Abstract
Little information is available about pediatric infections caused by extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli. We characterized an outbreak caused by a CTX-M-14-producing E. coli isolate in a neonatal intensive care unit (NICU) and studied other infections caused by ESBL-producing E. coli in non-NICU pediatric units. All children ≤4 years old who were infected or colonized by ESBL-producing E. coli isolates between January 2009 and September 2010 were included. Molecular epidemiology was studied by phylogroup analysis, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing. Antibiotic resistance genes were analyzed by PCR and sequencing. Plasmids were studied by PFGE with S1 nuclease digestion and by incompatibility group analysis using a PCR-based replicon-typing scheme. Of the ESBL-producing E. coli isolates colonizing or infecting the 30 newborns, identical PFGE results were observed for 21 (70%) isolates, which were classified as CTX-M-14-producing E. coli of ST23 phylogroup A. blaCTX-M-14a was linked to ISEcp1 and was carried on an ∼80-bp IncK plasmid. A smaller ongoing outbreak due to SHV-12-producing ST131 E. coli was also identified in the same NICU. Fifteen additional infections with ESBL-producing E. coli were identified in non-NICU pediatric units, but none was caused by the CTX-M-14-producing E. coli epidemic clone. Overall, CTX-M-14 (71.1%), CTX-M-15 (13.3%), and SHV-12 (13.3%) were the most important ESBLs causing pediatric infections in this study. Infections of newborns with CTX-M-14-producing E. coli were caused by both clonal and nonclonal isolates.
INTRODUCTION
Most countries have experienced rapid dissemination of extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae isolates, particularly Escherichia coli and Klebsiella pneumoniae isolates. This increase is due mainly to the dissemination of the CTX-M type, particularly CTX-M-14 and CTX-M-15 ESBLs (12).
The spread of ESBLs is frequently due to the dissemination of mobile genetic elements among a genetically diverse E. coli population. Although a community outbreak of clonally related CTX-M-14-producing E. coli in Canada has been reported (13), the dissemination of CTX-M-14 in E. coli is usually nonclonal (11).
In neonatal intensive care units (NICUs), few cases of clonal spread of ESBL-producing E. coli have been reported (8, 16), and thus far, significant nosocomial outbreaks among newborns due to the clonal dissemination of CTX-M-14-producing E. coli have not been described.
The aims of this study were (i) to characterize an outbreak caused by a CTX-M-14-producing E. coli strain in an NICU and (ii) to study other infections due to ESBL-producing E. coli in pediatric units other than the NICU.
MATERIALS AND METHODS
Study design and bacterial isolates.
During 2010, an unexpected increase in the frequency of isolation of ESBL-producing E. coli was observed in the NICU of the University Hospital Gregorio Marañón (UHGM), Madrid, Spain. This observation prompted the present investigation. All children admitted to this NICU, either infected or colonized by ESBL-producing E. coli isolates between January 2009 and September 2010, were included in the study. Additionally, all cases of infection due to ESBL-producing E. coli occurring in children ≤4 years old who were admitted to other, nonneonatal pediatric units during the study period were also studied.
Antibiotic susceptibility testing and ESBL detection were performed by using the MicroScan microdilution (Siemens Healthcare Diagnostics, Deerfield, IL) and Etest (AB Biodisk, Solna, Sweden) methods according to the Clinical and Laboratory Standards Institute guidelines (4). Antibiotic-susceptible E. coli ATCC 25922 and CTX-M-15-producing E. coli 222 (10) were used as quality control strains.
Molecular epidemiology.
The genetic relationships between the ESBL-producing E. coli isolates were determined by pulsed-field gel electrophoresis (PFGE) after digestion of total chromosomal DNA with XbaI (10).
E. coli isolates representing the different clusters detected by PFGE were studied further by multilocus sequence typing (MLST) according to the University College Cork (Cork, Ireland) scheme for E. coli (http://mlst.ucc.ie/mlst/dbs/Ecoli [date last accessed, 20 May 2011]).
E. coli phylogenetic groups were determined by a multiplex PCR assay as described previously (2).
To search for the ST131 phylogroup B2 serotype O25b E. coli clone, O25b type detection was performed using allele-specific PCR (3).
Characterization of antibiotic resistance genes.
Multiplex PCR conditions were used to amplify genes encoding ESBLs belonging to the CTX-M-1, CTX-M-9, SHV, and TEM groups. Entire ESBL genes were amplified and sequenced with primers specific for blaSHV and blaCTX-M (10). In addition, the aac(3)-IIa aminoglycoside resistance gene and the aac(6′)-Ib-cr aminoglycoside-quinolone resistance gene were characterized by PCR amplification with specific primers and DNA sequencing.
Furthermore, the genetic environment of blaCTX-M-14a was analyzed using specific primers to detect the linkage of blaCTX-M-14 alleles with ISEcp1-like or IS903-like elements (6).
Conjugation assay and plasmid characterization.
Conjugation experiments were performed using the kanamycin-azide-resistant E. coli strain BM101 as a recipient. Putative transconjugants were selected on Mueller-Hinton agar plates containing kanamycin (100 mg/liter) and cefotaxime (4 mg/liter).
The number and size of plasmids were determined by PFGE after S1 nuclease digestion of whole genomic DNA (7). Plasmids were classified according to their incompatibility groups by using a PCR-based replicon-typing scheme (1).
RESULTS AND DISCUSSION
Patients and bacterial isolates.
Between January 2009 and September 2010, 30 newborns admitted to the NICU were either infected (n = 14 [46.7%]) or colonized (n = 16 [53.3%]) by ESBL-producing E. coli. Their clinical diagnostics were as follows: blood infections, 6 cases (42.9%); wound infections, 3 cases (21.4%); urinary tract infections (UTIs), 2 cases (14.3%); respiratory tract infections, 2 cases (14.3%); and ear infection, 1 case (7.1%). For the 16 colonized children, ESBL-producing E. coli was isolated mainly from rectal swabs (n = 14 [87.5%]).
The NICU has a total of 66 beds, with average occupancy rates of 83.64% and 79.7% in 2009 and 2010, respectively. During the period of study, 3,114 newborns were admitted to this unit: 1,762 newborns in 2009 and 1,352 in 2010. The prevalence of ESBL-producing E. coli infection in the NICU was 0.3% (5 cases) in 2009 and 1.8% (25 cases) in 2010, a 6-fold increase. The highest prevalence was detected in August 2010 (6 cases out of 139 new admissions [4.3%]).
During the study period, ESBL-producing E. coli caused infections in 15 children ≤4 years old who were admitted to non-NICU pediatric units: 12 (80%) in the Emergency Unit and 3 (20%) in the Nephrology Unit. Most of these isolates (13 [86.7%]) caused UTIs.
ESBL types and molecular epidemiology of ESBL-producing E. coli from the NICU.
Of the 30 ESBL-producing E. coli specimens isolated from newborns, 25 (83.3%), 3 (10%), and 2 (6.7%) were identified as producing CTX-M-14, SHV-12, and CTX-M-15, respectively. All CTX-M-14-producers had the blaCTX-M-14a gene.
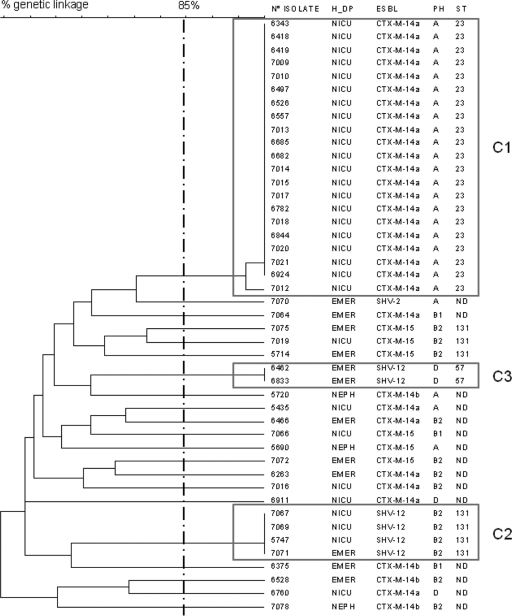
PFGE profiles consistently revealed two different clusters (Fig. 1). Cluster 1 (C1) included 21 indistinguishable CTX-M-14-producing isolates, and cluster 2 (C2) included 3 SHV-12-producing E. coli isolates. The remaining 6 isolates (4 with CTX-M-14 and 2 with CTX-M-15) were nonrelated according to PFGE (Fig. 1; Table 1).
Fig 1.
Dendrogram illustrating the genetic relationships among 45 ESBL-producing Escherichia coli isolates obtained from children ≤4 years old. H_DP, hospital department; PH, phylogroup; ST, sequence type by MLST; EMER, Emergency Unit; NEPH, Nephrology Unit; ND, not determined.
Table 1.
ESBL types and molecular epidemiology markers of 45 ESBL-producing Escherichia coli isolates obtained from children ≤4 years old
| Pediatric unita | No. of cases | PFGE clusterb | MLST typec | Phylogroup | ESBL |
|---|---|---|---|---|---|
| NICU | 21 | C1 | 23 | A | CTX-M-14a |
| NICU | 3 | C2 | 131 | B2 | SHV-12 |
| NICU | 1 | NR | ND | D | CTX-M-14a |
| NICU | 1 | NR | ND | D | CTX-M-14a |
| NICU | 1 | NR | ND* | B2 | CTX-M-14a |
| NICU | 1 | NR | ND | A | CTX-M-14a |
| NICU | 1 | NR | 131 | B2 | CTX-M-15 |
| NICU | 1 | NR | ND | B1 | CTX-M-15 |
| EMER | 2 | C3 | 57 | D | SHV-12 |
| EMER | 1 | C2 | 131 | B2 | SHV-12 |
| EMER | 1 | NR | ND* | B2 | CTX-M-14a |
| EMER | 1 | NR | ND* | B2 | CTX-M-14a |
| EMER | 1 | NR | ND* | B2 | CTX-M-14b |
| NEPH | 1 | NR | ND* | B2 | CTX-M-14b |
| NEPH | 1 | NR | ND | A | CTX-M-14b |
| EMER | 1 | NR | ND | B1 | CTX-M-14b |
| EMER | 1 | NR | ND | B1 | CTX-M-14a |
| EMER | 1 | NR | 131 | B2 | CTX-M-15 |
| EMER | 1 | NR | 131 | B2 | CTX-M-15 |
| EMER | 1 | NR | ND* | B2 | CTX-M-15 |
| NEPH | 1 | NR | ND | A | CTX-M-15 |
| EMER | 1 | NR | ND | A | SHV-2 |
NICU, neonatal intensive care unit; EMER, Emergency Unit; NEPH, Nephrology Unit.
NR, nonrelated.
ND, not determined. *, phylogroup B2 isolate negative for serotype O25b.
According to MLST and phylogroup characterization, C1 isolates producing CTX-M-14 were identified as ST23 phylogroup A, whereas SHV-12-producing C2 isolates were classified as ST131 phylogroup B2 serotype O25b.
Very few outbreaks due to ESBL-producing E. coli have been reported in NICUs. Recently, two outbreaks caused by E. coli producing TEM-12 and TEM-52 in the NICUs of hospitals in Switzerland (16) and France (8), respectively, have been reported. To our knowledge, this is the first documented report of the clonal spread of E. coli ST23 phylogroup A harboring CTX-M-14 in newborns. CTX-M-14 is one of the most prevalent ESBL types produced by E. coli (12) and usually belongs to a wide variety of clones (9, 11). Only a few CTX-M-14-producing E. coli strains have caused community outbreaks in the general population, as reported in Canada and Japan (13, 15).
Four isolates of CTX-M-14-producing E. coli belonging to the ST23 phylogroup A clone were previously identified in two different Spanish studies (11, 17), but this clone was not detected in a recent study in northwest Spain (9).
In addition, SHV-12-producing ST131 E. coli outbreaks have not been described previously in newborns, either.
Identification of ESBL-producing E. coli in non-NICU pediatric units.
Among the 15 children ≤4 years old who were infected by ESBL-producing E. coli and were admitted to non-NICU pediatric units, 7 isolates (46.7%) produced CTX-M-14 (3 produced blaCTX-M-14a, and 4 produced blaCTX-M-14b), whereas 4 (26.7%), 3 (20%), and 1 (6.7%) isolate produced CTX-M-15, SHV-12, and SHV-2, respectively (Table 1).
None of these 15 isolates belonged to the C1 ST23 epidemic clone, but 1 additional SHV-12-producing isolate belonging to the C2 ST131 phylogroup B2 cluster was detected. In addition, two SHV-12-producing E. coli isolates had a new common profile (cluster C3 [Fig. 1]), further identified as ST57 phylogroup D (Fig. 1).
CTX-M-15-producing E. coli ST131 in children ≤4 years old.
Overall, six CTX-M-15 isolates from children were identified: two among the NICU patients and four among the non-NICU pediatric patients (Table 1). According to PFGE data, these six isolates were genetically unrelated (Fig. 1), although three of them belonged to the international ST131 phylogroup B2 serotype O25b clone (12). According to PFGE, the genetic linkage of these three ST131 CTX-M-15 isolates (one from the NICU and two from the Emergency Room) was approximately 70%.
Four E. coli clones producing CTX-M-15, according to repetitive element-based PCR results, in the NICU of an Indian hospital have been described recently, although their MLST types were not reported (14).
Genetic environment and characterization of the plasmid carrying blaCTX-M-14a.
All 25 CTX-M-14-producing E. coli specimens isolated in the NICU neonates carried blaCTX-M-14a linked to ISEcp1, which was detected 42 nucleotides upstream of this ESBL gene.
PFGE after S1 nuclease digestion of whole genomic DNA obtained from clinical isolates revealed two plasmids of approximately 80 and 100 kb, but only the 80-kb plasmid was detected in the cefotaxime-resistant transconjugants. This plasmid belonged to incompatibility group K and carried blaCTX-M-14a.
Both mobile genetic elements—IncK conjugative plasmids and the ISEcp1 insertion sequence—were described previously as responsible for the spread of CTX-M-14a (17).
Antibiotic susceptibility of the C1 ST23 clone.
All isolates of the C1 ST23 clone had identical susceptibility patterns, including resistance to ampicillin (MIC, >16 μg/ml), cefazolin (MIC, >16 μg/ml), cefuroxime (MIC, >16 μg/ml), cefotaxime (MIC, 64 to 256 μg/ml), cefepime (MIC, 16 to >16 μg/ml), and gentamicin (MIC, >8 μg/ml) and susceptibility to ceftazidime (MIC, ≤1 μg/ml), amoxicillin-clavulanic acid (MIC, ≤4 to 8 μg/ml), piperacillin-tazobactam (MIC, ≤8 μg/ml), imipenem (MIC, ≤1 μg/ml), cotrimoxazole (MIC, ≤2 μg/ml), and ciprofloxacin (MIC, ≤0.12 μg/ml). Tobramycin and amikacin MICs ranged from ≤2 to >8 μg/ml and from ≤8 to 16 μg/ml, respectively.
Additionally, in all isolates of the C1 ST23 E. coli clone, the aac(3)-IIa aminoglycoside resistance gene was positively identified by PCR, whereas none of the isolates expressed the aac(6′)-Ib-cr aminoglycoside-quinolone resistance gene.
C1 ST23 outbreak evolution.
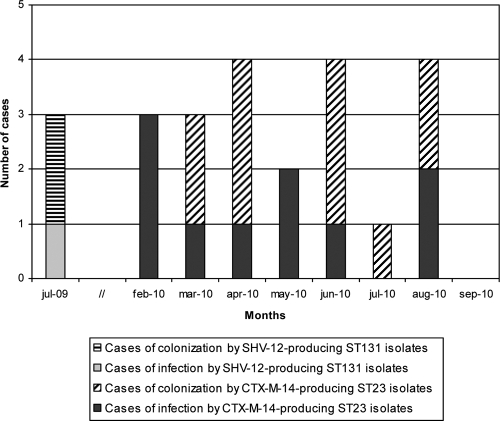
The monthly evolution of the 21 cases of infection (n = 10) or colonization (n = 11) caused by the C1 ST23 clone in the NICU is shown in Fig. 2. All neonates were admitted to the NICU between February and August 2010; during the same period, three additional newborns admitted to the NICU had infections due to CTX-M-14a-producing E. coli, but the isolates were genetically unrelated (Table 1).
Fig 2.
Monthly evolution of cases of infection or colonization of newborns by CTX-M-14- or SHV-12-producing Escherichia coli isolates belonging to C1 ST23 or C2 ST131, respectively.
The first case identified was that of a 7-day-old male who was transferred from another Spanish hospital to the NICU of UHGM because of congenital heart disease. Previously, from January 2009 to February 2010, only five cases of ESBL-producing E. coli infection or colonization had been detected in this NICU: four in July 2009 (two colonizations and one infection by SHV-12-producing ST131 isolates, and one colonization by a nonrelated CTX-M-15-producing isolate) and one in January 2009 (infection by a nonrelated CTX-M-14a-producing isolate).
Clinical risk factors, treatment, and outcome.
All 10 newborns infected by C1 ST23 isolates had underlying predisposing conditions, including congenital heart disease and prematurity (5) (Table 2). All 10 of these patients had received previous antibiotic treatment, and 7 (70%) had been treated with more than one antibiotic (Table 2). These 10 patients were mostly treated with meropenem as a monotherapy (n = 6 [60%]) or in combination with amikacin (n = 1 [10%]); 9 patients were cured with these treatments, but 1 newborn with bacteremia, who was treated with meropenem monotherapy, died (Table 2).
Table 2.
Clinical characteristics of patients infected with CTX-M-14-producing isolates belonging to the C1 ST23 clone
| Gendera | Localization of infectionb | Underlying condition(s)c | Previous antibiotic treatment(s) | Antimicrobial therapy | Outcome |
|---|---|---|---|---|---|
| M* | UTI | CC | Amikacin | Meropenem | Cure |
| M | LRTI | CC | Cefazolin | Meropenem | Cure |
| F | Wound infection | CC | Cefazolin | Meropenem | Cure |
| F | Bacteremia | PNI | Amikacin + vancomycin | Meropenem | Death |
| M | Wound infection | CC | Ampicillin + gentamicin + cefazolin | Amikacin + chlorhexidine washings | Cure |
| M | Bacteremia | PNI | Ampicillin + gentamicin | Meropenem + amikacin | Cure |
| M | LRTI | CC | Amikacin + vancomycin | Cotrimoxazole | Cure |
| F | Bacteremia | PNI | Ampicillin + gentamicin; amikacin + vancomycin | Meropenem | Cure |
| M | LRTI | PNI + MCM | Ampicillin + gentamicin; amikacin + vancomycin | Amikacin | Cure |
| F | Bacteremia | PNI | Ampicillin + gentamicin | Meropenem | Cure |
M, male; F, female. *, index case.
LRTI, lower respiratory tract infection.
CC, congenital cardiopathy; PNI, preterm newborn infant; MCM, multiple congenital malformations.
Infection control measures.
All children with ESBL-producing E. coli recovered from any clinical specimen according to the microbiology laboratory reports were identified. Active surveillance of all patients admitted to the NICU was performed at least once a week; rectal swabs were cultured for detection of ESBL-producing E. coli. Patients harboring ESBL-producing E. coli were assigned to contact precautions, including the use of disposable gowns and gloves. In addition, standard precautions were reinforced for all patients admitted to the NICU, including improvement of hand hygiene compliance by the use of alcohol rubs before and after the care of patients. None of the environmental cultures was positive for CTX-M-14-producing E. coli. Between September and December 2010, no cases of infection or colonization by ESBL-producing E. coli isolates were detected in the NICU.
Concluding remarks.
We report the characterization of an NICU outbreak caused by a CTX-M-14-producing E. coli isolate belonging to the ST23 phylogroup A clone, an ESBL type associated mainly with polyclonal community acquisition in adults. An additional, smaller outbreak, due to SHV-12-producing E. coli, was also identified in the same unit. Overall, CTX-M-14, CTX-M-15, and SHV-12 were the most important ESBLs causing pediatric infections in this study. Infections of newborns with CTX-M-14-producing E. coli were caused by both clonal and nonclonal isolates.
ACKNOWLEDGMENTS
This study was supported by the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, Spanish Network for Research in Infectious Diseases (REIPI C03/14 and RD06/0008), a research grant from the Fondo de Investigaciones Sanitarias (FIS PI09/917), an intramural research grant from the Instituto de Salud Carlos III (MPY 022/09), and the Dirección General de Salud Pública, Ministry of Health, Spain (reference DGVI 1909/10-TS-15).
J.O., O.C., V.B., and J.C. are members of the Spanish Network in Infectious Pathology (REIPI).
Footnotes
Published ahead of print 10 October 2011
REFERENCES
- 1. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 2. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clermont O, et al. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing, 20th informational supplement, M100–S21, vol 31, no. 1 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Dubois V, et al. 2010. CTX-M-producing Escherichia coli in a maternity ward: a likely community importation and evidence of mother-to-neonate transmission. J. Antimicrob. Chemother. 65:1368–1371 [DOI] [PubMed] [Google Scholar]
- 6. Eckert C, Gautier V, Arlet G. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14–23 [DOI] [PubMed] [Google Scholar]
- 7. García A, et al. 2007. Acquisition and diffusion of blaCTX-M-9 gene by R478-IncHI2 derivative plasmids. FEMS Microbiol. Lett. 271:71–77 [DOI] [PubMed] [Google Scholar]
- 8. Moissenet D, et al. 2010. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum β-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J. Clin. Microbiol. 48:2459–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mora A, et al. 2011. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4–B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int. J. Antimicrob. Agents 37:16–21 [DOI] [PubMed] [Google Scholar]
- 10. Oteo J, et al. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 44:2359–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oteo J, et al. 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173–176 [DOI] [PubMed] [Google Scholar]
- 12. Oteo J, Pérez-Vázquez M, Campos J. 2010. Extended-spectrum beta-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23:320–326 [DOI] [PubMed] [Google Scholar]
- 13. Pitout JD, Gregson DB, Church DL, Elsayed S, Laupland KB. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary Health Region. J. Clin. Microbiol. 43:2844–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shakil S, Akram M, Ali SM, Khan AU. 2010. Acquisition of extended-spectrum beta-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal intensive care unit: molecular epidemiology and analysis of risk factors. J. Med. Microbiol. 59:948–954 [DOI] [PubMed] [Google Scholar]
- 15. Suzuki S, et al. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72–79 [DOI] [PubMed] [Google Scholar]
- 16. Tschudin-Sutter S, Frei R, Battegay M, Hoesli I, Widmer AF. 2010. Extended-spectrum β-lactamase-producing Escherichia coli in neonatal care unit. Emerg. Infect. Dis. 16:1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valverde A, et al. 2009. Spread of blaCTX-M-14 is driven by IncK plasmids disseminated among Escherichia coli phylogoups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53:5204–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]