotoxic shock by modulating the release of inflammatory cytokines. Antimicrob. Agents Chemother. 45:2638–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joo HS, Chan JL, Cheung GY, Otto M. 2010. Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:4942–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaplan SL, et al. 2005. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin. Infect. Dis. 40:1785–1791 [DOI] [PubMed] [Google Scholar]
- 32. Karlstrom A, Boyd KL, English BK, McCullers JA. 2009. Treatment with protein synthesis inhibitors improves outcomes of secondary bacterial pneumonia after influenza. J. Infect. Dis. 199:311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Labandeira-Rey M, et al. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133 [DOI] [PubMed] [Google Scholar]
- 34. LaPlante KL, Leonard SN, Andes DR, Craig WA, Rybak MJ. 2008. Activities of clindamycin, daptomycin, doxycycline, linezolid, trimethoprim-sulfamethoxazole, and vancomycin against community-associated methicillin-resistant Staphylococcus aureus with inducible clindamycin resistance in murine thigh infection and in vitro pharmacodynamic models. Antimicrob. Agents Chemother. 52:2156–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewin CS, Amyes SG. 1991. The role of the SOS response in bacteria exposed to zidovudine or trimethoprim. J. Med. Microbiol. 34:329–332 [DOI] [PubMed] [Google Scholar]
- 36. Li M, et al. 2010. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J. Infect. Dis. 202:1866–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M, et al. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C, et al. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52:e18–55 [DOI] [PubMed] [Google Scholar]
- 39. Loffler B, et al. 2010. Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6:e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loughman JA, Fritz SA, Storch GA, Hunstad DA. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maguire BA. 2009. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol. Mol. Biol. Rev. 73:22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCormick JK, Yarwood JM, Schlievert PM. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77–104 [DOI] [PubMed] [Google Scholar]
- 43. Miller LG, et al. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 352:1445–1453 [DOI] [PubMed] [Google Scholar]
- 44. Montgomery CP, Daum RS. 2009. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for Panton-Valentine leukocidin? Infect. Immun. 77:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pantrangi M, Singh VK, Wolz C, Shukla SK. 2010. Staphylococcal superantigen-like genes, ssl5 and ssl8, are positively regulated by sae and negatively by agr in the Newman strain. FEMS Microbiol. Lett. 308:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perbet S, et al. 2010. Multifocal community-acquired necrotizing fasciitis caused by a Panton-Valentine leukocidin-producing methicillin-sensitive Staphylococcus aureus. Infection 38:223–225 [DOI] [PubMed] [Google Scholar]
- 47. Plaisance KI, Drusano GL, Forrest A, Townsend RJ, Standiford HC. 1989. Pharmacokinetic evaluation of two dosage regimens of clindamycin phosphate. Antimicrob. Agents Chemother. 33:618–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rose WE, Knier RM, Hutson PR. 2010. Pharmacodynamic effect of clinical vancomycin exposures on cell wall thickness in heterogeneous vancomycin-intermediate Staphylococcus aureus. J. Antimicrob. Chemother. 65:2149–2154 [DOI] [PubMed] [Google Scholar]
- 49. Rybak M, et al. 2009. Therapeutic monitoring of vancomycin in adult patients: a consensus review of the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists. Am. J. Health Syst. Pharm. 66:82–98 [DOI] [PubMed] [Google Scholar]
- 50. Sammons LR. 2010. In vitro regulation of the seq and sek staphylococcal enterotoxin genes among four allelic variants. M.S. thesis University of Wisconsin—La Crosse, La Crosse, WI [Google Scholar]
- 51. Schlievert PM, Case LC. 2007. Molecular analysis of staphylococcal superantigens. Methods Mol. Biol. 391:113–126 [DOI] [PubMed] [Google Scholar]
- 52. Shukla SK, et al. 2010. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J. Clin. Microbiol. 48:3582–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stevens DL, et al. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 195:202–211 [DOI] [PubMed] [Google Scholar]
- 54. Varoquaux O, et al. 1985. Pharmacokinetics of the trimethoprim-sulphamethoxazole combination in the elderly. Br. J. Clin. Pharmacol. 20:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Voyich JM, et al. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 56. Welling PG, Shaw WR, Uman SJ, Tse FL, Craig WA. 1975. Pharmacokinetics of minocycline in renal failure. Antimicrob. Agents Chemother. 8:532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yanagihara K, et al. 2009. Efficacy of linezolid against Panton-Valentine leukocidin (PVL)-positive methicillin-resistant Staphylococcus aureus (MRSA) in a mouse model of haematogenous pulmonary infection. Int. J. Antimicrob. Agents. 34:477–481 [DOI] [PubMed] [Google Scholar]