Abstract
Nonnucleoside reverse transcriptase inhibitors (NNRTIs) are one of the key components of antiretroviral therapy drug regimen against human immunodeficiency virus type 1 (HIV-1) replication. We previously described a newly synthesized small molecule, 10-chloromethyl-11-demethyl-12-oxo-calanolide A (F18), a (+)-calanolide A analog, as a novel anti-HIV-1 NNRTI (H. Xue et al., J. Med. Chem. 53:1397–1401, 2010). Here, we further investigated its antiviral range, drug resistance profile, and underlying mechanism of action. F18 consistently displayed potent activity against primary HIV-1 isolates, including various subtypes of group M, circulating recombinant form (CRF) 01_AE, and laboratory-adapted drug-resistant viruses. Moreover, F18 displayed distinct profiles against 17 NNRTI-resistant pseudoviruses, with an excellent potency especially against one of the most prevalent strains with the Y181C mutation (50% effective concentration, 1.0 nM), which was in stark contrast to the extensively used NNRTIs nevirapine and efavirenz. Moreover, we induced F18-resistant viruses by in vitro serial passages and found that the mutation L100I appeared to be the dominant contributor to F18 resistance, further suggesting a binding motif different from that of nevirapine and efavirenz. F18 was nonantagonistic when used in combination with other antiretrovirals against both wild-type and drug-resistant viruses in infected peripheral blood mononuclear cells. Interestingly, F18 displayed a highly synergistic antiviral effect with nevirapine against nevirapine-resistant virus (Y181C). Furthermore, in silico docking analysis suggested that F18 may bind to the HIV-1 reverse transcriptase differently from other NNRTIs. This study presents F18 as a new potential drug for clinical use and also presents a new mechanism-based design for future NNRTI.
INTRODUCTION
Despite more than 28 years of effort, neither a protective vaccine nor a therapeutic cure exists for HIV/AIDS. The current clinical management of HIV-1 infections relies heavily on life-long antiretroviral therapy (ART). Upon virus entry into host cells, single-stranded HIV-1 RNA is converted into double-stranded proviral DNA by the virally encoded reverse transcriptase (RT). Due to the pivotal role of HIV-1 RT, this enzyme is one of the major therapeutic targets in impeding the replication of HIV-1 (15, 33). At present, two classes of RT inhibitors are available as treatment for HIV-1 infections: nucleoside/nucleotide RT inhibitors (NRTIs) and nonnucleoside RT inhibitors (NNRTIs). A standard regimen of ART consists of two NRTIs plus one NNRTI or one NRTI and one NNRTI plus one protease inhibitor (PI). NNRTIs remain a key component in drug regimens against HIV-1 replication and infection.
Unlike the range of available NRTIs and PIs, only three NNRTIs, namely, nevirapine (NVP), efavirenz (EFV), and etravirine (ETR), are currently available for the treatment of AIDS patients in the clinical setting. For ART-naïve patients, NVP and EFV are usually first included in the drug regimen; however, the side effects of these two drugs often result in poor adherence in these patients as well as failure of the drug treatment, as these patients exhibit a low genetic barrier to drug resistance and cross-resistance (4, 11). In fact, drug-resistant mutations can readily emerge after 1 week of NVP monotherapy (36). K103N is one of the prevalent mutations in ART-experienced patients that cause a high degree of resistance to both NVP and EFV, while another frequent mutation, Y181C, also causes a significant resistance to NVP (29, 41). On the other hand, ETR, which is a new NNRTI approved by the U.S. Food and Drug Administration (FDA) in 2007 often used to treat ART-experienced patients, retains high efficacy against NVP- and EFV-resistant viruses both in vitro and in vivo (1, 26, 38). Although recent clinical trials did not report any serious side effects associated with ETR (21), its safety in long-term usage is yet to be determined. A disadvantage of ETR is that it is administered twice a day, and this contributes to the inconvenience for patients, whereas EFV requires only a single dose per day (19). Therefore, the discovery of new NNRTIs remains an ongoing priority to ensure that effective treatment is available for HIV/AIDS patients.
(+)-Calanolide A is a natural product initially extracted from the tropical rainforest tree Calophyllum lanigerum that was recently identified as an attractive NNRTI against HIV-1 despite virus strains' containing drug-resistant K103N/Y181C mutations (6, 9, 20, 43). Unlike conventional NNRTIs, (+)-calanolide A was postulated to compete with deoxynucleoside triphosphates (dNTPs) in binding to the HIV-1 RT active site (10) and thereby hindering its activity. Although it shows promising results, (+)-calanolide A is difficult to purify from its natural source in a sufficient amount for clinical use. In addition, its low therapeutic index (TI; range, 16 to 279) and nonideal antiviral activity have contributed to the delay of its clinical development (9, 14). We previously reported the successful construction of a small molecule library of (+)-calanolide A analogs based on tetracyclic dipyranocoumarin in vitro, which should allow largely scaled-up production in the future and the opportunity for structure-based drug optimization to improve antiviral activity (24, 25, 44). One of these analogs, 10-chloromethyl-11-demethyl-12-oxo-calanolide A (designated F18) (Fig. 1A), possesses high potency against wild-type HIV-1 (50% effective concentration [EC50], 7.4 nM) in a TZM-bl cell-based assay (44). We therefore investigated F18 further in regard to its antiviral range, drug resistance profile, and underlying mechanism of action. The data presented here provide a basis not only for the clinical development of F18 but also for the mechanism-based design of novel NNRTIs.
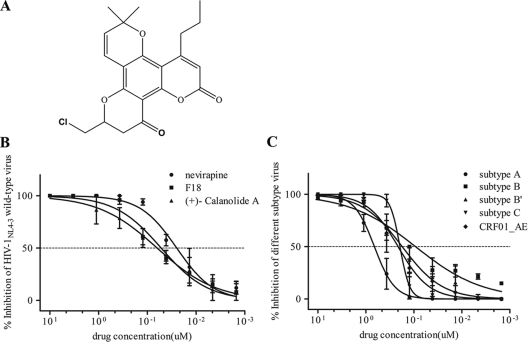
Fig 1.
Activity of F18 against HIV-1NL4-3 wild-type strains and five major subtypes in PBMCs. (A) Structure of 10-chloromethyl-11-demethyl-12-oxo-calanolide A (F18). (B and C) Serially diluted F18, NVP, and (+)-calanolide A were added to PBMCs infected with wild-type HIV-1NL4-3 (B) or five major subtypes (C). The p24 level was measured 7 days PI. Each drug was tested in three PHA-stimulated fresh healthy donor PBMCs. All results are means ± standard errors of the means from three independent experiments.
MATERIALS AND METHODS
Viruses and compounds.
All laboratory-adapted HIV strains (see Fig. 3C and Table 3) and antiretrovirals (ARVs) (see Tables 2 and 3) were obtained from the NIH AIDS Research and Reference Reagent Program (Germantown, MD). Primary HIV-1 isolates include group M subtypes A (TK1135), B (10HKQEH1661), B′(02HNsmx), and C (93IN109), as well as circulating recombinant form (CRF) 01_AE (10HKQEH1447 and 10HKQEH1479). Patient specimens used by us for isolating 10HKQEH1661, 10HKQEH1447, and 10HKQEH1479 were provided by Queen Elizabeth Hospital (Hong Kong SAR, People's Republic of China). Both F18 and (+)-calanolide A were synthesized in-house, and no cytotoxicity was observed with any concentrations used in our experiments.
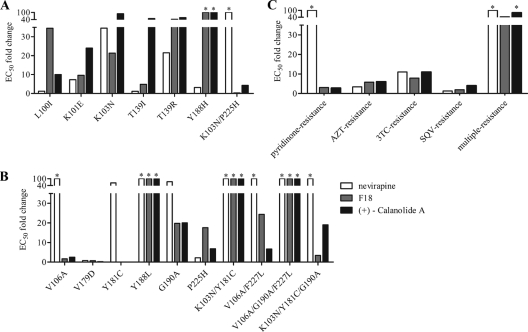
Fig 3.
Resistance of SDM-constructed HIVNL4-3 and laboratory-adapted virus strains to different NNRTIs. GHOST(3)-CCR5 cells were infected in the presence of NVP, F18, or (+)-calanolide A with NNRTI mutant pseudoviruses constructed by SDM for mutations identified from Table 1 (A) and mutations known to resistant different classes of ARVs (B). (C) PBMCs were infected in the presence of NVP, F18, or (+)-calanolide A with laboratory-adapted virus strains that show resistance to ARVs. Data are averages from triplicate experiments. *, the EC50 was not reached at the concentration tested.
Table 3.
Antiviral efficacy results for F18 in combination with eight FDA-approved antiretroviral compounds against drug-resistant virus in PBMC assays
| Drug | Mean vol (effect) on virusa |
|||||
|---|---|---|---|---|---|---|
| NNRTI resistant |
NRTI resistant |
PI (SQV) resistant |
Multiresistant |
|||
| NVP resistant 181C | HIV-1IIIB A17, 103N/181C | AZT resistant, 41L/67N/69N/70R/215F/219Q | 3TC resistant, 184V | 48V/90 M | Δ67/69G/70R/74I/103N/215F/219Q | |
| NRTI | ||||||
| AZT | 502.94/−175.96 (highly synergistic) | 251.20/−82.10 (highly synergistic) | 316.43/−8.59 (highly synergistic) | 312.73/−105.60 (highly synergistic) | 2.43/−86.59 (additive) | 219.68/−152.19 (highly synergistic) |
| ddI | 595.13/0.00 (highly synergistic) | 126.80/−57.63 (highly synergistic) | 287.59/−102.88 (highly synergistic) | 665.78/−22.84 (highly synergistic) | 784.27/−41.37 (highly synergistic) | 542.96/−4.62 (highly synergistic) |
| d4T | 515.28/−150.56 (highly synergistic) | 267.79/−10.87 (slightly synergistic) | 8.70/−30.20 (additive) | 162.66/−184.84 (highly synergistic) | 209.30/−29.01 (highly synergistic) | 263.00/−281.44 (highly synergistic) |
| 3TC | 31.25/−229.93 (additive) | 130.45/−29.03 (highly synergistic) | 334.98/−138.74 (highly synergistic) | 339.17/−9.15 (highly synergistic) | 248.87/−30.61 (highly synergistic) | 289.70/−173.05 (highly synergistic) |
| NNRTI | ||||||
| NVP | 648.63/−14.76 (highly synergistic) | 173.14/−210.29 (highly synergistic) | 344.95/−94.52 (highly synergistic) | 212.14/−12.90 (highly synergistic) | 308.10/−11.27 (highly synergistic) | 289.52/−44.27 (highly synergistic) |
| EFV | 39.12/−64.89 (additive) | 78.93/−82.52 (highly synergistic) | 42.18/−6.82 (additive) | 50.98/−14.52 (additive) | 41.63/−12.94 (additive) | 742.48/−84.42 (highly synergistic) |
| PI | ||||||
| NFV | 0.60/−2.27 (additive) | 13.79/−0.89 (additive) | 288.95/−19.76 (highly synergistic) | 43.95/−6.32 (additive) | 37.01/−9.88 (additive) | 292.56/−47.33 (highly synergistic) |
| IN | ||||||
| RAL | 413.26/−148.00 (highly synergistic) | 52.12/−34.89 (additive) | 56.47/−9.78 (slightly synergistic) | 5.39/−0.13 (additive) | 43.24/−68.98 (additive) | 236.80/−25.89 (highly synergistic) |
Given as mean synergy volume/antagonist volume. Positive and negative values show synergistic and antagonistic interaction between F18 and other ARVs, respectively. SQV, saquinavir.
Table 2.
Antiviral efficacy of F18 in combination with eight FDA-approved antiretroviral compounds against HIV-1NL4-3 in PBMC assays
| Drug | Mean synergy vol/antagonism vola | Antiviral effect |
|---|---|---|
| NNRTI | ||
| NVP | 73.1/−134.39 | Slightly synergistic |
| EFV | 4.01/0.00 | Additive |
| NRTI | ||
| AZT | 87.62/−45.52 | Slightly synergistic |
| 3TC | 17.32/−134.28 | Additive |
| d4T | 27.71/−68.38 | Additive |
| ddI | 297.47/−92.99 | Highly synergistic |
| PI | ||
| NFV | 116.85/−65.37 | Highly synergistic |
| IN | ||
| RAL | 54.12/−9.53 | Slightly synergistic |
Positive and negative values are represent synergistic and antagonistic interactions between F18 and other ARVs, respectively.
Cells.
293T, TZM-bl, GHOST(3)-CCR5, and MT-2 cells were obtained from the NIH AIDS Research and Reference Reagent Program. 293T and TZM-bl cells were maintained in culture medium (Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum [FBS] plus 100 U/ml penicillin and 100 μg/ml streptomycin [Invitrogen, Carlsbad, CA]). GHOST(3)-CCR5 cells were maintained in culture medium supplemented with 500 μg/ml G418, 1 μg/ml puromycin, and 100 μg/ml hygromycin B (Invitrogen). MT-2 cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin. Fresh peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy volunteer blood donors (Hong Kong Red Cross) using a Ficoll-Hypaque gradient centrifugation method. Total PBMCs were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin 100 μg/ml streptomycin, and 10 U/ml recombinant human interleukin-2 (Roche, Indianapolis, IN) and stimulated with 5 μg/ml phytohemagglutinin (PHA) (Sigma, St. Louis, MO) for 72 h. All cells were cultured in a humidified incubator with a 5% CO2 atmosphere at 37°C.
Site-directed mutagenesis of HIV-1 RT.
All NNRTI mutant constructs were generated based on a molecular clone backbone, pNL4-3Luc+Env−Vpr−(pNL4-3R−E−Luc+). Site-directed mutagenesis (SDM) was performed by using a QuikChange RII XL site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Generation of pseudovirus.
NNRTI-resistant pseudoviruses were generated by cotransfecting (using polyethylenimine [PEI]; Polysciences Inc., Warrington, PA) 293T cells with RT mutants pNL4-3R−E−Luc+ and the monotropic HIV-1ADA envelope. Cell-free supernatant was collected 48 h posttransfection and frozen at −80°C. The 50% tissue culture infective dose (TCID50) was calculated as described previously (35).
Time-of-drug-addition study.
GHOST(3)-CCR5 cells (1 × 104) were infected with 200 TCID50 of HIV-1NL4-3R−E−Luc+ pseudovirus. Compounds were added to parallel cultures every 90 min. The final concentrations of the compounds in the cultures were 1 μM for F18, NVP, zidovudine (AZT), ritonavir (RTV), and raltegravir (RAL) and 10 μM for TAK-779. At 48 h postinfection (PI), cells were washed with phosphate-buffered saline (PBS) and lysed (1× cell culture lysis reagent; Promega, Madison, WI). A 40-μl portion of cell lysate was mixed with 100 μl of luciferase assay reagent (Promega), and luciferase intensity was measured with an Inspire Victor3 multilabel counter, model 1420 (Perkin Elmer, Waltham, MA).
Drug susceptibility assay using RT in lysed viruses.
HIV-1NL4-3 viral particles were concentrated by centrifugation at 20,000 × g at 4°C for 1 h. RT was released by lysis of the viral pellets with PBS containing 2% Triton X-100 on ice for 40 min. Supernatants that contain RT were harvested after another centrifugation at 20,000 × g at 4°C for 15 min. Simian immunodeficiency virus (SIV) Gag RNA was used as the template to generate cDNA by conventional reverse transcription reaction. A total of 105 copies of SIV Gag RNA was mixed with random hexamers (2.5 ng/μl) and dNTPs (0.5 mM), incubated at 65°C for 5 min, and then chilled on ice for 1 min before 1 μl of serially diluted F18 or other drugs (e.g., NVP as a positive control) was added to each reaction mixture containing 5 μl of RT-containing supernatant or 10 U/μl SuperScript III in reaction buffer (containing 1× first-strand buffer, 5 mM dithiothreitol, and 2 U/μl RNaseOUT) in a total volume of 20 μl. The RT reaction was carried out at 25°C for 5 min, 50°C for 1 h, and 70°C for 15 min. The copy numbers of newly synthesized SIV cDNA were subsequently determined by a SIV proviral load assay, as we previously described (8). Briefly, SYBR green-based real-time PCR was performed using FastStart universal SYBR green master (ROX) reaction mix (Roche, Indianapolis, IN) with the SIV-specific primers gag3 (5′-GTAGTATGGGCAGCAAATGAAT-3′) and gag4 (5′-CACCAGATGACGCAGACAGTAT-3′). Amplification was initiated at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s. The susceptibility of RT to F18 was calculated with GraphPad Prism software v5.0 (GraphPad Software Inc., La Jolla, CA). The IC50 is defined as the drug concentration achieving 50% reduction of cDNA copy number.
Antiviral-activity assay.
The activity of each compound against NNRTI-resistant pseudoviruses and F18-induced resistant viruses was tested by measuring luciferase intensity in GHOST(3)-CCR5 cells or TZM-bl cells. Briefly, 1 × 104 cells per well in 100 μl medium were seeded in 96-well plates the day before infection. The next day, 200 TCID50 of virus with serially 3-fold-diluted compounds (in 100 μl medium, ranging from 6 μM to 0.9 nM) was added to the cells. The luciferase intensity was measured 48 h PI.
The activity of each compound against wild-type HIV-1NL4-3 virus, laboratory-adapted HIV strains (resistant to NRTI, NNRTI, and PI), and different subtype isolates was determined in a PBMC-based assay. Briefly, PHA-stimulated PBMCs were infected with 50 TCID50 of virus in the absence of compounds and were incubated at 37°C in a 5% CO2 atmosphere for 24 h, washed, resuspended in fresh medium, and seeded into 96-well plates at 2 × 105 cells per well in 160 μl medium. Serially 3-fold-diluted compounds (in 40 μl medium, ranging from 10 μM to 1.5 nM) were added to the wells. Cells were incubated for 7 days, and a 50% medium change occurred at day 4 while the same concentration of compounds was maintained. The production of virus in the individual well was quantified at day 7 by measuring the level of production of viral core p24 antigen in the culture supernatant with a Retro-Tek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation, Buffalo, NY).
The antiviral activity of each compound was expressed as an EC50 calculated by GraphPad Prism software v5.0.
Selection of HIV-1NL4-3 mutant virus.
F18-induced drug-resistant virus was selected after consecutive rounds of viral passages in the presence of increasing doubling concentrations of F18 (0.14 nM to 20 μM). Briefly, 4 × 105 MT-2 cells were infected with 104 TCID50 of HIV-1NL4-3 wild-type virus in the presence of F18. The course of infection was evaluated by syncytium formation until the HIV-induced cytopathic effect observed was more than 75% (approximately 7 days). Supernatants containing virus were infected with fresh MT-2 cells, and the drug concentration was increased twofold with each successive passage of virus to enhance the selective pressure on the virus. Selected viruses were grown in fresh MT-2 cells in the absence of F18, followed by titration and genotypic analysis. These newly obtained viruses were used for susceptibility testing in an in vitro TZM-bl cell-based assay according to the method described for the antiviral-activity assay above.
Genotypic analysis of mutant HIV-1NL4-3 reverse transcriptase.
Virus RNA was extracted and purified from 1 ml drug-resistant-virus-containing supernatant with a QIAamp viral RNA minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol. For synthesis of cDNA, an RT reaction was carried out with a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen), before amplification of DNA and sequencing of the 1,283-bp pol gene fragment, including the complete protease gene and the first 313 codons of the RT gene, as described previously (23). Two rounds of PCR thermal cycling were performed with the primer pairs ZcrprtF0 (sense, 5′-TTTAGCCTCCCTCAAATCACTCTTT-3′)/ZcrtB4 (antisense, 5′-GCCTCTGTTAATTGTTTTACATCATTAGTGTG-3′) and ZcprtF1 (sense, 5′-CTTCCCTCAGATCACTCTTTGGC-3′)/ZcrtB3 (antisense, 5′-GCTCTTGATAAATTTGGTATGTCCATTG-3′). The remaining 247 codons of the RT gene region were amplified using the primers RT3300F (sense, 5′-GCTGGACTGTCAATGACATAC-3′)/RT4300B (antisense, 5′-GGTTAAAATCACTAGCCATTG-3′). Thermal cycling conditions were 95°C for 2 min followed by 35 cycles of 95°C for 15 s, 55°C for 45 s, and 72°C for 1.5 min, with a last extension step of 72°C for 10 min. Amplified PCR products were purified from agarose gels by using a QIAquick gel extraction kit (Qiagen) before sequenced with an ABI 3730xl DNA analyzer (Life Technologies, Carlsbad, CA). Sequencing results were reported as amino acid changes relative to the HIV-1NL4-3 wild-type virus sequence as determined with BioEdit software (Ibis Biosciences, Carlsbad, CA).
Drug combination studies.
Combinations of F18 with other FDA-approved antiviral agents against HIV-1NL4-3 wild-type virus and other laboratory-adapted drug-resistant isolates were tested in PBMCs. Drugs tested in combination with F18 included the following: the NRTIs AZT, lamivudine (3TC), didanosine (ddI), and stavudine (d4T); the NNRTIs NVP, EFV, ETR; the PI nelfinavir (NFV); and the integrase inhibitor (IN) RAL. PBMCs were infected with wild-type or drug-resistant HIV isolates for 7 days, and a 50% medium changed occurred at day 4 while the same concentration of compounds was maintained. The production of virus was quantified at day 7 using the Retro-Tek HIV-1 p24 antigen ELISA kit. The synergistic effect of F18 with other drugs was determined with MacSynergy II software as previously described (34). In this study, the synergy volume in a synergy plot (95%) was used to define the synergistic, additive, or antagonistic effect of drug-drug interaction (22). A synergy volume between −50 and 50 was defined as an additive effect. Slightly and highly synergistic effects were defined as synergy volumes of 50 to 100 and of >100, respectively. A synergy volume of less than −50 was defined as an antagonistic effect.
Molecular docking of F18 to HIV-1 reverse transcriptase and its mutants.
Molecules of F18 were docked into the NNRTI binding site of wild-type (1VRT), L100I mutant (1S1U), and Y181C mutant (1JLB) HIV-1 RT structures (PDB database) using Autodock 4.2 software (30, 31). The Lamarckian genetic algorithm (LGA) was utilized to search for the conformations using the following docking parameters: a population size of 150 individuals, a maximum number of generations of 27,000, a maximum energy evaluation of 25 million, 50 docking runs, and random initial positions and conformations. The binding energy of each conformation was calculated using the AMBER force field, and a root mean square deviation (RMSD) tolerance of 0.7 Å was used to cluster the conformations using the Autodock Tools software (http://autodock.scripps.edu/). The docking of NVP to HIV-1 RT was used as a positive control for comparison.
RESULTS
Anti-HIV activity of F18.
To determine the antiviral activity of F18, PBMCs isolated from three healthy individuals were infected with live HIV-1NL4-3 (subtype B, X4) wild-type virus in the presence of F18. As controls, the widely used NVP and in-house-synthesized (+)-calanolide A were used in parallel experiments. As shown in Fig. 1B, F18 displayed an EC50 of 52.0 nM (95% confidence interval [CI], 39.7 to 68.3 nM) in the inhibition of HIV-1 replication in infected PBMCs, which is slightly lower than that of (+)-calanolide A (EC50 of 63.0 nM; 95% CI, 38.3 to 103.3 nM). NVP showed an EC50 of 25.0 nM (95% CI, 17.9 to 34.9 nM), which was 2-fold lower than that of F18. When F18 was tested against simian immunodeficiency virus (SIV) infection of PBMCs, no inhibition was observed, suggesting that F18 acts specifically in inhibiting the replication of HIV-1 (data not shown).
Activity of F18 against major subtypes of HIV-1 strains.
Considering the broad genetic diversity of HIV-1, we further investigated the anti-HIV activity of F18 against a panel of clinical isolates of various subtypes. These isolates represent HIV-1 group M subtypes A (TK1135), B (10HKQEH1661), B′ (02HNsmx), and C (93IN109), as well as CRF 01_AE (10HKQEH1447 and 10HKQEH1479). All of these isolates were confirmed by sequence analysis, and no drug-resistant mutations were found in either protease or RT genes (data not shown). PBMCs from at least three healthy individuals were used in independent experiments. Surprisingly, F18 displayed consistent inhibitory activities against HIV-1 isolates of all five subtypes (Fig. 1C), with EC50s of approximately 0.2 μM for subtypes B′ and C and CRF 01_AE and 92.0 nM for subtype B. However, based on the EC50s, F18 was approximately 5-fold more potent against these subtypes than against subtype A and 4-fold less potent against these subtypes than against the HIV-1NL4-3 strain described above. This is likely due to the genetic variation of the HIV RT gene. Nonetheless, F18 shows activity against the replication of HIV-1 in the various isolates tested.
Time-of-drug-addition study of F18.
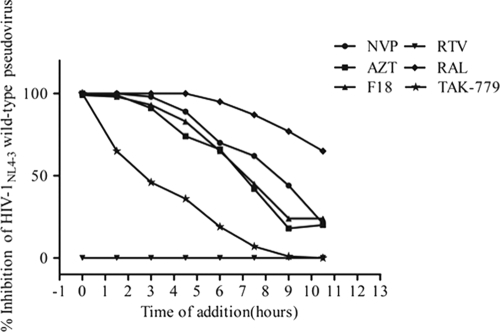
To define the stage in the HIV-1 life cycle at which F18 plays its antiviral role, a time-of-addition study was conducted in GHOST(3)-CCR5 cells infected with an HIV-1NL4-3R−E−Luc+ wild-type pseudovirus. Here, a panel of drugs—NVP (NNRTI), AZT (NRTI), RTV (PI), RAL (IN), and TAK-779 (CCR5 antagonist)—was added to the cells in parallel with F18 every 90 min. Figure 2 shows the timing effect of F18 on its anti-HIV-1 activity compared to those of other ARVs. TAK-779 had a significant loss of antiviral activity when added 1.5 h PI, while RTV, which acts at the end of viral replication, showed no activity, as the expression of luciferase is initiated following viral integration. On the other hand, RAL retained antiviral activity even when added 11 h PI. The significant loss of AZT activity was observed at 4.5 h PI. The antiviral activity profile of F18 was similar to that of the NVP, with an apparent loss of activity at 7 h PI. Based on these results, F18 could very likely act as an HIV-1 RT inhibitor. To further verify this finding, a drug susceptibility assay was performed to determine the anti-RT activity of F18. An IC50 of 12.0 nM for F18 was obtained (data not shown). This finding indicated that F18 is indeed an RT inhibitor, in line with its parent analog (+)-calanolide A (14, 20).
Fig 2.
Antiviral activity of F18 based on time of addition, compared with that of other ARVs. The antiviral activity of F18 was measured at different times following addition of the compound in order to determine the stage in the HIV-1 replication cycle at which F18 inhibits. The ARV inhibitors NVP, AZT, RTV, RAL, and TAK-779 were used in parallel as controls.
In vitro selection of F18-resistant HIV-1.
MT-2 cells were infected with 104 TCID50 of HIV-1NL4-3 wild-type virus and cultured in the presence of increasing twofold concentrations of F18, ranging from 0.14 nM to 20 μM, for 120 days. Virus replication was monitored by observing the syncytium formation. Ten resistant strains were selected, and the genotypic profiles are presented in Table 1. All drug-resistant viruses emerged after 90 days. The breakthrough concentration at which new HIV-1 variants emerged was 625 nM. Genotypic analysis showed that 7 of 10 variants contained a single mutation of Leu to Ile at position 100 in the RT. In addition, mutations of V292I, K103N, Y188H, V106I, T139R, and P225H also emerged in three other strains following F18 treatment. Interestingly, the T139I mutation, which is the dominant mutation induced by (+)-calanolide A (6), was not found even under high selective pressure. Instead, T139R emerged during this selection process as a minor variant.
Table 1.
Selection scheme and genotypic analysis of the reverse transcriptase of mutant HIV-1 strains that emerged under dose-escalating treatment of HIV-1NL4-3 wild-type virus with F18
| Strain | No. of daysa | F18 concnb | Mutation(s) in HIV-1 RTc | EC50 change (fold)d |
|||
|---|---|---|---|---|---|---|---|
| F18 | NVP | EFV | ETR | ||||
| 1, 4, 6, 7, 8, 9, 10 | 0 | 0.14 nM | |||||
| 91 | 625 nM | L100I | |||||
| 128 | 20 μM | L100I | >128 | 1.67 | 2.05 | 1.37 | |
| 2 | 0 | 0.14 nM | |||||
| 91 | 625 nM | Y188H/WT | |||||
| 105 | 2.5 μM | Y188H | |||||
| 128 | 20 μM | Y188H | >128 | 3.75 | 0.004 | 0.34 | |
| 3 | 0 | 0.14 nM | |||||
| 98 | 1.25 μM | T139R/WT | |||||
| 120 | 10 μM | V106I/WT, T139R | |||||
| 128 | 20 μM | V106I/WT, T139R | 121 | 36.5 | 18.06 | 20.15 | |
| 5 | 0 | 0.28 nM | |||||
| 91 | 1.25 μM | P225H | |||||
| 98 | 2.5 μM | K103N/WT, P225H/WT | |||||
| 105 | 5 μM | K103N | |||||
| 112 | 10 μM | K103N, P225H/WT | |||||
| 120 | 20 μM | K103N, P225H/WT, V292I | >128 | >357 | 568.45 | 2.79 | |
Number of days in cell culture.
F18 concentration at which the selection was initiated and the resistant strain was selected.
The complete protease and reverse transcriptase genes were sequenced. The predominant mutation is in bold.
Calculated as the ratio of the EC50 of the compound for the mutant strain to that for the HIV-1NL4-3 wild-type strain obtained in the same experiment at the end of culture. EC50s (means ± standard errors of the means) of F18, NVP, EFV, and ETR against HIV-1NL4-3 wild-type virus were 41.0 ± 2.0 nM, 42.0 ± 1.7 nM, 0.1 ± 0.003 nM, and 0.1 ± 0.002 nM, respectively. Data are the means from a single experiment conducted in triplicate.
In order to confirm the resistance and cross-resistance profiles of these 10 variants, we assessed them in MT-2 cells in the presence of F18, NVP, EFV, and ETR. As shown in Table 1, all mutant viruses were resistant to F18. The mutant viruses with the L100I or Y188H mutation retained high sensitivity to the other three NNRTIs. The K103N mutation resulted in a >350-fold change in EC50 of EFV and NVP, but the mutant virus was sensitive to ETR, whereas the HIV-1 variant with V106I/WT and T139R showed moderate resistance to EFV, NVP, and ETR. Therefore, F18-resistant HIV-1 mutations occurred in vitro in long-term culture of MT-2 cells infected with wild-type virus in the presence of F18, and these mutations conferred some cross-resistance to other NNRTIs.
Activity of F18 against NNRTI-resistant pseudovirus.
To confirm the relevance of these in vitro induced resistant mutations, we established a panel of seven NNRTI-resistant SDM-generated pseudoviruses for a GHOST(3)-CCR5 cell based assay. Mutations that were induced by in vitro F18 selection as described above were engineered into a clean HIV-1NL4-3R−E−Luc+ pseudovirus backbone by SDM. The sensitivities of these mutant viruses were then determined by infection of GHOST(3)-CCR5 cells in the presence of F18, NVP, or (+)-calanolide A, and the degree of inhibition achieved in comparison to infection with a wild-type virus was measured (Fig. 3A). Viruses constructed with the minor Y188H mutation (Table 1) displayed >100-fold-higher resistance to F18 and (+)-calanolide A than NVP. Virus containing the dominant mutation (7 of out 10 strains) L100I induced by F18 in vitro exhibited 30- and 10-fold changes in EC50s of F18 and (+)-calanolide A, respectively. Viruses with the T139I mutation, the dominant mutation induced by (+)-calanolide A (2, 6, 9), displayed a modest change (4-fold) in the EC50 of F18 but remained highly resistant to (+)-calanolide A, with a nearly 50-fold difference in the EC50. NVP susceptibility was noted with L100I and T139I mutant viruses. The novel mutation T139R, found by F18 selection in vitro, resulted in cross-resistance to NVP, F18, and (+)-calanolide A. To a lesser extent, viruses with K101E and K103N mutations also displayed cross-resistance to F18, NVP, and (+)-calanolide A. Interestingly, viruses with the double mutation K103N-P225H exhibited high resistance to NVP, with a >100-fold change in EC50, but resistance to F18 and (+)-calanolide A was abolished, in contrast to viruses with the single mutation K103N. On the whole, we found that F18 could be effective against certain NNRTI-resistant pseudoviruses.
Considering that cross-resistant mutations may exist among various NNRTIs, we sought to investigate further the susceptibilities of NNRTI-resistant HIV-1 carrying 10 additional mutations generated by SDM, either alone or in combination, to F18, NVP and (+)-calanolide A (Fig. 3B). These mutations were previously reported based on the current NNRTIs in clinical use. One of the most prevalent mutations among ART-experienced patients is Y181C, and our experiments found that virus with this mutation was sensitive to F18, with an EC50 of 1.0 nM, and had reduced susceptibility to NVP, with a >100-fold difference in EC50. Interestingly, a pseudoviral variant with both K103N and Y181C was highly resistant to all three NNRTIs tested; however, an additional mutation, G190A, abolished F18 resistance and partially restored sensitivity to (+)-calanolide A (Fig. 3B). In contrast, a single G190A mutation resulted in strong resistance to NVP and partial resistance to F18 and (+)-calanolide A. Moreover, although the pseudovirus containing both K103N and P225H was sensitive to F18 (Fig. 3A), virus with the single mutation P225H was moderately resistant to F18 (Fig. 3B). In addition to the K103N/Y181C mutant, the Y188L and V106A/G190A/F227L mutants were also cross-resistant to the antiviral effects of the three NNRTIs. These experiments furthered our understanding of the mutations in the RT gene underlying resistance to F18 treatment.
Activity of F18 against laboratory-adapted drug-resistant HIV-1 isolates.
To determine the drug resistance profile of F18, drug-resistant strains of laboratory-adapted HIV-1 isolates were used to infect PBMCs in the presence of F18. These drug-resistant viruses include the following: strains resistant to NRTIs, AZT-resistant 7324-1 (M41L/D67N/T69N/K70R/T215F/K219Q) and 3TC-resistant HIV-1LAI-M184V (M184V); pyridinone (NNRTI)-resistant HIV-1IIIB A17 (K103N/Y181C); saquinavir (PI)-resistant strain (G48V/L90M); and a multiresistant virus containing NRTI and NNRTI resistance mutations (Δ67/T69G/K70R/L74I/K103N/T215F/K219Q). Figure 3C shows the changes in EC50 between mutants and wild-type cultured with NNRTIs. The activity of F18 against these isolates was similar to that of (+)-calanolide A. To our surprise, the pyridinone-resistant virus (K103N/Y181C) showed an F18 EC50 of 133.0 nM (95% CI, 82.6 to 214.0 nM), which contradicted the results of the pseudovirus assay presented in Fig. 3B. Similarly, all NRTI- and PI-resistant viruses retained sensitivity to F18, with less than an 8-fold change in EC50s. However, cross-resistance to F18, NVP, and (+)-calanolide A was observed in the multiresistant virus, with changes in EC50 of approximately 50-, 250-, and 80-fold, respectively. In summary, resistance to NVP, F18, and (+)-calanolide A was investigated in a panel of HIV-1 pseudoviruses, viruses with SDM mutations, and drug-resistant isolates, and it appears that sensitivity to F18 occurred in a majority of HIV-1 genotypes.
Antiviral activity of F18 in combination with some FDA-approved ARVs.
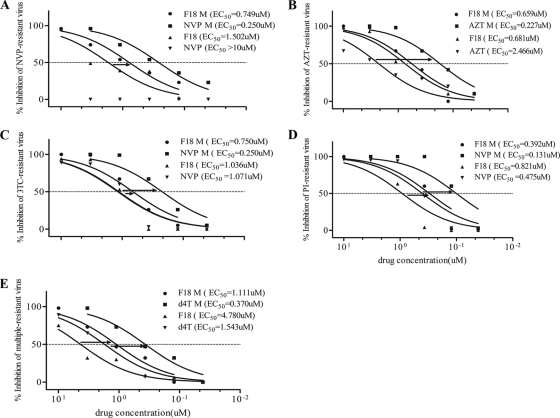
Given the unique antiviral features of F18, we next sought to determine its efficacy in combination with each of eight FDA-approved drugs, by using PBMC assays. The results (Table 2) demonstrated that there were no antagonistic effects in any combination of F18 plus another drug against HIV-1NL4-3 in infected PBMCs. Most combinations showed a slightly or highly synergistic effect, as calculated by the Prichard and Shipman MacSynergy II software. When we tested combinations of F18 and other ARVs against drug-resistant viruses, no antagonistic effects were observed (Table 3). On the contrary, we found highly synergistic interactions between F18 and NVP, AZT, 3TC, ddI, and d4T. For example, for NVP-resistant virus (Y181C), the combination of F18 with NVP resulted in a 1.5-fold dose reduction for F18 and a >38-fold reduction for NVP relative to the individual EC50s (Fig. 4A). The combination of F18 with AZT led to a 1.3-fold dose reduction for F18 and a >13-fold reduction for AZT against AZT-resistant virus (M41L/D67N/T69N/K70R/T215F/K219Q) (Fig. 4B). A similar trend was also observed for 3TC-resistant virus (M184V), where the combination of F18 with NVP resulted in a 1.7-fold reduction in the F18 dose and a 5-fold decrease in the NVP dose (Fig. 4C). A potent synergistic effect was also observed when F18 was used together with NVP or d4T against a PI-resistant virus (G48V/L90M) or multiresistant virus (Δ67/T69G/K70R/L74I/K103N/T215F/K219Q), with 4-fold, 4-fold, and 5-fold reductions in EC50s of F18, NVP, and d4T, respectively (Fig. 4D and E). Taken together, using F18 in conjunction with other ARVs against HIV-1 did not give rise to any antagonistic effect but did result in slight to high synergy, which resulted in a reduction of the dose required to achieve the antiviral effects.
Fig 4.
Synergistic effect of combinations of F18 with ARVs in inhibition of drug-resistant HIV-1 infection in PBMCs. The concentration ratio between F18 and other ARVs was 3:1. (A) F18 plus NVP against NVP-resistant virus; (B) F18 plus AZT against AZT-resistant virus; (C) F18 plus NVP against 3TC-resistant virus; (D) F18 plus NVP against PI-resistant virus; (E) F18 plus d4T against multiresistant virus. M refers to drug combinations.
Docking of NVP and F18 to the HIV-1 reverse transcriptase.
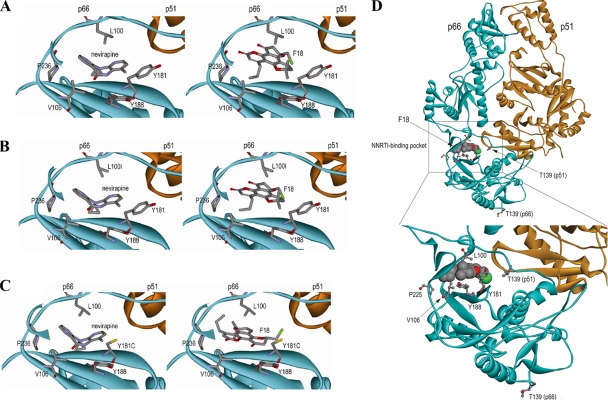
Several lines of biological evidence indicated that NVP and F18 might bind to a distinct binding motif on HIV-1 RT to block RT activity. For this reason, we sought to understand the molecular interactions between the small molecules and HIV-1 RT in a macrostructural setting using Autodock software. The docking of NVP (control) and F18 to the structures of wild-type (1VRT), L100I mutant (1S1U), and Y181C mutant (1JLB) HIV-1 RT proteins was examined. Fifty conformations of each molecule were calculated based on the binding energy and clustered within 0.7 RMSD. In three different HIV RT structures, over 95% of the F18 and NVP conformations were clustered together, suggesting that the orientation of these small molecules results in a highly stable and low-energy state at the NNRTI binding pocket of HIV-1 RT. As shown in Fig. 5A and B, the conformations of NVP and F18 are the same in binding to wild-type and L100I mutant. The aromatic residue L100 appeared to be more essential for the interaction with F18 ring structures than with those of NVP. Moreover, both Y181 and Y188 stabilized the ring structures of the two compounds in the binding site. Critically, with the structure of the Y181C mutant, the orientation of F18 for binding to HIV RT is completely different from that for binding to the wild type and the L100I mutant (Fig. 5C). The chloro group of F18 is orientated close to cysteine 181 for a tighter interaction, although the aromatic moiety fits in the same NNRTI binding pocket. Inspection of the structure at the point of interaction between F18 and wild-type RT (Fig. 5D) reveals that the orientation of T139 in the p51 subunit places it in close proximity to the NNRTI binding site, suggesting that this residue probably contributes to the interaction with F18.
Fig 5.
Binding of F18 to HIV-1 RT in conformational space. Using computer software, structural binding of F18 or NVP to the HIV-1 RT binding pocket was determined for wild-type (A), the L100I mutant (B), and the Y181C mutant (C). (D) Interaction between the T139 residue and F18. Amino acid residues important for binding interactions are labeled. Indicated elements: carbon (gray), oxygen (red), nitrogen (purple), sulfur (yellow), F18 chloro group (green).
DISCUSSION
This study comprehensively characterized a novel NNRTI, F18, for its range of antiviral activity, drug resistance, and underlying mechanism of action against the prominent HIV-1 RT. We report new findings regarding this new agent. (i) F18 demonstrated consistent specific anti-HIV-1 activity in PBMC assays against a panel of primary HIV-1 isolates derived from subtypes A, B, B′, and C and CRF 01_AE. (ii) F18 successfully inhibited several drug-resistant pseudotype viruses and laboratory-adapted viruses. In particular, F18 has excellent potency against the clinically prevalent Y181C-containing NNRTI-resistant virus (EC50 = 1.0 nM), with activity superior to that of other NNRTIs. (iii) Instead of the (+)-calanolide A-resistant mutation T139I, L100I appeared to be the dominant F18-induced resistant mutation. Since both NVP and EFV are effective against L100I mutant virus, their mechanism of action may differ from that of F18. (iv) Results from two-drug-combination studies strongly suggested that F18 is not antagonistic with any ARVs tested against either wild-type or drug-resistant viruses. The strong synergistic effects of F18 with NVP, AZT, ddI, and 3TC against corresponding drug-resistant viruses suggest that it may be an attractive new choice of NNTRI for novel ART drug regimens. (v) Lastly, protein docking analysis suggested that F18 likely acts to block HIV-1 RT in two different ways depending on the distinct binding moieties of wild-type and drug-resistant viruses. These new findings not only provide strong implications for the clinical development of F18 but also present a new structural-based direction for the design and development of future NNRTIs.
Although there are nearly 30 ARVs and regimens routinely used in developed countries, only a limited number of drugs are available in developing countries (13, 16, 27). To reduce mortality in HIV-infected individuals, clinical efforts have been made to provide ART to AIDS patients in all parts of the world (16, 45). Moreover, ART can act as a preventive measure against HIV-1 in developing countries to reduce the incidence rate (12, 28). Therefore, affordable ART regimens to combat HIV-1 infection, in addition to other preventive measures, remain highly desirable, especially in developing countries, such as China (7, 45). F18 appears to be a cost-effective candidate: good oral bioavailability and no adverse effects were detected in animal testing, and it is synthesized in vitro from a plant by using medicinal chemistry techniques, which is feasible for large-scale production for clinical application (44). To this end, understanding the complete anti-HIV profile of F18 is a necessity for use in patients. In this study, F18 was tested against HIV-1 primary isolates of subtype A, B, B′, and C and CRF 01_AE, which are predominantly prevalent in China and surrounding countries (40, 46, 47). We found that F18 displayed antiviral activity comparable to that of existing ARVs against several regional clinic isolates, including primary strains of subtypes B and B′ and CRF 01_AE, which were isolated from patients of Henan and Hong Kong, China, while against subtype A, which is prevalent in West and Central Africa, F18 caused a fivefold decrease in the level of viral replication. Further investigation is warranted to test F18 against more primary isolates from different regions of the globe to understand its potential efficacy worldwide.
One of the major obstacles for the development of NNRTIs is the extensive cross-resistance within this class of ARVs. However, from our experiments, we demonstrated that F18 displayed a unique profile of cross-resistance. In striking contrast to NVP, F18 remains effective against HIV-1 containing the SDM-generated NNTRI-resistant mutations T139I, Y181C, V179D, V106A, K103N/P225H, and K103N/Y181C/G190A. Moreover, although K103N/Y181C conferred a high degree of resistance to F18 or to (+)-calanolide A in the pseudoviral assay (Fig. 3B), the live A17 strain, which contains the same K103N/Y181C mutations, was found to be highly sensitive to F18 and to (+)-calanolide A (Fig. 3C). This result is consistent with previous findings that (+)-calanolide A inhibits A17 effectively (20, 32). By comparing sequence similarity between the K103N/Y181C-containing pseudovirus and A17, we found nine additional amino acid differences (K102, E122, S162, L214, P272, R277, I292, V295, and I310) besides K103N and Y181C in A17. It is possible that one or more of these amino acid variations rendered A17 sensitive to F18, as was shown by introducing the G190A mutation into another NNRTI-resistant pseudoviral variant already containing the K103N/Y181C mutations (Fig. 3B). It would be of interest to investigate these amino acid polymorphisms and how they contribute to a change in sensitivity to F18 and other NNRTIs. From our F18 resistance induction study, we found that F18 did not readily induce NVP-resistant nor (+)-calanolide A-resistant viruses, but the resultant dominant L100I mutation is attributed to F18 resistance, which is similar to another (+)-calanolide A stereoisomer, dihydrocostatolide (6). This finding suggested that F18 may function more similarly to dihydrocostatolide than to (+)-calanolide A. In addition, due to structural similarity, F18 and (+)-calanolide A have a similar profile against most NNRTI-cross-resistance viruses (Fig. 3A and B). These two drugs, however, also display unique characteristics. For example, F18 was a potent inhibitor against a (+)-calanolide A-induced T139I mutant. Conversely, (+)-calanolide A inhibited an F18-induced L100I mutant more effectively. These results suggested that the major binding moiety of F18 engages HIV-1 RT at a different binding motif relative to L100, compared to the T139 usage of (+)-calanolide A. In addition, most of the resistant viruses, including those with the L100I mutation selected by F18, can be inhibited by NVP, EFV, and ETR, indicating a low level of cross-resistance between F18 and these three FDA-approved NNRTIs (Table 1). Thus, we conclude that the drug resistance profile of F18 is distinctly different from that of other NNRTIs (3, 36, 42), possibly due to a distinct motif of F18 binding to HIV-1 RT which is not identical to that of other NNRTIs.
Upon in silico analysis of the possible binding moieties of F18 to HIV-1 RT, we found that F18 could block HIV-1 RT in two distinct ways in wild-type and drug-resistant viruses. Structural differences between F18 and NVP lead to the difference in resistance to them, based on the wild-type HIV-RT binding site (Fig. 5A, B, and C). F18 has a rigid structure and restricted binding compared to NVP, which has a natural bend and fits well in the binding cleft. Thus, NVP could bind to the important amino acid residue Y188, which allows aromatic interaction with the compounds and renders wild-type virus more sensitive to NVP than to F18. However, the mutation Y188H alters the binding platform, making it more difficult for both NVP and F18 to bind to. Indeed, our cell assays confirmed this and showed that strains with Y188H have high resistance to NVP and F18. As reported previously, this mutation abrogates aromatic ring interactions between NNTRI and the RT binding pocket, leading to curtailed binding affinity (37). With another mutation, L100I, the shape of the binding pocket is altered and there is less hydrophobic interaction with the aromatic moiety of F18. The binding of F18 to L100I RT is less than that to wild-type HIV-1 RT, and therefore, L100I confers moderate resistance to F18. On the other hand, Y181 interacts with the side ring structure of NVP, and a change of tyrosine to cysteine at this location abolished binding of NVP to RT due to a lack of aromatic interactions. However, this is favorable for F18, as the lack of bulky aromatic tyrosine in the RT binding site permits more spatial flexibility for F18 to rotate and fit into the binding pocket; thus, increased antiviral activity was observed (Fig. 5C). This improvement is likely due to the enhanced binding affinity between the chlorine residue of F18 and the RT residue 181C through the formation of a new hydrogen bond. This mode of binding is completely different from the interaction of (+)-calanolide A with HIV-1 RT (43). This pocket, however, is rather fragile, because an additional K103N mutation easily rendered Y181C-containing HIV-1 highly resistant to F18, probably by interfering with the entry of F18 into the RT binding pocket (17). Interestingly, when a G190A mutation was introduced into K103N/Y181C-containg HIV-1, the virus became sensitive to F18 again (Fig. 3B); thus, the effects of these mutations in the quaternary space of ARV binding to RT remain to be elucidated. Computational analysis of the motifs of binding of NVP and F18 to the HIV-1 wild-type and drug-resistant mutated RT correlates with the cell-based results obtained in this study. However, the real picture of drug binding mechanism would require future crystal structural analysis.
Our analysis of possible drug-RT interaction shows that like to other NNRTIs, F18 could bind to the common pocket of the p66 subunit (Fig. 5A, B, and C), whereas the interaction with the p51 subunit was primarily via the β7-β8 loop, similar to (+)-calanolide A (Fig. 5D) (2). The β7-β8 loop (residues 132 to 140) of the p51 but not the p66 subunit is located at the RT heterodimer interface and contributes to the formation and stability of the NNRTI binding pocket (NNRTI) (18). For instance, residues I132, I135, N136, E138, and T139 in the p51 subunit contributed specifically to NNRTI resistance (2, 5, 39). Moreover, previous data generated with the recombinant subunit-specific RT systems suggested that residue 139 in the p66 subunit is not involved in resistance to the F18 analogue (+)-calanolide A (2, 6, 9). In our study, the experimental finding of an F18-induced T139R mutation has, therefore, provided direct evidence of the involvement of the p51 subunit in interaction with F18 (Table 1). To confirm this finding, we also generated T139R-containing virus and demonstrated that this mutation indeed conferred resistance to F18 (Fig. 3A). Guided by our mutagenesis data, further evidence from our docking analysis illustrates that F18 binds to the HIV-1 RT p51 subunit T139 residue with low energy (Fig. 5D). We therefore speculate that the p51 subunit may play a role in affecting the antiviral activities of F18.
Since the discovery of NNRTIs, combination therapy with at least three ARVs has become the gold standard for clinical treatment of HIV-1 infection in the last 20 years. To avoid issues of drug-drug interaction, we tested the combined anti-HIV activity of F18 with each of eight commonly used ARVs. As a natural-product-derived small molecule, F18 had no antagonistic effect when used in two-drug combinations against both wild-type and drug-resistant viruses (Tables 2 and 3). Interestingly, the lack of antagonistic effects between F18 and either NVP or EFV provided further evidence that the motif of F18 binding to HIV-1 RT is not identical to that of either NNRTI. On the other hand, we observed that F18 had synergistic effects with NVP against NVP-resistant (Y181C) virus (Fig. 4A). The combination of F18 and NVP reduced the dose required for NVP, which could potentially reduce NVP-related in vivo toxicity. The underlying mechanism for this finding remains elusive and requires future investigation. One possibility is that binding of F18 to HIV-1 RT may trigger a conformational change in the binding site favorable for NVP interaction. Another possibility is that the high affinity of F18 for Y181C (Fig. 5C) could result in dominant binding, leading to a better fit of NVP with HIV-1 RT to exert its antiviral activity, and thereby leading to the highly synergistic effect we observed with F18 against NVP-resistant virus. Also, F18 could act similarly to (+)-calanolide A by competing with dNTPs for binding to the active site of HIV-1 RT, which is a unique, yet-to-be-confirmed mechanism compared to mechanisms employed by conventional NNRTIs (10). In addition, we found that F18 acts synergistically with NVP, AZT, and d4T against 3TC-resistant, AZT-resistant, PI-resistant, and multiresistant viruses (Fig. 4B, C, D, and E). Taken together, these unique features of F18 and its synergy with other ARVs establish it as an attractive ARV candidate for clinical regimens for treating HIV-1-infected patients.
We present a new NNRTI agent, F18, as a promising drug derived from a natural product for clinical development and use in combination with existing regimens for patients infected with HIV-1 to thwart progression to AIDS. Although we report in this study substantial data regarding its potential effectiveness, further basic and clinical investigation of its mode of action, efficacy, and adverse effects, as well as potential new derivatives, would provide a stronger case for its application in treating HIV-1 patients.
ACKNOWLEDGMENTS
We thank Allen K. L. Cheung and Jenny Ng for their editorial assistance.
This work was supported by Hong Kong Research Fund for the Control of Infectious Diseases (RFCID09080772 to ZC) and the China's 11th Five-Year Mega Project on the prevention and treatment of AIDS, viral hepatitis and other infectious disease (2009ZX09501-012 to GL). We also thank the HKU-UDF and LSK Faculty of Medicine Matching Fund for financial supports to HKU AIDS Institute.
We declare no financial or commercial conflict of interest.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Andries K, et al. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Auwerx J, et al. 2005. The role of Thr139 in the human immunodeficiency virus type 1 reverse transcriptase sensitivity to (+)-calanolide A. Mol. Pharmacol. 68:652–659 [DOI] [PubMed] [Google Scholar]
- 3. Azijn H, et al. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bacheler L, et al. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balzarini J, et al. 2005. The amino acid Asn136 in HIV-1 reverse transcriptase (RT) maintains efficient association of both RT subunits and enables the rational design of novel RT inhibitors. Mol. Pharmacol. 68:49–60 [DOI] [PubMed] [Google Scholar]
- 6. Buckheit RW, Jr., et al. 1999. Unique anti-human immunodeficiency virus activities of the nonnucleoside reverse transcriptase inhibitors calanolide A, costatolide, and dihydrocostatolide. Antimicrob. Agents Chemother. 43:1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao YZ, Lu HZ. 2005. Care of HIV-infected patients in China. Cell Res. 15:883–890 [DOI] [PubMed] [Google Scholar]
- 8. Chen Z, et al. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J. Virol. 74:6501–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Currens MJ, et al. 1996. Antiviral activity and mechanism of action of calanolide A against the human immunodeficiency virus type-1. J. Pharmacol. Exp. Ther. 279:645–651 [PubMed] [Google Scholar]
- 10. Currens MJ, Mariner JM, McMahon JB, Boyd MR. 1996. Kinetic analysis of inhibition of human immunodeficiency virus type-1 reverse transcriptase by calanolide A. J. Pharmacol. Exp. Ther. 279:652–661 [PubMed] [Google Scholar]
- 11. Delaugerre C, et al. 2001. Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. J. Med. Virol. 65:445–448 [PubMed] [Google Scholar]
- 12. Edmonds A, et al. 2011. The effect of highly active antiretroviral therapy on the survival of HIV-infected children in a resource-deprived setting: a cohort study. PLoS Med. 8:e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Safadi Y, Vivet-Boudou V, Marquet R. 2007. HIV-1 reverse transcriptase inhibitors. Appl. Microbiol. Biotechnol. 75:723–737 [DOI] [PubMed] [Google Scholar]
- 14. Flavin MT, et al. 1996. Synthesis, chromatographic resolution, and anti-human immunodeficiency virus activity of (±)-calanolide A and its enantiomers. J. Med. Chem. 39:1303–1313 [DOI] [PubMed] [Google Scholar]
- 15. Herschhorn A, Hizi A. 2010. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 67:2717–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho DD, Bieniasz PD. 2008. HIV-1 at 25. Cell 133:561–565 [DOI] [PubMed] [Google Scholar]
- 17. Hsiou Y, et al. 2001. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 309:437–445 [DOI] [PubMed] [Google Scholar]
- 18. Hsiou Y, et al. 1996. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4:853–860 [DOI] [PubMed] [Google Scholar]
- 19. Hughes CA, Robinson L, Tseng A, MacArthur RD. 2009. New antiretroviral drugs: a review of the efficacy, safety, pharmacokinetics, and resistance profile of tipranavir, darunavir, etravirine, rilpivirine, maraviroc, and raltegravir. Expert Opin. Pharmacother. 10:2445–2466 [DOI] [PubMed] [Google Scholar]
- 20. Kashman Y, Gustafson KR, Fuller RW, Cardellina JH, II, et al. 1992. The calanolides, a novel HIV-inhibitory class of coumarin derivatives from the tropical rainforest tree, Calophyllum lanigerum. J. Med. Chem. 35:2735–2743 [DOI] [PubMed] [Google Scholar]
- 21. Katlama C, et al. 2010. Efficacy and safety of etravirine at week 96 in treatment-experienced HIV type-1-infected patients in the DUET-1 and DUET-2 trials. Antivir. Ther. 15:1045–1052 [DOI] [PubMed] [Google Scholar]
- 22. Lai MT, et al. 2009. Antiviral activity of MK-4965, a novel nonnucleoside reverse transcriptase inhibitor. Antimicrob. Agents Chemother. 53:2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo M, et al. 2009. Prevalence of drug-resistant HIV-1 in rural areas of Hubei province in the People's Republic of China. J. Acquir. Immune Defic. Syndr. 50:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma T, Gao Q, Chen Z, Wang L, Liu G. 2008. Chemical resolution of (±)-calanolide A, (±)-cordatolide A and their 11-demethyl analogues. Bioorg. Med. Chem. Lett. 18:1079–1083 [DOI] [PubMed] [Google Scholar]
- 25. Ma T, et al. 2008. Chemical library and structure-activity relationships of 11-demethyl-12-oxo calanolide A analogues as anti-HIV-1 agents. J. Med. Chem. 51:1432–1446 [DOI] [PubMed] [Google Scholar]
- 26. Madruga JV, et al. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:29–38 [DOI] [PubMed] [Google Scholar]
- 27. Menendez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124–146 [DOI] [PubMed] [Google Scholar]
- 28. Miiro G, et al. 2009. Reduced morbidity and mortality in the first year after initiating highly active anti-retroviral therapy (HAART) among Ugandan adults. Trop. Med. Int. Health 14:556–563 [DOI] [PubMed] [Google Scholar]
- 29. Milinkovic A, Martinez E. 2004. Nevirapine in the treatment of HIV. Expert Rev. Anti-Infect. Ther. 2:367–373 [DOI] [PubMed] [Google Scholar]
- 30. Morris GM, et al. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639–1662 [Google Scholar]
- 31. Morris GM, et al. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nunberg JH, et al. 1991. Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J. Virol. 65:4887–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pomerantz RJ, Horn DL. 2003. Twenty years of therapy for HIV-1 infection. Nat. Med. 9:867–873 [DOI] [PubMed] [Google Scholar]
- 34. Prichard MN, Shipman C., Jr 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181–205 [DOI] [PubMed] [Google Scholar]
- 35. Richards KH, Clapham PR. 2006. Human immunodeficiency viruses: propagation, quantification, and storage. Curr. Protoc. Microbiol. Chapter 15, Unit 15J.1 [DOI] [PubMed] [Google Scholar]
- 36. Richman DD, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarafianos SG, et al. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schiller DS, Youssef-Bessler M. 2009. Etravirine: a second-generation nonnucleoside reverse transcriptase inhibitor (NNRTI) active against NNRTI-resistant strains of HIV. Clin. Ther. 31:692–704 [DOI] [PubMed] [Google Scholar]
- 39. Sluis-Cremer N, et al. 2007. Characterization of novel non-nucleoside reverse transcriptase (RT) inhibitor resistance mutations at residues 132 and 135 in the 51 kDa subunit of HIV-1 RT. Biochem. J. 404:151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su B, et al. 2003. HIV-1 subtype B′ dictates the AIDS epidemic among paid blood donors in the Henan and Hubei provinces of China. AIDS 17:2515–2520 [DOI] [PubMed] [Google Scholar]
- 41. Tambuyzer L, et al. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 14:103–109 [PubMed] [Google Scholar]
- 42. Vingerhoets J, et al. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu ZQ, Flavin MT, Jenta TR. 2000. Calanolides, the naturally occurring anti-HIV agents. Curr. Opin. Drug Discov. Dev. 3:155–166 [PubMed] [Google Scholar]
- 44. Xue H, et al. 2010. Highly suppressing wild-type HIV-1 and Y181C mutant HIV-1 strains by 10-chloromethyl-11-demethyl-12-oxo-calanolide A with druggable profile. J. Med. Chem. 53:1397–1401 [DOI] [PubMed] [Google Scholar]
- 45. Zhang FJ, Pan J, Yu L, Wen Y, Zhao Y. 2005. Current progress of China's free ART program. Cell Res. 15:877–882 [DOI] [PubMed] [Google Scholar]
- 46. Zhang L, et al. 2004. Molecular characterization of human immunodeficiency virus type 1 and hepatitis C virus in paid blood donors and injection drug users in china. J. Virol. 78:13591–13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y, et al. 2006. Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med. 3:e443. [DOI] [PMC free article] [PubMed] [Google Scholar]