Abstract
A major challenge to clinical therapy of Burkholderia cepacia complex (Bcc) pulmonary infections is their innate resistance to a broad range of antimicrobials, including polycationic agents such as aminoglycosides, polymyxins, and cationic peptides. To identify genetic loci associated with this phenotype, a transposon mutant library was constructed in B. multivorans ATCC 17616 and screened for increased susceptibility to polymyxin B. Compared to the parent strain, mutant 26D7 exhibited 8- and 16-fold increases in susceptibility to polymyxin B and colistin, respectively. Genetic analysis of mutant 26D7 indicated that the transposon inserted into open reading frame (ORF) Bmul_2133, part of a putative hopanoid biosynthesis gene cluster. A strain with a mutation in another ORF in this cluster, Bmul_2134, was constructed and named RMI19. Mutant RMI19 also had increased polymyxin susceptibility. Hopanoids are analogues of eukaryotic sterols involved in membrane stability and barrier function. Strains with mutations in Bmul_2133 and Bmul_2134 showed increased permeability to 1-N-phenylnaphthylamine in the presence of increasing concentrations of polymyxin, suggesting that the putative hopanoid biosynthesis genes are involved in stabilizing outer membrane permeability, contributing to polymyxin resistance. Results from a dansyl-polymyxin binding assay demonstrated that polymyxin B does not bind well to the parent or mutant strains, suggesting that Bmul_2133 and Bmul_2134 contribute to polymyxin B resistance by a mechanism that is independent of lipopolysaccharide (LPS) binding. Through this work, we propose a role for hopanoid biosynthesis as part of the multiple antimicrobial resistance phenotype in Bcc bacteria.
INTRODUCTION
Burkholderia multivorans is a member of the B. cepacia complex (Bcc), a group of 17 closely related Gram-negative species with extreme genetic capacity and metabolic diversity (11, 26). The members of the Bcc have emerged as opportunistic pulmonary pathogens of particular importance to patients with cystic fibrosis (CF) or chronic granulomatous disease (CGD). Bcc bacteria are intrinsically resistant to a broad spectrum of antimicrobials, including polycationic agents such as polymyxins, impeding treatment efforts in both of these patient populations (32, 35, 45).
Bcc infections in CF patients are associated with enhanced morbidity and mortality compared to infections caused by the more common pathogen Pseudomonas aeruginosa (8), and they have the potential to cause rapid clinical deterioration with septicemia that leads to death (20, 50). Although B. cenocepacia has widely been considered the most prevalent and virulent Bcc species in CF infections, the proportion of B. multivorans infections is increasing (16), with its incidence in CF patients exceeding that of B. cenocepacia in the United States and Canada (4, 52). An outbreak of B. multivorans causing severe morbidity and mortality in CF patients has occurred (48).
Bcc bacteria cause serious invasive infections in patients with CGD. Neutrophils from CGD patients are unable to elicit an oxidative burst in response to invading microbes and are therefore dependent on nonoxidative killing mechanisms by cationic peptides. Bcc organisms resist killing in vitro by CGD neutrophils but not normal neutrophils (43). This correlates with the ability of Bcc organisms to resist killing in vitro by high concentrations of cationic peptides and polymyxin B (25).
With the escalating emergence of multidrug-resistant organisms, physicians are increasingly dependent upon polymyxins, especially inhaled colistin, for therapy of recalcitrant respiratory P. aeruginosa infections (5). Although cationic peptides and antibiotics appear to be unable to kill Bcc organisms, they may enhance the antimicrobial activity of other antibiotics (37, 38), providing physicians with a broader spectrum of therapeutic options for Bcc infections.
Polymyxin B and colistin (polymyxin E) are the two polymyxins used clinically. These are rapid-acting bactericidal agents with detergent-like properties that accumulate in the bacterial membrane, affecting selective permeability. Polymyxins bind to negatively charged sites in the lipopolysaccharide (LPS) layer of the membrane to initiate “self-promoted uptake” (31). Constitutive bacterial mechanisms of resistance to polymyxins are therefore thought to be based on features of LPS and/or membrane permeability (51).
The intrinsic resistance of Bcc bacteria to polymyxins is due to unique features of the cytoplasmic membrane, as Bcc spheroplasts are resistant to lysis by polymyxin B (27). Investigations into the mechanism of polymyxin resistance in the Bcc have previously focused on the effects of LPS modifications. Polymyxin B binds poorly to the LPS layer on intact Bcc cells (29). There is a constitutive presence of 4-amino-4-deoxy-l-arabinose (Ara4N) linked to the lipid A phosphate groups in the LPS of Bcc bacteria, eliminating the negative charge required for polymyxin binding (9) and presumably blocking the self-promoted uptake utilized by polycationic antimicrobials (17). The synthesis and composition of the LPS core oligosaccharide are also involved in resistance to polymyxin B in the Bcc bacterium B. cenocepacia (23, 33). Recent work by Loutet et al. (24) suggests a two-tier model of polymyxin B resistance in B. cenocepacia, with primary mechanisms involving intrinsic features of the cytoplasmic membrane and secondary mechanisms that confer less dramatic changes in resistance and are observed when the membrane barrier has been compromised.
In this study, we identified a B. multivorans transposon mutant with 8- and 16-fold increases in susceptibility to polymyxin B and colistin, respectively, independent of polymyxin B binding. In this mutant, the transposon was found to insert into a gene in the putative hopanoid biosynthesis cluster. Hopanoids are pentacyclic triterpenoid lipids that are sterol analogues in prokaryotic membranes and are presumed to be involved in membrane stability and barrier function (21). Not all bacteria produce hopanoids, yet those that do occupy diverse environmental niches (39). While details on the structure and chemical diversity of bacterial hopanoids produced have been described, little is known about their biosynthesis and function in the bacterial membrane (21, 39). We suggest a role for hopanoids in membrane stability that contributes to constitutive antibiotic resistance of bacteria of the Bcc.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were stored at −70°C in Mueller-Hinton (MH) broth with 8.0% (vol/vol) dimethyl sulfoxide (DMSO). For routine bacterial growth, Luria-Bertani (LB) broth was used. Unless otherwise stated, bacteria were incubated at 37°C, and liquid cultures were grown with aeration. Bacteriological medium components were purchased from Becton Dickinson and Company (Sparks, MD), and all chemicals were purchased from Sigma-Aldrich Canada Ltd. (Oakville, Ontario, Canada), unless stated otherwise. Minimal salts medium supplemented with 2.0% Casamino Acids and 5.0% mannitol was used to screen the transposon library for polymyxin susceptibility, as it reduced the background growth of spontaneously resistant clones of B. multivorans. MH broth without cation supplementation was used for MIC, dansyl-polymyxin (DPX) binding, and membrane permeability assays. The following media were used for growth experiments: LB broth, YP (0.3% yeast extract, 0.3% peptone), MH broth, LB broth with 2.5 to 10% ethanol, MH broth with 0.16 to 2.5% sodium dodecyl sulfate (SDS), MH broth with 0.5 to 5% sodium chloride, YP with 0.87 to 10% bile salts (Oxoid Ltd., England), and acid broth (1) buffered to pH 5, 6, 7, and 8. Plasmids were maintained by growth in the presence of the appropriate antibiotic. All antibiotics were purchased from Sigma-Aldrich Canada Ltd. The antibiotics used were as follows: for Escherichia coli, tetracycline at 15 μg/ml, kanamycin at 50 μg/ml, gentamicin at 50 μg/ml, ampicillin at 100 μg/ml, and trimethoprim at 750 μg/ml; and for B. multivorans, tetracycline at 75 μg/ml and trimethoprim at 100 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| B. multivorans strains | ||
| ATCC 17616 | Type strain | ATCC |
| 26D7 | Polymyxin B-susceptible derivative of ATCC 17616 with Tn5-751S disrupting Bmul_2133; TMPr | This study |
| RMI17 | ΔBmul_2134 derivative of ATCC 17616; TETs | This study |
| RMI18 | ΔBmul_2134 derivative of ATCC 17616; TETs | This study |
| RMI19 | ΔBmul_2134 derivative of ATCC 17616; TETr | This study |
| P. aeruginosa strains | ||
| ATCC 27853 | MIC quality control strain | ATCC |
| M2 | Mouse intestinal isolate susceptible to neutrophil killing | 44 |
| E. coli strains | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| HB101 | supE44 hsdS20(rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 40 |
| XL1-Blue MR | Cosmid library host strain | Agilent Technologies |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; AMCr | Agilent Technologies |
| Super Cos 1 | Cosmid cloning vector; AMCr NEMr | Agilent Technologies |
| pCR-BluntII-TOPO | Cloning vector; KANr ZEOr | Invitrogen |
| pTGL166 | Broad-host-range temperature-sensitive IncP1 plasmid containing Tn5-751S; KANr TMPr TETr PENr | 7 |
| pRK2013 | ColE1 Tra (RK2)+ KANr | 14 |
| pEX18Tc | Suicide vector; TETr | 18 |
| pUCP26 | Broad-host-range vector; TETr | 47 |
| pUCP28T | Broad-host-range vector; TMPr | 42 |
| pBS1 | pBluescript II KS(+) containing Tn5-751 and flanking DNA from mutant 26D7; AMPr KANr | This study |
| pBS4 | pBluescript II KS(+) carrying a 4.0-kb BamHI fragment containing Bmul_2134; AMCr | This study |
| pRM301 | pEX18Tc carrying the 4.0-kb BamHI fragment from pBS4 containing Bmul_2134; TETr | This study |
| pRM302 | pRM301 with a 718-bp NotI fragment deletion in Bmul_2134; TETr | This study |
| pRM305 | pUCP28T with a 1.6-kb AfeI/ApaI fragment from pBS4 blunt ligated into SmaI with Bmul_2134; TMPr | This study |
| pRM306 | pCR-BluntII-TOPO with a 1.6-kb Bmul_2133 PCR fragment; TETr | This study |
| pRM308 | pUCP26 with a 1.6-kb XbaI/HindIII fragment from pRM306 containing Bmul_2133; TETr | This study |
AMC, ampicillin; KAN, kanamycin; NEM, neomycin; PEN, penicillin; TET, tetracycline; TMP, trimethoprim; ZEO, zeocin.
Determination of MICs.
The MICs for all antibiotics were determined by the standard microtiter broth dilution method following the protocols outlined by The Clinical and Laboratory Standards Institute (http://www.clsi.org). MICs for polycationic antimicrobials were determined in polypropylene 96-well plates (Corning Inc., Corning, NY).
Molecular techniques.
DNA manipulations were executed using standard techniques as described by Ausubel et al. (3). General molecular biology techniques were preformed according to the methods of Sambrook et al. (40). Plasmid DNA was extracted with QIAprep Spin miniprep kits (Qiagen Inc., Mississauga, Ontario, Canada). Restriction endonucleases, DNA polymerases, T4 DNA polymerase, and T4 DNA ligase were purchased from New England BioLabs (Mississauga, Ontario, Canada) and used according to the manufacturer's instructions. DNA fragments used in cloning procedures were purified with a QIAquick gel extraction kit (Qiagen Inc.). Oligonucleotide primers were purchased from Integrated DNA Technologies (San Diego, CA) or the Nucleic Acid-Protein Service Unit at the University of British Columbia (Vancouver, British Columbia, Canada). Radiolabeled probes for Southern hybridizations were labeled with [α-32P]dGTP (American Radiolabeled Chemicals Inc., St. Louis, MO) by random priming using the Klenow fragment of DNA polymerase I (Invitrogen Canada Inc., Burlington, Ontario, Canada) per the manufacturer's instructions. DNA sequencing was performed with the assistance of the Nucleic Acid-Protein Service Unit at the University of British Columbia (Vancouver, British Columbia, Canada) or the Center for Molecular Medicine and Therapeutics (Vancouver, British Columbia, Canada). Sequences and contigs were assembled using Lasergene for Windows (DNAStar Inc., Madison, WI). The nucleotide sequence data for the hopanoid biosynthesis locus were provided by the B. multivorans ATCC sequencing project at the U.S. Department of Energy Joint Genome Institute (JGI) (http://genome.jgi-psf.org/burmu/burmu.home.html). Sequence analysis with the provided sequence was performed using DNAMAN sequence analysis software (Lynnon Biosoft, Vaudreuil, Quebec, Canada). Additional in silico promoter and terminator predictions were performed with BPROM and FindTerm (Softberry, Inc., Mount Kisco, NY). Sequence similarity searches were performed using the National Center for Biotechnology Information Basic Local Alignment Search Tool (NCBI BLAST) (2).
Isolation of a polymyxin B-susceptible mutant of B. multivorans ATCC 17616 by random transposon mutagenesis.
The transposon-carrying vector pTGL166 (7) was transferred into B. multivorans ATCC 17616 by conjugation with pRK2013 as the mobilizing vector (14). Tetracycline- and trimethoprim-resistant colonies, containing pTGL166, were grown for 60 h at 47°C in LB broth to encourage loss of the vector. Resulting trimethoprim-resistant clones, carrying Tn5-751S incorporated into the B. multivorans genome, were screened for susceptibility to tetracycline to ensure that they had lost the vector DNA.
Six thousand trimethoprim-resistant, tetracycline-susceptible colonies were picked into 96-well plates. These Tn5-751S insertion mutants were screened by replica plating onto supplemented minimal salts medium without antibiotic or containing either 600 or 1,200 units of polymyxin B. This screen resulted in 12 putative mutants that did not grow on either concentration of polymyxin B. To ensure that only one transposon insertion occurred in the putative mutants, Southern hybridizations were performed on SalI-digested genomic DNA, using a transposon-specific probe. The probe was a 32P-labeled PCR product amplified from pTGL166 by using primers N727-3 (5′-ATCGACAAGACCGGCTTCCATCCGA-3′) and N725-5 (5′-TCAGCGCAGGGGCGCCCGGTTCTTT-3′). Each of the 12 mutants had only one insertion. The probe bound to genomic fragments of different sizes, indicating that the transposon inserted at different locations in the genome.
Identification of the genomic region disrupted by Tn5-751S in the polymyxin B-susceptible mutant 26D7.
To identify the genes responsible for the phenotype of mutant strain 26D7, the genomic region around the transposon was cloned and sequenced. A SalI minilibrary of 26D7 genomic DNA was constructed in pBluescript II SK(+) (Agilent Technologies, Mississauga, Ontario, Canada) and transformed into E. coli DH5α by electroporation. A kanamycin-resistant clone containing Tn5-751S was isolated and named pBS1. The nucleotide sequence of the genomic region surrounding the transposon on pBS1 was obtained using primers T3 (5′-ATTAACCCTCACTAAAGGGA-3′) and TN5-1 (5′-GAGGTCACATGGAAGTCAG-3′). The B. multivorans ATCC 17616 sequence data were not available for the majority of this study, and the genomic region disrupted by the transposon was initially characterized by traditional molecular biology techniques. Briefly, the sequence obtained from pBS1 was used to design the primers Seq1R (5′-GCGCTTCTTATGCTCGAT-3′) and Seq1L (5′-ACTAGCTCGCCGACGATT-3′) to amplify a 1.2-kb product specific for the sequence adjacent to the transposon insertion site. This product was used as a probe to screen a B. multivorans ATCC 17616 cosmid library by colony hybridization (49). The cosmid library was constructed from partially digested Sau3AI fragments ranging in size from 30 to 40 kb in a SuperCos I cosmid according to the manufacturer's instructions (Agilent Technologies). The insert sequence-specific probe bound to five cosmids. These cosmids contained the desired genomic region. The cosmid DNA was isolated and digested with various restriction enzymes. A 4.0-kb BamHI fragment was identified and subcloned into pBluescript II KS(+) (Agilent Technologies), resulting in vector pBS4. Sequence analysis of the genomic sequence in these vectors was carried out by random primer walking. Once the B. multivorans ATCC 17616 genome sequence became available (http://genome.jgi-psf.org/burmu/burmu.home.html), the initial sequence obtained from pBS1 was used as a query to confirm the location of the disrupted locus.
Construction of mutant RMI19 in B. multivorans ATCC 17616.
A Bmul_2134 allelic exchange vector, pRM302, was constructed by first subcloning the 4.0-kb BamHI fragment containing Bmul_2134 from pBS4 into pEX18Tc (18), resulting in pRM301. The open reading frame (ORF) was disrupted by deletion of an internal 718-bp NotI fragment and religation of the vector (Fig. 1). Vector pRM302 was introduced into B. multivorans ATCC 17616 by triparental mating using pRK2013 as the mobilization vector (14). Transconjugants were plated onto LB agar with gentamicin (to inhibit growth of the E. coli strains) and tetracycline. Tetracycline-resistant single-crossover mutants were confirmed by a PCR using primers HpnIF (5′-CGGCTTTGCGGGAATC-3′) and HpnIR (5′-TGCCCGCTACTCCGTGA-3′). Selected single-crossover mutants were grown in liquid libraries and plated onto LB agar to encourage loss of the vector sequence. Screening for tetracycline sensitivity identified the double-crossover ΔBmul_2134 mutant, designated RMI19, and the mutant genotype was confirmed by PCR with the HpnIF and HpnIR primers. Later, after the majority of the analysis with this mutant was completed, MIC experiments indicated that the mutant was tetracycline resistant. PCR with primers M13F (5′-GTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′) to amplify a fragment from pEX18Tc indicated that at least a portion of the vector sequence was present in this strain. Genotyping of the mutant was repeated with the HpnIF and HpnIR primers, and only a mutated copy of Bmul_2134 was amplified. Two additional ΔBmul_2134 mutant colonies, RMI17 and RMI18, isolated during the same round of allelic exchange as RMI19 but not containing the vector sequence, were screened for acquired tetracycline resistance by MIC analysis. These mutant isolates yielded identical PCR products with the HpnIF and HpnIR primers to those amplified from RMI19, yet they were tetracycline sensitive.
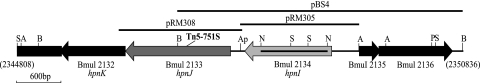
Fig 1.
Map of the B. multivorans ATCC 17616 locus affected by Tn5-751S in mutant 26D7. The region from base pairs 2344808 to 2350836 of the B. multivorans ATCC 17616 genome sequence (http://genome.jgi-psf.org/burmu/burmu.home.html) is shown in the published orientation with ORFs Bmul_2132, Bmul_2133, and Bmul_2134. The locations of the transposon insertion in the 26D7 mutant and key restriction sites are marked. The DNA fragments used for complementation experiments are indicated by the black bars above the sequence. The region of Bmul_2134 deleted in the RMI19 mutant construct is indicated by the black bar in the ORF. A, AfeI; Ap, ApaI; B, BamHI; N, NotI; S, SalI; P, PstI.
Cloning of Bmul_2133 and Bmul_2134 for complementation of mutants 26D7 and RMI19 in trans.
A 1.6-kb AfeI/ApaI fragment from pBS4, containing Bmul_2134 and the predicted promoter region (Fig. 1), was blunt end ligated into the SmaI site of pUCP28T (42) to construct pRM305. To construct the Bmul_2133 complementation vector, pRM308, a 1,661-bp PCR fragment containing the Bmul_2133 ORF and predicted promoter region (Fig. 1) was amplified using primers hpnJF (5′-GCCGACGGAATACGG-3′) and hpnJR (5′-ACCGCCGCATTCACA-3′) and initially cloned into pCR-Blunt II-TOPO (Invitrogen), yielding pRM306. A 1.6-kb XbaI/HindIII fragment from pRM306 was subcloned into pUCP26 (47), yielding pRM308. The pUCP28T-based vectors were conjugated into the parent and the RMI19 mutant by triparental mating using pRK2013 (14) as the mobilization strain, and the pUCP26-based vectors were transferred into the parent and the 26D7 mutant by electroporation (12).
Permeabilization of whole cells of B. multivorans with NPN.
Membrane permeability in the presence of polymyxin B was assayed using the hydrophobic fluorescent probe 1-N-phenylnaphthylamine (NPN) similarly to the manner previously described (22, 24). Overnight seed cultures were subcultured 1/100 and grown to mid-log phase (optical density at 600 nm [OD600], between 0.4 and 0.6). Bacteria were collected and washed twice in 5 mM HEPES buffer (pH 7.4) with 10 mM sodium azide and then resuspended to an OD600 of 0.5. The assay was carried out in a 100-μl total volume with 90 μl of bacterial suspension in a transparent 96-well plate (Corning, Inc.). NPN was added to a final concentration of 10 μM. Polymyxin B was subsequently added in increasing final concentrations from 0 to 200 μg/ml. Excitation and emission wavelengths were set at 350 and 420 nm, respectively, and fluorescence was measured using a Tecan Infinite M200 spectrophotometer (Tecan, Durham, NC).
DPX binding assay.
The binding of DPX to the parent and mutant strains was examined as previously described (28, 33), with the following modifications. The fluorescent dansyl derivative of polymyxin B was purchased from Invitrogen. The assay was read in clear-welled 96-well plates (Corning Inc.) with a Tecan Infinite M200 spectrophotometer (Tecan), using an excitation wavelength of 340 nm and an emission wavelength of 485 nm.
Bacterial growth.
The growth of the parent and mutant strains was monitored over time by measuring the OD600 with a Bioscreen C spectrophotometer (Oy Growth Curves Ab Ltd., Finland). Overnight seed cultures were normalized to an initial OD600 of 0.1 in fresh medium. A 1/50 subculture in 100 μl of medium for each condition was prepared in a 96-well Bioscreen plate. Each condition was tested in triplicate for each culture. The cultures were grown for 18 h at 37°C with continuous shaking, and OD600 measurements were taken every 15 min.
Neutrophil killing assay.
The CGD neutrophil killing assay was performed as previously described (43). Complement component five-deficient serum was used because B. multivorans is susceptible to complement-mediated lysis.
RESULTS
Description of the genomic region flanking Tn5-751S in the polymyxin B-susceptible transposon mutant 26D7.
To identify a mutant of B. multivorans rendered susceptible to polymyxin B, a random transposon insertion library of the B. multivorans strain ATCC 17616 was constructed using the transposon Tn5-751S on the temperature-susceptible plasmid pTGL166 (7). A library of 6,000 mutants was screened for increased susceptibility to polymyxin B. Of the 12 susceptible mutants that were isolated, mutant 26D7 had the greatest increase in susceptibility (data not shown) and was selected for further characterization. The DNA sequences immediately flanking Tn5-751 in 26D7 were obtained by shotgun cloning of the mutant genomic DNA into pBluescript II SK(+). The captured DNA in the resulting plasmid, pBS1, was sequenced and was compared to the B. multivorans ATCC 17616 genome sequence to identify the disrupted locus. The transposon inserted into ORF Bmul_2133 (Fig. 1). This ORF has been annotated as hpnJ, encoding a putative hopanoid biosynthesis-associated radical S-adenosylmethionine (SAM) protein. ORF Bmul_2133 is part of a predicted hopanoid biosynthesis gene cluster with Bmul_2134 (annotated as hpnI), encoding a putative hopanoid biosynthesis-associated glycosyltransferase, located directly upstream, and Bmul_2132 (annotated as hpnK), encoding a putative hopanoid biosynthesis protein, located directly downstream (Fig. 1). In silico promoter prediction indicated a promoter upstream of each ORF, and a transcriptional terminator was identified in the intergenic region between Bmul_2133 and Bmul_2134 (data not shown), suggesting that these genes are not cotranscribed.
Confirming a role for Bmul_2133 and Bmul_2134 in B. multivorans polymyxin B resistance.
Mutant 26D7 was determined to be 8-fold more susceptible to polymyxin B than the parental strain, B. multivorans ATCC 17616 (Table 2). In order to confirm the role of Bmul_2133 in polymyxin B resistance, the predicted Bmul_2133 promoter region and ORF were added back to mutant 26D7 in trans. The addition of vector pRM308 increased the polymyxin B resistance phenotype to within 2-fold that of the parent strain (Table 2). To further associate the ORFs in the predicted hopanoid biosynthesis gene cluster with polymyxin susceptibility, a Bmul_2134 allelic exchange mutant, RMI19, was constructed in B. multivorans ATCC 17616. The RMI19 mutant had a 4-fold increase in polymyxin B susceptibility compared to the parent, and the original phenotype was restored upon complementation with Bmul_2134 in trans on vector pRM305 (Table 2).
Table 2.
Polymyxin B and colistin MICs for the parent strain B. multivorans ATCC 17616 and mutants 26D7 and RMI19a
| Strain | ORF provided in trans | MIC (μg/ml) |
|
|---|---|---|---|
| Polymyxin B | Colistin | ||
| B. multivorans strains | |||
| ATCC 17616 | 256 | 512 | |
| ATCC 17616(pUCP26) | 256 | 512 | |
| ΑTCC 17616(pUCP28T) | 256 | 512 | |
| 26D7 | 32 | 32 | |
| 26D7(pUCP26) | 32 | 32 | |
| 26D7(pRM308) | Bmul_2133 | 128 | 256 |
| RMI19 | 64 | 64 | |
| RMI19(pUCP28T) | 64 | 64 | |
| RMI19(pRM305) | Bmul_2134 | 256 | 512 |
| P. aeruginosa strain | |||
| ATCC 27853 | 1 | 1 | |
Data represent the modes for at least four replicate experiments after 48 h of growth.
The antibiotic susceptibility phenotype of the mutant strains is specific to polymyxins.
To determine the spectrum of antimicrobial resistance conferred by the identified hopanoid biosynthesis gene cluster, MICs of both close and distant families of antibiotics were determined (Table 2 and data not shown). Sixteen- and 8-fold increases in susceptibility to colistin (polymyxin E) were observed for the 26D7 and RMI19 mutants, respectively, compared to the MIC of the parent strain (B. multivorans ATCC 17616) (Table 2). The parental level of the colistin MIC was restored (within 2-fold) when the respective wild-type genes were added in trans (Table 2). There was no significant (i.e., >2-fold) decrease in MIC between the parent and mutant strains for the aminoglycoside gentamicin, indicating that the mutated genes are not involved in resistance to all polycationic antimicrobials (data not shown). No differences in resistance to antimicrobials of other classes were observed (data not shown). The tetracycline MIC of 26D7 was not significantly different from that of the parent strain. The RMI19 mutant developed tetracycline resistance; however, two additional ΔBmul_2134 mutant isolates, RMI17 and RMI18, did not. Therefore, this phenotype cannot be attributed to the mutation in Bmul_2134. MIC values for erythromycin, ciprofloxacin, ceftazidime, and ampicillin were the same between the parent and mutant strains. These data suggest that Bmul_2133 and Bmul_2134 confer resistance specifically to polymyxins in B. multivorans.
Mutations in Bmul_2133 and Bmul_2134 affect membrane permeability.
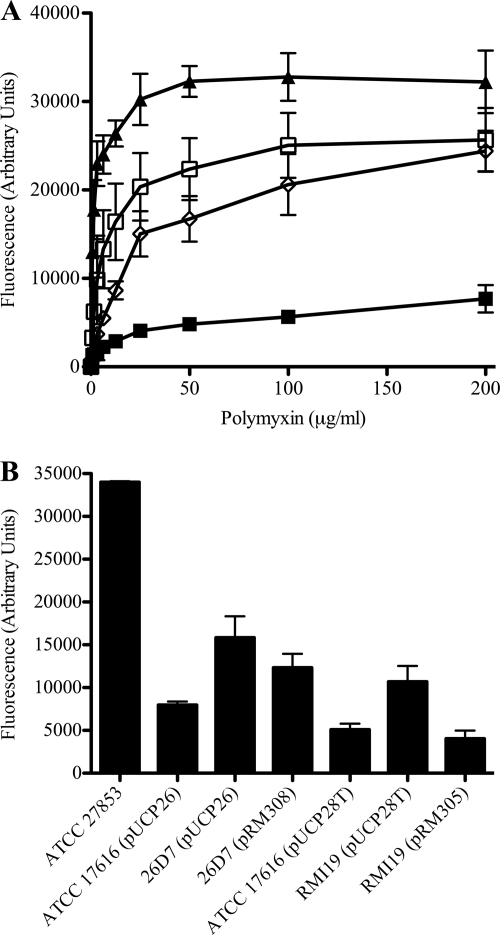
Hopanoids are thought to be involved in maintaining membrane fluidity and permeability (21, 39). To investigate the role of the putative hopanoid biosynthesis genes Bmul_2133 and Bmul_2134 in membrane permeability in the presence of polymyxin B, NPN uptake assays were performed. NPN is a hydrophobic compound that fluoresces weakly in a hydrophilic environment but strongly in a hydrophobic environment, as in the periplasm of Gram-negative bacteria. An increase in membrane permeability and uptake of NPN is measured as an increase in fluorescence (22). Using this method, it has been shown that Burkholderia spp. are resistant to membrane permeabilization by polymyxin B and other polycations (30). The putative hopanoid biosynthesis mutants and the parent strain were exposed to increasing concentrations of polymyxin B in the presence of NPN. Both mutants 26D7 and RMI19 showed significant increases in permeability to NPN in the presence of 0.78 μg/ml to 200 μg/ml polymyxin B compared to the parental strain (P < 0.02; t test) (Fig. 2A). Mutant strains 26D7 and RMI19 transformed with their respective empty vectors remained significantly more permeable to NPN at 100 μg/ml polymyxin B than the parent strain transformed with the same vector (P < 0.05; analysis of variance [ANOVA]) (Fig. 2B). Adding Bmul_2133 back to mutant 26D7 on vector pRM308 decreased the permeability of the strain to NPN in the presence of 100 μg/ml polymyxin, but not entirely to the level of the parent (Fig. 2B). The effect of the deletion mutation in Bmul_2134 on outer membrane permeability was restored to parental levels when Bmul_2134 was provided in trans on pRM305 (Fig. 2B). These results suggest that mutations in the predicted hopanoid biosynthesis genes result in an increase in outer membrane permeability, presumably due to abrogation of hopanoid biosynthesis, with enhancement of polymyxin B and colistin susceptibility.
Fig 2.
Effect of increasing concentrations of polymyxin B on the outer membrane permeability of B. multivorans ATCC 17616 compared to mutants 26D7 and RMI19. Polymyxin B was titrated into suspensions of whole bacteria at an OD600 of 0.5 and 10 μM NPN. An increase in fluorescence corresponded to an increase in membrane permeability and uptake of NPN into the hydrophobic periplasm. Data were calculated as the fluorescence with polymyxin B minus the fluorescence without it and represent means for three biological replicates ± standard errors of the means. (A) Mutants 26D7 and RMΙ19 were each significantly more permeable to NPN than the parent strain in 0.78 to 200 μg/ml of polymyxin B (P < 0.02; t test). ■, B. multivorans ATCC 17616; □, B. multivorans 26D7; ◊;, B. multivorans RMI19; ▴, P. aeruginosa ATCC 27853. (B) Outer membrane permeability of mutant strains in the presence of 100 mg/ml polymyxin B is restored when the mutations are complemented in trans. P. aeruginosa ATCC 27853 was used as a positive control. B. multivorans 26D7(pUCP26) and B. multivorans RMI19(pUCP28T) were significantly more permeable to NPN at 100 mg/ml polymyxin B than the B. multivorans ATCC 17616 parent strain with the respective empty vector (P < 0.05; ANOVA). B. multivorans 26D7(pRM308) was not significantly different from the parent or mutant strain with pUCP26. The permeability of B. multivorans RMI19(pRM305) to NPN was significantly less than that of RMI19(pUCP28T) at 100 mg/ml of polymyxin B (P < 0.05; ANOVA).
Enhanced susceptibility to polymyxin B is independent of LPS binding.
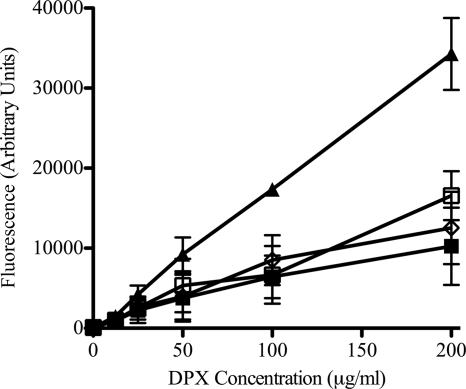
To determine if the increase in polymyxin B susceptibility observed in the mutants was due to an increase in binding of the antibiotic, the level of polymyxin B binding was evaluated in the parent and mutant strains. Interaction of the fluorescent compound DPX with LPS enhances the fluorescence of DPX. Whole cells of Bcc strains bind DPX poorly (30). There was no difference in fluorescence when whole cells of the parent strain (ATCC 17616) and polymyxin B-susceptible mutants were exposed to DPX (Fig. 3). P. aeruginosa ATCC 27853 was used as a positive control for DPX binding. These data suggest that mutating the putative hopanoid biosynthesis genes leads to increased susceptibility to polymyxin B by a mechanism that is independent of LPS binding.
Fig 3.
Binding of DPX to whole parent and mutant cells. Bacterial cells were treated with increasing concentrations of DPX, and fluorescence was determined by emission at 485 nm upon excitation at 340 nm. ■, B. multivorans ATCC 17616; □, B. multivorans 26D7; ◊, B. multivorans RMI19; ▴, P. aeruginosa ATCC 27853. The background fluorescence of DPX with buffer alone was subtracted at each concentration. The data represent the means for three biological replicates ± standard errors of the means.
There are no differences in growth under stress conditions in putative hopanoid biosynthesis mutants.
There were no apparent defects in growth of the 26D7 or RMI19 mutant under nutrient-rich conditions (data not shown). Hopanoids have been shown to be involved in resistance to bile salts and low pH (46), in ethanol tolerance (19), and in membrane stability during growth at a high temperature (41). There was no significant difference in growth between the parent and mutant strains with increasing concentrations of ethanol, SDS, bile salts, or sodium chloride, at either acidic or basic pH, or at 20°C, 37°C, or 42°C (data not shown).
Mutant 26D7 remains resistant to nonoxidative killing by neutrophils.
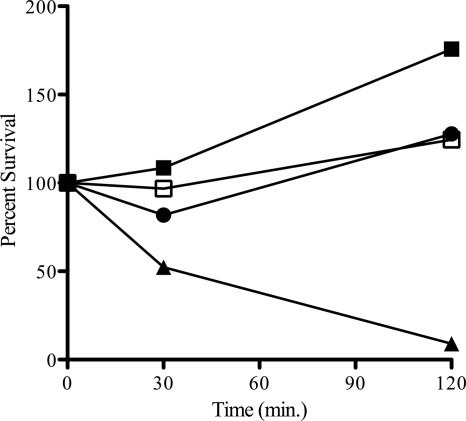
The possibility that the increase in polymyxin susceptibility exhibited by the Bmul_2133 and Bmul_2134 mutants could translate into the strains being susceptible to cationic peptides during nonoxidative killing by neutrophils was investigated by neutrophil killing assays. For these experiments, only the more susceptible mutant, 26D7, was used, as primary human CGD neutrophils are infrequently available. CGD neutrophils are unable to kill bacteria via reactive oxygen species and rely exclusively on nonoxidative bactericidal mechanisms (43). CGD neutrophils were challenged at a neutrophil-to-bacterium ratio of 1:1. Both the parent and 26D7 mutant strains grew in the presence of CGD neutrophils (Fig. 4), as did B. multivorans JTC, a clinical CGD isolate. The susceptible control strain of P. aeruginosa exhibited a marked decrease in survival over the course of the 2-h challenge. All four bacterial strains were susceptible to killing by normal neutrophils (data not shown).
Fig 4.
Neutrophil bacterial killing by cells from a patient with CGD. CGD neutrophils were incubated with bacteria (1:1) at 37°C. Viable bacterial counts were determined at 0, 30, and 120 min postinfection. Data presented are percentages of bacteria relative to the starting inoculum at each time point. ■, B. multivorans ATCC 17616; □, B. multivorans 26D7; ●, B. multivorans JTC; ▴, P. aeruginosa M2 as a susceptible control (43).
DISCUSSION
The intrinsic resistance of Bcc species to polycationic antimicrobials is due to features of their LPS and outer membrane structure (23, 25, 30, 33). This study suggests a role for membrane hopanoids in modulating B. multivorans membrane permeability that contributes to high levels of polymyxin B and colistin resistance in an LPS-independent manner. LPS-independent mechanisms of polymyxin resistance present in other organisms include the K. pneumoniae capsule (6) and the outer membrane porin OmpU in Vibrio cholerae. The isoprenoid biosynthesis gene BCAL2710ispH (also called lytB) in both B. pseudomallei and B. cenocepacia contributes to polymyxin resistance (5, 24). In E. coli, LytB converts (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMB-PP) to isopentenyl pyrophosphate (IPP) as part of the mevalonate biosynthesis pathway (13). Interestingly, IPP is a key precursor of hopanoid biosynthesis (21). With the two predicted B. multivorans hopanoid biosynthesis genes identified in this study and two reports of the upstream isoprenoid precursor involved in polymyxin B resistance in Burkholderia spp. (5, 24), hopanoids can now be included as part of the Burkholderia innate polymyxin B resistance repertoire.
The key hopanoid biosynthesis enzyme is the squalene-hopene cyclase (shc), which converts a squalene precursor into the cyclic form of hopene (34). The hopanoid gene cluster characterized in this study does not contain all of the genes identified in other hopanoid biosynthesis gene clusters (36, 46) and does not contain the key shc gene. In the current annotation of B. multivorans ATCC 17616, one of the squalene-hopane cyclase genes is designated Bmul_3175 and is located on the second chromosome. Directly upstream of Bmul_3175 are a putative squalene synthase (Bmul_3173) and a putative squalene-associated FAD-dependent desaturase (Bmul_3174). These three genes are homologs of hpnF, hpnD, and hpnE of the characterized hopanoid biosynthesis gene clusters of Zymomonas mobilis and Bradyrhizobium japonicum (36). There is an additional squalene-hopene cyclase homologue in the annotated genome: Bmul_5992. Hopanoid biosynthesis is thought to be under tight control and is expressed in response to stress conditions (21, 39). The fragmented organization of these genes suggests that a complex regulatory network is likely involved in B. multivorans hopanoid biosynthesis. We are currently conducting bacterial genetic experiments to elucidate the regulatory network orchestrating the hopanoid biosynthesis pathway as well as biochemical experiments to characterize the roles of the predicted hopanoid biosynthesis genes that lead to the production of membrane hopanoids.
Cholesterol in eukaryotic membranes protects the host cell from damage caused by polymyxin and antimicrobial peptide therapies (15). Thus, it is not surprising that the analogous hopanoids are involved in polymyxin resistance in prokaryotes. The observations presented in this study suggest that the mutations in Bmul_2133 and Bmul_2134 influenced membrane permeabilization, yet the effect was specific to polymyxins and was not a general permeability defect. Testing against the macrolide erythromycin showed no difference in MIC between the parent and mutant strains, confirming that permeability to hydrophobic antibiotics and efflux of these substances were not affected by the mutations. Testing with other antibiotics, including tetracycline, the penicillin ampicillin, the cephalosporin ceftazidime, and the fluoroquinolone ciprofloxacin, also did not show enhanced antimicrobial activity in the mutants, confirming that the hydrophilic (porin-mediated) uptake pathway was not affected. The predicted hopanoid biosynthesis mutants also did not show enhanced susceptibility to gentamicin. Aminoglycosides and cationic peptides enter Gram-negative bacterial cells by the same pathway, self-promoted uptake (17, 31), and have similar modes of action. Therefore, more than one mechanism must be involved in the resistance of B. multivorans to different cationic antimicrobial agents.
Cationic antimicrobial peptides are key elements for nonoxidative bactericidal activity by neutrophils. CGD neutrophils are dependent on nonoxidative killing mechanisms, and resistance of B. multivorans and other members of the Bcc to CGD neutrophils is considered a possible mechanism of pathogenesis. The 26D7 mutant was equally resistant to killing by CGD neutrophils as the parent strain and the control B. multivorans strain JTC. This suggested that although mutation of the putative hopanoid biosynthesis genes results in reductions in the MICs of polymyxins, it does not confer susceptibility to killing by CGD neutrophils. MICs of 32 μg/ml and 64 μg/ml for mutants 26D7 and RMI19, respectively, are still classified as resistant in the clinical sense. The reduction in resistance to polymyxins in mutant strain 26D7 is not enough to translate into susceptibility to nonoxidative killing by CGD neutrophils. Also, our data show that the increase in susceptibility of the mutants is specific to polymyxin B and colistin and not to other polycationic agents, such as gentamicin. Nonoxidative killing by neutrophils is mediated by a milieu of proteases, lysozymes, and antimicrobial peptides, including polycationic antimicrobial peptides. The mutations in Bmul_2133 must not confer resistance to these host defenses. Our observations provide proof of principle that hopanoids contribute to resistance of the members of the Bcc to cationic antimicrobial activity, but the roles of other features (such as LPS modifications) are clearly complementary.
Hopanoid biosynthesis genes are implicated in antibiotic resistance in Rhodopseudomonas palustris, in which a mutation in the shc gene confers susceptibility to erythromycin and rifampin (46). However, the effect of hopanoids on antibiotic resistance in clinically relevant organisms beyond Burkholderia spp. has yet to be characterized. In fact, few bacteria associated with human infection possess the genetic machinery necessary for the biosynthesis of hopanoids. A BLAST search (2) with the amino acid sequence of the squalene-hopane cyclase, Shc (Bmul_3175), did not align with any of the sequences encoded in the complete genome sequences of other known obligate and opportunistic respiratory pathogens, including Streptococcus pneumoniae, Neisseria meningitidis, Yersinia pestis, Legionella pneumophila, and P. aeruginosa. Yet the shc gene is distributed throughout the sequenced Bcc species and B. pseudomallei. Hopanoids have been isolated from a variety of Burkholderia species but not from the closely related and typically polymyxin B-sensitive Pseudomonas and Ralstonia organisms (10). The extraordinary and predictable resistance of Bcc bacteria to cationic antimicrobial agents, typically useful in treating many other human pathogens, may be due in part to the production of hopanoids. It is therefore necessary to understand all aspects of antimicrobial resistance in the Bcc. There are few therapeutic options for Bcc infections; therefore, understanding why members of the Bcc are additionally resistant to antimicrobials that are often used to treat P. aeruginosa infections in CF patients is crucial. Work is ongoing to determine if the presence of hopanoids in the Burkholderia membrane confers an advantage for the organism's survival and infection of patients with CF or CGD.
ACKNOWLEDGMENTS
This work was supported by a Cystic Fibrosis Canada operating grant. B.R.S.-K. received a studentship from Cystic Fibrosis Canada. R.J.M. received support from Canadian Institutes of Health Research and Cystic Fibrosis Canada fellowships.
We thank J. Zlosnik for a critical review of the initial manuscript as well as S. Farmer and T. Hird for technical assistance.
Footnotes
Published ahead of print 17 October 2011
REFERENCES
- 1. Altas RM. 1993. Acid broth, p 52 In Parks LC. (ed), Handbook of microbiological media. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Ausubel F, et al. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 4. Bumford AA, Spilker T, LiPuma J. 2010. Epidemiology of Burkholderia infection in U.S. CF patients. International Burkholderia cepacia Working Group, Seattle, WA [Google Scholar]
- 5. Burtnick MN, Woods DE. 1999. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 43:2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campos MA, et al. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72:7107–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng HP, Lessie TG. 1994. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J. Bacteriol. 176:4034–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corey M, Farewell V. 1996. Determinants of mortality from cystic fibrosis in Canada, 1970–1989. Am. J. Epidemiol. 143:1007–1017 [DOI] [PubMed] [Google Scholar]
- 9. Cox AD, Wilkinson SG. 1991. Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol. Microbiol. 5:641–646 [DOI] [PubMed] [Google Scholar]
- 10. Cvejic JH, et al. 2000. Bacterial triterpenoids of the hopane series as biomarkers for the chemotaxonomy of Burkholderia, Pseudomonas and Ralstonia spp. FEMS Microbiol. Lett. 183:295–299 [DOI] [PubMed] [Google Scholar]
- 11. Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16:821–830 [DOI] [PubMed] [Google Scholar]
- 12. Dubarry N, Du W, Lane D, Pasta F. 2010. Improved electrotransformation and decreased antibiotic resistance of the cystic fibrosis pathogen Burkholderia cenocepacia strain J2315. Appl. Environ. Microbiol. 76:1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eberl M, et al. 2002. Accumulation of a potent gammadelta T-cell stimulator after deletion of the lytB gene in Escherichia coli. Immunology 106:200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glukhov E, Stark M, Burrows LL, Deber CM. 2005. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J. Biol. Chem. 280:33960–33967 [DOI] [PubMed] [Google Scholar]
- 16. Govan JR, Brown AR, Jones AM. 2007. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2:153–164 [DOI] [PubMed] [Google Scholar]
- 17. Hancock RE, Raffle VJ, Nicas TI. 1981. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 19:777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 19. Horbach S, Neuss B, Sahm H. 1991. Effect of azasqualene on hopanoid biosynthesis and ethanol tolerance of Zymomonas mobilis. FEMS Microbiol. Lett. 79:347–350 [Google Scholar]
- 20. Isles A, et al. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206–210 [DOI] [PubMed] [Google Scholar]
- 21. Kannenberg EL, Poralla K. 1999. Hopanoid biosynthesis and function in bacteria. Naturwissenschaften 86:168–176 [Google Scholar]
- 22. Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 188:2073–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loutet SA, Mussen LE, Flannagan RS, Valvano MA. 2011. A two-tier model of polymyxin B resistance in Burkholderia cenocepacia. Environ. Microbiol. Rep. 3:278–285 [DOI] [PubMed] [Google Scholar]
- 25. Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front. Microbiol. 2:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156 [DOI] [PubMed] [Google Scholar]
- 27. Manniello JM, Heymann H, Adair FW. 1978. Resistance of spheroplasts and whole cells of Pseudomonas cepacia to polymyxin B. Antimicrob. Agents Chemother. 14:500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore RA, Bates NC, Hancock RE. 1986. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 29:496–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore RA, Chan L, Hancock RE. 1984. Evidence for two distinct mechanisms of resistance to polymyxin B in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore RA, Hancock RE. 1986. Involvement of outer membrane of Pseudomonas cepacia in aminoglycoside and polymyxin resistance. Antimicrob. Agents Chemother. 30:923–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicas TI, Hancock RE. 1983. Alteration of susceptibility to EDTA, polymyxin B and gentamicin in Pseudomonas aeruginosa by divalent cation regulation of outer membrane protein H1. J. Gen. Microbiol. 129:509–517 [DOI] [PubMed] [Google Scholar]
- 32. Nzula S, Vandamme P, Govan JR. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 50:265–269 [DOI] [PubMed] [Google Scholar]
- 33. Ortega X, et al. 2009. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J. Biol. Chem. 284:21738–21751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pearson A, Flood Page SR, Jorgenson TL, Fischer WW, Higgins MB. 2007. Novel hopanoid cyclases from the environment. Environ. Microbiol. 9:2175–2188 [DOI] [PubMed] [Google Scholar]
- 35. Peeters E, Nelis HJ, Coenye T. 2009. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 64:801–809 [DOI] [PubMed] [Google Scholar]
- 36. Perzl M, et al. 1998. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim. Biophys. Acta 1393:108–118 [DOI] [PubMed] [Google Scholar]
- 37. Rajyaguru JM, Muszynski MJ. 1997. Enhancement of Burkholderia cepacia antimicrobial susceptibility by cationic compounds. J. Antimicrob. Chemother. 40:345–351 [DOI] [PubMed] [Google Scholar]
- 38. Rajyaguru JM, Muszynski MJ. 1998. Sensitization of Burkholderia cepacia to antibiotics by cationic drugs. J. Antimicrob. Chemother. 41:277–280 [DOI] [PubMed] [Google Scholar]
- 39. Rezanka T, Siristova L, Melzoch K, Sigler K. 2010. Hopanoids in bacteria and cyanobacteria-their role in cellular biochemistry and physiology, analysis and occurrence. Mini Rev. Org. Chem. 7:300–313 [Google Scholar]
- 40. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Schmidt A, Bringer-Meyer S, Poralla K, Sahm H. 1986. Effect of alcohols and temperature on the hopanoid content of Zymomonas mobilis. Appl. Microbiol. Biotechnol. 25:32–36 [Google Scholar]
- 42. Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195–1204 [DOI] [PubMed] [Google Scholar]
- 43. Speert DP, Bond M, Woodman RC, Curnutte JT. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524–1531 [DOI] [PubMed] [Google Scholar]
- 44. Stieritz DD, Holder IA. 1975. Experimental studies of the pathogenesis of infections due to Pseudomonas aeruginosa: description of a burned mouse model. J. Infect. Dis. 131:688–691 [DOI] [PubMed] [Google Scholar]
- 45. Vermis K, Vandamme PA, Nelis HJ. 2003. Burkholderia cepacia complex genomovars: utilization of carbon sources, susceptibility to antimicrobial agents and growth on selective media. J. Appl. Microbiol. 95:1191–1199 [DOI] [PubMed] [Google Scholar]
- 46. Welander PV, et al. 2009. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191:6145–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 48. Whiteford ML, et al. 1995. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 50:1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woods D. 1984. Oligonucleotide screening of cDNA libraries. Focus 6:1–2 [Google Scholar]
- 50. Zahariadis G, Levy MH, Burns JL. 2003. Cepacia-like syndrome caused by Burkholderia multivorans. Can. J. Infect. Dis. 14:123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206–1215 [DOI] [PubMed] [Google Scholar]
- 52. Zlosnik JE, Henry DA, Speert DP. 2011. Burkholderia cepacia complex infections in Vancouver and British Columbia from 1981 to 2010. International Burkholderia cepacia Working Group, Prague, Czech Republic. [Google Scholar]