Abstract
An experiment was conducted in animal facilities to compare the impacts of four avian colibacillosis treatments—oxytetracycline (OTC), trimethoprim-sulfadimethoxine (SXT), amoxicillin (AMX), or enrofloxacin (ENR)—on the susceptibility of Escherichia coli in broiler intestinal tracts. Birds were first orally inoculated with rifampin-resistant E. coli strains bearing plasmid genes conferring resistance to fluoroquinolones (qnr), cephalosporins (blaCTX-M or blaFOX), trimethoprim-sulfonamides, aminoglycosides, or tetracyclines. Feces samples were collected before, during, and after antimicrobial treatments. The susceptibilities of E. coli strains were studied, and resistance gene transfer was analyzed. An increase in the tetracycline-resistant E. coli population was observed only in OTC-treated birds, whereas multiresistant E. coli was detected in the dominant E. coli populations of SXT-, AMX-, or ENR-treated birds. Most multiresistant E. coli strains were susceptible to rifampin and exhibited various pulsed-field gel electrophoresis profiles, suggesting the transfer of one of the multiresistance plasmids from the inoculated strains to other E. coli strains in the intestinal tract. In conclusion, this study clearly illustrates how, in E. coli, “old” antimicrobials may coselect antimicrobial resistance to recent and critical molecules.
INTRODUCTION
Antimicrobials are frequently administered to poultry to control bacterial diseases such as colibacillosis, mycoplasmosis, or other respiratory or intestinal diseases (11). These treatments are usually given to the whole flock via the drinking water or feed over several days and may thus impact the equilibrium and susceptibility of bacteria present in the intestinal flora. The aim of this study was to analyze, under experimental conditions, the impacts of different antimicrobials, such as oxytetracycline, trimethoprim-sulfadimethoxine, amoxicillin, or enrofloxacin, on the antimicrobial resistance of Escherichia coli strains from the digestive tract when birds are already colonized with a multidrug-resistant (MDR) E. coli strain, particularly a strain harboring transmissible fluoroquinolone resistance, extended-spectrum beta-lactamases, or plasmid AmpC beta-lactamase genes.
MATERIALS AND METHODS
Bacterial strains.
Two fecal multidrug-resistant E. coli strains and one avian-pathogenic E. coli (APEC) strain were used for the inoculation of birds (Table 1). The multidrug-resistant E. coli strains 177pMG252 and 43pMG298 were obtained as described previously (23). Both strains were rifampin resistant and contained, respectively, plasmids pMG252 and pMG298, kindly provided by G. Jacoby, Lahey Clinic, Burlington, MA (21, 22). The pMG252 plasmid contains the qnrA1 and blaFOX-5 genes, which confer reduced susceptibility to fluoroquinolones and cephamycins, respectively, as well as resistance genes against streptomycin, sulfonamide, trimethoprim, chloramphenicol, gentamicin, and kanamycin. The pMG298 plasmid contains the qnrB1 and blaCTX-M15 genes, which confer reduced susceptibility to fluoroquinolones and resistance to cephalosporins, respectively, as well as resistance genes against sulfonamide, chloramphenicol, streptomycin, kanamycin, gentamicin, tetracycline, and trimethoprim.
Table 1.
Characteristics of E. coli strains
| E. coli strain | Serotypea | Phylogenetic group | Resistance genes | Inc groupb | MIC (mg/liter)c |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | TIO | FOX | NAL | CIP | TET | SXT | CHL | STR | KAN | GEN | RIF | |||||
| 177pMG252 | Neg | A | blaFOX, qnrA, tetA | A/C, F | >32 | >32, 16 | >8 | >32 | 32 | 1 | >32 | >4, 76 | >32 | >64 | 64 | 16 | >128 |
| 43pMG298 | Neg | D | blaCTX-M, qnrB, tetA | NT | >32 | 32, 16 | >8 | 32 | 8 | 2 | >32 | >4, 76 | >32 | >64 | >64 | >16 | >128 |
| O78K80 | O78:K80 | A | I1, FIB | 4 | 4, 8 | 0.5 | 8 | 4 | 0.03 | >4 | ≤0.12, 2.38 | 8 | ≤32 | 16 | 1 | ≤16 | |
Neg, negative with anti-O78:K80 antisera.
NT, not typeable.
AMP, ampicillin; AMC, amoxicillin-clavulanic acid; TIO, ceftiofur; FOX, cefoxitin; NAL, nalidixic acid; CIP, ciprofloxacin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; STR, streptomycin; KAN, kanamycin; GEN, gentamicin; RIF, rifampin.
The APEC strain was an O78:K80 E. coli isolate susceptible to tetracycline, trimethoprim-sulfadimethoxazole, amoxicillin, and enrofloxacin, kindly provided by R. Froyman (Bayer AG, Mannheim, Germany). The MICs for the three inoculated strains, determined by a broth microdilution test using CMV1AGNF Sensititre plates (Biocentric, Bandol, France) and Mueller-Hinton broth, are given in Table 1.
Experimental model.
Assays were performed in compliance with French animal welfare regulations (authorization B-22-741-1, given by the County Veterinary Services Department). The experimental model has been described previously (15) and is summarized in Table 2. Briefly, 360 specific-pathogen-free (SPF) chicks were randomly allocated to six groups that were placed in six pressure-controlled animal rooms, with filtered air and controlled temperature. The different groups of birds were designated NINT (non-APEC infected, nontreated), APEC-NT (APEC infected, nontreated), APEC-AMX (APEC infected, amoxicillin treated), APEC-SXT (APEC infected, trimethoprim-sulfadimethoxine treated), APEC-OTC (APEC infected, oxytetracycline treated), and APEC-ENR (APEC infected, enrofloxacin treated). At the ages of 14, 15, and 16 days, the birds from the six groups were orally administered a daily 0.1-ml dose of cultures of E. coli 177pMG252 and E. coli 43pMG298. The titers obtained for E. coli 177pMG252 on the three inoculation days ranged from 2 × 109 to 7 × 109 CFU/ml, and those for E. coli 43pMG298 ranged from 4 × 109 to 9 × 109 CFU/ml. At the age of 17 days, all birds were vaccinated with an infectious bronchitis vaccine (Bioral H120; Merial, Lyon, France) in order to increase the clinical signs of colibacillosis. On day 20, birds from five groups (APEC-NT, APEC-AMX, APEC-OTC, APEC-SXT, and APEC-ENR) were inoculated in the left air sac with 0.1 ml of a culture of the APEC O78:K80 strain, and one group, NINT, was left unchallenged. The titer of the APEC inoculum was 4.2 × 108 CFU/ml. Medication was initiated 1 day later (day 21). Birds from the APEC-OTC, APEC-SXT, APEC-AMX and APEC-ENR groups were treated, respectively, with therapeutic doses of 20 mg/kg of body weight of oxytetracycline (Terramycin; Pfizer, Paris, France), 28 and 6 mg/kg of sulfadimethoxine-trimethoprim (Trisulmix liquid; Coophavet, Ancenis, France), 10 mg/kg of amoxicillin (Suramox 10%; Virbac, Carros, France), or 10 mg/kg of enrofloxacin (Baytril 10%; Bayer, Puteaux, France), as recommended by the suppliers. Treatment doses were calculated daily according to body weight and the group's water consumption (Table 2). The antibiotics were given in the drinking water. The medicated solutions were freshly prepared daily. Oxytetracycline was given on three consecutive days, whereas the other medications were administered for 5 days, as indicated in the summary of product characteristics. The actual mean drug consumption calculated for each day according to body weight and water consumption for that day was 22.5 mg/kg for oxytetracycline, 33.4 and 7.15 mg/kg for sulfadimethoxine-trimethoprim, 9.95 mg/kg for amoxicillin, and 9.05 mg/kg for enrofloxacin.
Table 2.
Experimental design
| Group | Inoculation of digestive strainsa on days 14–16 | Infectious bronchitis vaccine on day 17 | Inoculation of the air sac with the APEC O78:K80 strain on day 20 | Treatment |
|---|---|---|---|---|
| NINT | Yes | Yes | No | None |
| APEC-NT | Yes | Yes | Yes | None |
| APEC-OTC | Yes | Yes | Yes | Oxytetracycline (20 mg/kg), days 21–23 |
| APEC-SXT | Yes | Yes | Yes | Sulfadimethoxine-trimethoprim (28 and 6 mg/kg), days 21–25 |
| APEC-AMX | Yes | Yes | Yes | Amoxicillin (10 mg/kg), days 21–25 |
| APEC-ENR | Yes | Yes | Yes | Enrofloxacin (10 mg/kg), days 21–25 |
E. coli 177pMG252 and E. coli 43pMG298.
Fecal samples were collected from 10 birds in each group either before treatment (days 18 and 21), during treatment (days 21 and 24), or after treatment (days 35, 42, 49, and 63). For each group and each day, the 10 samples were pooled into five pools of 2 fecal samples. The pools were then inoculated onto MacConkey medium alone or supplemented with gentamicin (8 mg/liter; Sigma-Aldrich, Saint-Quentin Fallavier, France) or cefoxitin (16 mg/liter; Sigma-Aldrich) for detection of the resistant inoculated E. coli strains. The numbers of E. coli colonies appearing on the different media were scored as follows: 1, fewer than 15 colonies; 2, 15 to 200 colonies; 3, >200 colonies; 4, confluent culture. The mean score per group and per period was calculated. One isolate per pool and per supplemented medium was randomly selected and was immediately tested for susceptibility by a disk diffusion assay according to CA-SFM recommendations (7), using disks of nalidixic acid (30 μg), oxolinic acid (10 μg), flumequine (30 μg), ciprofloxacin (5 μg), enrofloxacin (5 μg), difloxacin (10 μg), amoxicillin (25 μg), amoxicillin-clavulanic acid (20 and 10 μg), cefotaxime (30 μg), cefoxitin (30 μg), chloramphenicol (30 μg), gentamicin (15 μg), rifampin (30 μg), sulfonamides (200 μg), trimethoprim (5 μg), and tetracycline (30 IU) (Bio-Rad, Marnes la Coquette, France). Interpretation was carried out according to the CA-SFM 2009 recommendations (7). Unfortunately, isolates obtained directly on MacConkey medium were neither tested immediately nor stored. Thus, isolation on a nonsupplemented medium was performed again after thawing of pooled feces samples stored at −65°C, and the susceptibility of the E. coli isolates obtained on the nonsupplemented medium was tested by disk diffusion as described above. PCR was carried out to search for the presence of the qnr, blaCTX-M, and blaFOX genes (8, 31). The isolates' phylogenetic groups were determined according to the method of Clermont et al. (13). DNA profiles were compared by pulsed-field gel electrophoresis (PFGE), after restriction with XbaI (12). The presence of plasmids in selected isolates was checked by conjugation with the rifampin-resistant E. coli strain J5 or the rifampin- and gentamicin-resistant E. coli strain UA6190 (24) using an agar medium supplemented with rifampin alone (200 mg/liter) or with rifampin (150 mg/liter), gentamicin (4 mg/liter), and either ampicillin (10 mg/liter) or tetracycline (8 mg/liter). The susceptibility profiles of the transconjugants were then studied, and the qnr and blaFOX or blaCTX-M resistance genes were detected by PCR. The presence of the tetA to tetD genes was determined by PCR (27), and replicon typing was performed according to the method of Carattoli et al. (6).
Quantification of the copies of qnr genes.
Total DNA was prepared from fecal samples using commercial extraction kits (QIAamp DNA stool minikit; Qiagen, France). Quantitative PCR was performed using primers targeting the DNA sequences of the qnrA and qnrB genes. These primers were defined on the basis of the qnrA1 (GenBank accession no. DQ831140) or qnrB1 (GenBank accession no. DQ351241) gene of plasmid pMG252 or pMG298, respectively. The amplified products were detected with TaqMan probes (Sigma-Aldrich, France) (Table 3) labeled at the 5′ end with the reporter dye 6-carboxyfluorescein and at the 3′ end with Black Hole Quencher. Primers and probes were designed with Beacon Designer software, version 6.00 (Premier Biosoft International, Palo Alto, CA). The reaction mixture consisted of 5 μl DNA, 300 nM each primer of either the qnrA or the qnrB gene, 200 nM TaqMan probes, and 12.5 μl of iQ Supermix (20 mmol/liter Tris-HCl, 50 mmol/liter KCl, 3 mmol/liter MgCl2 [pH 8.4], 800 μmol/liter of each deoxyribonucleoside triphosphate, 0.625 U Taq polymerase, and stabilizers) (Bio-Rad, France) in a final volume of 25 μl per reaction. PCR amplification was performed in duplicate with a Chromo4 real-time PCR detection system (Bio-Rad) as follows: 1 cycle at 95°C for 3 min, followed by 45 cycles at 95°C for 15 s and 60°C for 60 s. Each primer pair was validated by verifying that PCR efficiency was above 0.90. To generate a standard curve for real-time PCR, 10-fold serial dilutions ranging from 102 to 108 fg of qnrA or qnrB DNA were included in each experiment. The threshold cycle (CT) was determined for the samples and was compared with the standard curves in order to quantify the samples. Samples in which both duplicates had a CT value below 45 were regarded as positive.
Table 3.
Primers and probes for the quantification of qnr genes
| Primer or probe | Sequence (5′ → 3′)a | Target | Amplicon length (bp) |
|---|---|---|---|
| Primers | |||
| QnrA1-F | CTTCCTTGATGGGCTCAGATC | qnrA | 375 |
| QnrA1-R | CCAGATCGGCAAAGGTTAGG | ||
| QnrB1-F | GTAGCGCATATATCACGAATACC | qnrB | 481 |
| QnrB1-R | AGATCTGAACCACTGAACGTC | ||
| Probes | |||
| QnrA | FAM-CAGCCCCGCAGATTGACCTGTTG-BHQ1 | qnrA | |
| QnrB | FAM-ACCTGGGCACCTATCCAACGGTTT-BHQ1 | qnrB |
FAM, 6-carboxyfluorescein (a fluorescence reporter dye); BHQ, Black Hole Quencher.
Statistical tests.
The distributions of the isolation scores and of the resistance profiles of the groups were compared to those of the NINT group by a chi-square test or Fisher's exact test. Results were considered significantly different if the P value was <0.05.
RESULTS
The different oral inoculations with E. coli strains did not affect the birds' health, whereas inoculation of APEC into the air sac resulted in severe clinical signs and mortality. Thus, treatment began on the day following APEC inoculation.
Isolation of E. coli from fecal samples.
The mean isolation scores on MacConkey medium from all groups and all periods were similar and higher than 2.4 except for the APEC-ENR group during treatment. The distributions of the scores in this group were statistically different, with a mean score of 0.5. However, from day 35 on, the mean scores of this group rose again to >3.
After oral inoculation of the birds with the resistant E. coli strains 177pMG252 and 43pMG298 but before treatment, the mean scores obtained for the different groups on gentamicin-supplemented medium were not significantly different, mostly below 2. During treatment, the isolation scores for the APEC-AMX group were significantly higher than those for the noninfected, nontreated group. After treatment, the scores of the APEC-AMX, APEC-SXT, and APEC-ENR groups were significantly higher than those of the noninfected, nontreated group. The same phenomenon was observed for the same groups with the cefoxitin-supplemented medium.
Analysis of E. coli isolates recovered on nonsupplemented medium.
After thawing of samples, E. coli isolates were recovered on nonsupplemented medium from feces samples collected from all chicken groups except the APEC-ENR group during the treatment period. Disk diffusion assays revealed different profiles among the isolates: they were either susceptible to all molecules tested (SUS), resistant to tetracycline only (Tetr), or resistant to amoxicillin only (Amxr). Many isolates, referred to below as multidrug resistant (MDR), were resistant at least to amoxicillin, chloramphenicol, and trimethoprim-sulfamethoxazole, with resistance either to cefotaxime (MDR-CTX) or to cefoxitin (MDR-FOX) and with either no rifampin inhibition zone (Rifr) or rifampin inhibition zones of >10 mm (Rifs). The mean inhibition zones for most MDR isolates with oxolinic acid, nalidixic acid, flumequine, ciprofloxacin, difloxacin, or enrofloxacin were 5.3 mm (nalidixic acid) to 9 mm (difloxacin) smaller than those obtained for SUS or Tetr isolates. The percentages of resistance profiles obtained for the different groups during or after treatment are given in Fig. 1.
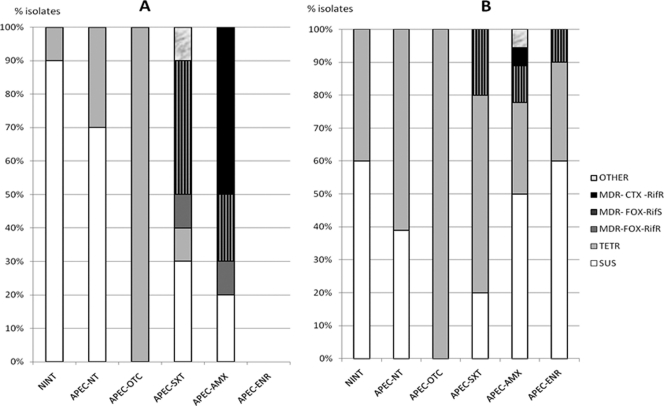
Fig 1.
Percentages of E. coli isolates exhibiting the different profiles that were recovered on nonsupplemented medium during (A) or after (B) treatment.
Before treatment, 55 out of 83 isolates obtained on nonsupplemented medium had a SUS profile; the others were Tetr (22 isolates), Amxr (1 isolate), and MDR-CTX-Rifr (5 isolates). No isolate with an MDR-FOX profile could be detected on nonsupplemented medium before treatment. For the two nontreated groups, the isolates recovered exhibited either a SUS profile (35/58 isolates) or a Tetr profile (23/58). No other profiles were observed.
In the APEC-OTC group, all the isolates obtained from day 24 to day 63 were Tetr. The ratios of Tetr isolates during and after treatment were significantly higher in the APEC-OTC group than in the other groups.
In the APEC-SXT group, during treatment, 5 out of 10 isolates were MDR, most frequently with cefoxitin resistance and a wide rifampin diameter (MDR-FOX-Rifs). This proportion of MDR isolates was significantly higher than those in the nontreated and APEC-OTC groups. After treatment, this profile was still present, but most isolates were SUS or Tetr. During treatment, one isolate with no rifampin inhibition zone showed resistance to chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole but not to amoxicillin or fluoroquinolones.
In the APEC-AMX group, during treatment, 8 out of 10 isolates were MDR, and the most frequently observed profile was MDR with resistance to cefotaxime and a small rifampin inhibition zone (MDR-CTX-Rifr). This proportion of MDR isolates was significantly higher than those in the nontreated and APEC-OTC groups. After treatment, MDR isolates were still observed, but the profiles found most frequently were SUS or Tetr.
In the APEC-ENR group, after treatment, 2 E. coli isolates were MDR-FOX-Rifs, and the other 18 were either SUS or Tetr. The inhibition diameters of quinolones and fluoroquinolones for isolates from this enrofloxacin-treated group were similar to those for isolates from other groups.
No MDR-CTX-Rifs isolate was detected among the isolates obtained on nonsupplemented medium.
Analysis of phylogenetic groups showed that most E. coli isolates obtained on nonsupplemented medium belonged to group B1 (182/249 isolates) as opposed to group D (47 isolates) or group A (20 isolates). No isolate belonging to group B2 was detected. For all groups and all periods, the B1 isolates were dominant, except in the APEC-AMX group during treatment, when of the 10 isolates tested, only 1 belonged to group B1, 3 to group A, and 6 to group D, with 5 of these 6 exhibiting an MDR-CTX-Rifr profile. Indeed, all the isolates with MDR-CTX-Rifr profiles belonged to group D, as does E. coli 43pMG298. The 2 isolates with an MDR-FOX-Rifr profile belonged to group A, as does E. coli 177pMG252, whereas isolates with MDR-FOX-Rifs profiles were in group B1 (11 isolates) or D (3 isolates).
Analysis of 115 strains resistant to tetracycline—with or without resistance to other agents—showed that all contained the tetA gene, and 1 contained both the tetA and tetB genes. PFGE analysis revealed that Tetr isolates belonging to phylogenetic group B1 exhibited various PFGE profiles, whereas one profile was frequently observed in isolates belonging to phylogenetic group D, whatever the experimental group or sampling date (Fig. 2). This particular profile was already present before treatment. Transconjugants prepared from 17 Tetr isolates indicated that the tetA gene was located on conjugative IncF, IncK, IncFIA, IncFIB, or IncI1 plasmids.
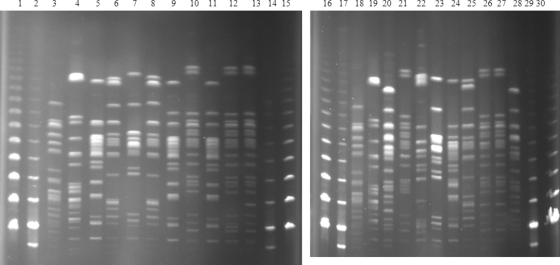
Fig 2.
PFGE analysis of XbaI-digested genomic DNAs of Tetr strains. Lanes: 1, 15, 16, and 30, Lambda PFGE marker (New England Biolabs); 2,14, 17, and 29, Low Range PFGE marker (New England Biolabs); 3 and 18, E. coli 177pMG252; 4 and 19, E. coli 43pMG298; 5 to 10, Tetr E. coli isolates obtained before treatment; 11, 12, 13, and 18, Tetr isolates obtained during treatment; 20 to 28, Tetr isolates obtained after treatment. Isolates whose patterns are shown in lanes 5, 6, 7, 8, 9, 11, 20, 22, 23, 24, 25, and 28 belong to phylogenetic group B1; those whose patterns are shown in lanes 10, 12, 13, 21, 26, and 27 belong to phylogenetic group D and have profiles similar to each other.
PCR results showed that the 11 tested strains with MDR-CTX profiles had a group1 blaCTX-M gene and a qnrB gene (except for 1 isolate that showed larger inhibition diameters for quinolones), and the 16 tested isolates with MDR-FOX profiles harbored a blaFOX and a qnrA gene (except for 1 isolate that showed larger inhibition diameters for quinolones). PFGE revealed various profiles, suggesting different transfer events (Fig. 3). Transconjugants prepared from 14 MDR-FOX-Rifs E. coli isolates acquired the different resistances of pMG252. Moreover, PCR showed that all transconjugants harbored an IncA/C replicon, as did pMG252 and their respective donor strains. The donor strains sometimes also harbored an IncF replicon (also present in 177pMG252) or IncFIA/FIB or IncI1 replicons.
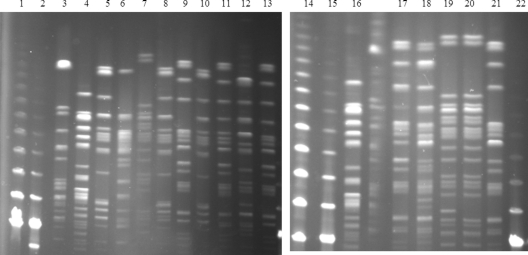
Fig 3.
PFGE analysis of XbaI-digested genomic DNAs of MDR-FOX-Rifs strains. Lanes: 1 and 14, Lambda PFGE marker (New England Biolabs); 2 and 15, Low Range PFGE marker (New England Biolabs); 3, E. coli 43pMG298; 4 and 16, E. coli 177pMG252; 5, an APEC-SXT group isolate on day 24; 6 and 7, APEC-AMX group isolates on day 24; 8, 9, and 10, APEC-SXT group isolates on day 27; 11, an APEC-SXT group isolate on day 35; 12 and 13, APEC-ENR group isolates on day 35; 17 and 18, APEC-SXT group isolates on day 42; 19 and 20, APEC-AMX group isolates on day 42; 21, an APEC-SXT group isolate on day 63.
Analysis of E. coli isolates obtained on supplemented medium.
Disk diffusion assays of isolates obtained on supplemented medium revealed that most isolates were multidrug resistant, with either an MDR-FOX (209 isolates) or an MDR-CTX (64 isolates) profile. Before treatment or in nontreated groups, 65/66 MDR isolates were highly resistant to rifampin. Similarly, after treatment with OTC, the seven MDR-FOX isolates detected were highly resistant to rifampin. In contrast, during or after treatment with sulfadimethoxine-trimethoprim, amoxicillin, or enrofloxacin, only 6/55, 8/38, and 1/41 MDR-FOX isolates, respectively, were highly resistant to rifampin. The proportions of highly rifampin resistant isolates in the APEC-SXT, APEC-AMX, and APEC-ENR groups were significantly different from those in nontreated groups.
Of the 64 MDR-CTX isolates, only 1—obtained after amoxicillin treatment—had a large rifampin inhibition diameter. This isolate (090629-EC14G) belonged to phylogenetic group B1, and its PFGE profile differed from that of E. coli 43pMG298. Transconjugants prepared from this isolate on CTX- and rifampin-supplemented medium exhibited the resistance profile corresponding to the transfer of pMG298, and the replicon, like pMG298, was not typeable by PCR. Besides the MDR-FOX and MDR-CTX profiles, a few isolates obtained had various profiles resulting from different combinations of resistance to amoxicillin, cefoxitin, chloramphenicol, trimethoprim-sulfonamide, tetracycline, streptomycin, fluoroquinolones, or aminoglycosides. Most of these isolates were not highly resistant to rifampin. For all these isolates obtained on supplemented media, PCR results for the presence of blaFOX, blaCTX-M, or qnr genes were consistent with their resistance profiles, and they had IncA/C replicons, such as pMG252.
Number of qnr gene copies.
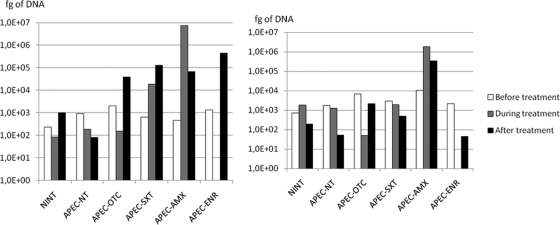
A total of 492 fecal samples were analyzed for the number of qnr gene copies (Fig. 4). For the qnrA gene, in nontreated groups, samples contained less than 103 fg, whereas after the different treatments, more than 3.9 × 104 fg was present, with a maximum of 4.4 × 105 fg for the APEC-ENR group. During treatment, the maximum level was obtained for the APEC-AMX group, with 7.4 ×106 fg. For the qnrB gene, the only significant increase was observed for the APEC-AMX group, with levels above 105 to 106 fg, whereas values for the other groups during or after treatment were below 2.1 × 103 fg.
Fig 4.
Quantification of qnr genes (expressed in femtograms of DNA). (Left) qnrA genes; (right) qnrB genes. Open bars, before treatment; shaded bars, during treatment; filled bars, after treatment.
DISCUSSION
As observed previously (23), the oral inoculation of the two E. coli strains 177pMG252 and 43pMG298 was successful. Strains exhibiting the same resistance profiles were isolated from nontreated birds until the end of the experiment. In these two groups, the two inoculated strains were maintained in the intestinal flora as a subpopulation, from a few days after inoculation onwards. They were never detected on nonsupplemented medium but could be obtained on a cefoxitin- or gentamicin-supplemented medium. In these nontreated groups, only 1 of 66 MDR-FOX isolates had a rifampin inhibition diameter of >10 mm. This suggests that in nontreated groups, the plasmids were rarely transferred to other E. coli strains.
In APEC-OTC birds, the treatment resulted in an important selection of isolates resistant to tetracycline only. Such isolates resistant to tetracycline only were already observed in the dominant flora of the feces of the SPF birds; their proportions ranged from 10% to 40% in the noninfected, nontreated chicken group during the experiment. These strains resistant to tetracycline only seemed particularly competitive in the birds' intestinal flora, since both during and after OTC administration, no other profile could be detected on nonsupplemented medium. This important intestinal population of Tetr E. coli may also explain the selection of tetracycline-resistant APEC observed in internal organs, as reported previously (15). Several Tetr isolates of phylogenetic group D present before treatment were observed in different groups from before treatment up to the last sampling day. Although 177pMG252 and 43pMG298 were tetracycline resistant, the scores obtained for APEC-OTC birds on supplemented medium were not significantly different from those for noninfected, nontreated birds, suggesting that selection of MDR E. coli strains was limited in APEC-OTC birds. However, a slight increase in the quantity of qnrA genes was observed after the end of OTC treatment, suggesting that fluoroquinolone resistance may have been transferred to other bacteria. Other experiments concerning the impact of tetracycline treatments of animals have been reported. In 1995, Guillot and Boucoud (20) observed that tetracycline treatment could enhance the transfer of conjugative plasmid pGB99, encoding resistance to chloramphenicol, tetracycline, sulfonamide, and trimethoprim, in the guts of chickens carrying an avian flora. However, van Essen-Zandbergen et al. (30) observed that doxycycline (or trimethoprim-sulfamethoxazole) administration to chickens had neither a positive nor a negative effect on the in vivo transfer of an IncFIB plasmid harboring both tetracycline resistance genes and a class 1 integron with the dfrA1-aadA1 gene cassettes. Furthermore, according to the observations of Fairchild et al. (16), the tetracycline exposure history of the chickens studied did not modify the variety of tet resistance determinants harbored by the birds. Indeed, in commercial poultry, the tetracycline resistance level of commensal E. coli may be very high (1), and it is probable that, under such conditions, tetracycline administration cannot induce much variation in this resistance rate.
In the APEC-SXT group, MDR-FOX E. coli strains were detected in the dominant flora. Again, this high number of E. coli strains containing the pMG252 plasmid may have contributed to the acquisition of the pMG252 plasmid by one APEC isolate recovered from the air sacs of a chicken (15). During and after treatment, eight out of nine isolates were probably transconjugants, since their rifampin inhibition diameters exceeded 10 mm and they belonged to phylogenetic group B1 or D, unlike E. coli strain 177pMG252, which had been inoculated. The results obtained on supplemented media confirmed that there were more colonies of MDR-FOX transconjugants than of the strain inoculated, E. coli 177pMG252, and various PFGE profiles could be observed. Few authors have studied the effect of trimethoprim-sulfonamide administration on intestinal flora. According to the results of a study carried out under field conditions, published by da Costa et al. (14), SXT treatment resulted in increases in the levels of resistance to kanamycin, ampicillin, tetracycline, sulfonamide, trimethoprim, streptomycin, gentamicin, and enrofloxacin, but the mechanisms of these resistances and their genetic bases were not reported.
In the APEC-AMX group, both during and after treatment, MDR-FOX and MDR-CTX isolates could be detected in the dominant flora. It is noteworthy that MDR-CTX isolates in the dominant flora were detected only in the amoxicillin-treated group, especially during treatment, when they apparently reached a proportion of 50% of the E. coli population. Analysis of isolates obtained on supplemented media showed that among the MDR-CTX isolates from this group, only one isolate had a large rifampin inhibition diameter. Its phylogenetic group and PFGE profile confirmed that it differed from the strain inoculated, 43pMG298. In contrast, in this group, after treatment, the MDR-FOX isolates were predominantly transconjugants. In this group only, increases in the copy numbers of both the qnrA and qnrB genes (the latter was borne on pMG298 as blaCTX-M) were observed. Field trials evaluating the influence of various medications on intestinal E. coli in growing broilers showed that amoxicillin increased the prevalence of resistance to ampicillin, cephalothin, streptomycin, kanamycin, and chloramphenicol in E. coli (14). Bibbal et al. (4, 5) showed that after intramuscular or oral administration of ampicillin to conventional pigs, the percentage of ampicillin-resistant Enterobacteriaceae and the number of blaTEM copies increased in the feces. Moreover, their observations revealed the coselection of resistance to tetracycline, streptomycin, trimethoprim, sulfonamides, and chloramphenicol, as well as the selection of preexisting multiresistant E. coli isolates belonging to one specific PFGE genotype. The effect of beta-lactam treatments on the in vivo selection or transfer of cephalosporin resistance genes in Enterobacteriaceae was also studied by Cavaco et al. (10), who observed that administration of amoxicillin—and, to a greater extent, ceftiofur or cefquinome—to SPF pigs resulted in higher titers of cephalosporin-resistant E. coli in the pig intestinal flora and reported a possible emergence of strains that had acquired blaCTX-M genes by horizontal transfer. Faure et al. (17, 18) reported experiments with axenic or human-flora-associated rats showing that the recipient and transconjugant populations decreased rapidly in size after medication but that the transfer of plasmid-mediated blaCTX-M-9 from Salmonella enterica serotype Virchow to Enterobacteriaceae was not enhanced by cefixime treatment.
In the APEC-ENR group, a dramatic drop in the number of E. coli isolates was observed during the treatment period, and no E. coli isolate could be obtained from the frozen samples. Within a few days after treatment, the E. coli population increased, and 18/20 isolates obtained on nonsupplemented medium had SUS or Tetr profiles, whereas 2 isolates were probably transconjugants. On supplemented media, many MDR profiles were detected, with a very large proportion of pMG252 transconjugants. Their inhibition diameters for quinolones and fluoroquinolones did not seem compatible with any resistance mechanism other than the presence of qnr genes (9). A drop in the E. coli population to below the detection limit was reported previously by van Boven et al. (29) for individually housed chickens after enrofloxacin treatment at a dose of 50 ppm for 10 days. The E. coli population reappeared rapidly after the cessation of enrofloxacin administration, and no modification of its susceptibility to fluoroquinolones was observed. Similarly, Medders et al. (25) reported that enrofloxacin treatment of chickens did not result in enrofloxacin-resistant E. coli, confirming their in vitro observations concerning the mutation rates of avian intestinal coliform bacteria pressured with different fluoroquinolones. However, according to Barrow et al. (2), enrofloxacin treatment induced the replacement of the initial nalidixic acid-susceptible E. coli population by a nalidixic acid-resistant population, which persisted for several weeks after the end of drug administration. A similar result was reported by Miranda et al. (26). This phenomenon was not observed in the present study. The impact of different fluoroquinolones on the ecological balance of human microflora was reviewed by Sullivan et al. (28), who explained that the very high concentration of antimicrobials achieved in feces, although probably bound to fecal material, and the susceptibility of Enterobacteriaceae are the reasons for the drastic drop in their levels during treatment. An increased ciprofloxacin MIC for E. coli could be observed after treatment. However, Gismondo et al. (19) could not detect any selection of bacterial resistance following ciprofloxacin administration to human-gastrointestinal-flora-associated germfree mice. In pigs, according to Belloc et al. (3), a noticeable but transitory increase in the percentage of quinolone-resistant E. coli strains was detected after flumequine treatment.
Finally, the main finding of this study was that the presence of fluoroquinolone, cephalosporin, trimethoprim, or sulfonamide resistance genes on plasmids resulted in the selection of many multiresistant isolates during or after treatment, most of them resulting from the transfer of plasmid pMG252 to other E. coli isolates, whereas plasmid pMG298 was detected only once in a non-rifampin-resistant isolate. However, in contrast to our previous experiment (23), there was no attempt to analyze Enterobacteriaceae other than E. coli, and it is possible that the pMG298 plasmid could be transferred to other Enterobacteriaceae.
Conclusion.
Analysis of the intestinal flora of birds during and following administration of common antimicrobials could illustrate several important points. We could highlight the important coselective pressures of the different antimicrobials (SXT, AMX, and ENR), since the broiler flora already contained multiresistant Enterobacteriaceae. The multiresistant E. coli strains were present in the dominant flora mainly during treatment, but a few could be isolated in the dominant flora several weeks after the end of treatment, at an age compatible with slaughter, thus constituting a potential risk for the food chain. Interestingly, no such coselection was observed in the tetracycline-treated birds, and only tetracycline-monoresistant isolates could be isolated in the dominant flora of these birds up to the end of the trial. These different results must be considered in evaluating the risks involved in antibiotic usage in food animals.
ACKNOWLEDGMENTS
This work is part of the EVALU-FQ-VOL project, supported by the French Research Agency (ANR) (grant ANR-07-PNRA-007) and the Conseil Général des Côtes d'Armor.
We are very grateful to G. Jacoby of Lahey Clinic, Burlington, MA, for providing E. coli strains J53pMG252 (21) and J53pMG298. We thank M. Llagostera, UAB, Barcelona, Spain, for E. coli strain HB101 UA6190 and A. Carattoli for providing reference strains and protocols for replicon typing. Corinne Marois (ANSES, Ploufragan, France) is acknowledged for helpful discussions and Michel Amelot (ANSES, Ploufragan, France) for technical assistance with the animal facilities.
Footnotes
Published ahead of print 10 October 2011
REFERENCES
- 1. Agence Française de Sécurité Sanitaire des Aliments September 2009. FARM 2005–2006: French antimicrobial resistance monitoring program in bacteria of animal origin. http://www.afssa.fr/Documents/SANT-Ra-FARM2006.pdf
- 2. Barrow PA, Lovell MA, Szmolleny G, Murphy CK. 1998. Effect of enrofloxacin administration on excretion of Salmonella enteritidis by experimentally infected chickens and on quinolone resistance of their Escherichia coli flora. Avian Pathol. 27:586–590 [DOI] [PubMed] [Google Scholar]
- 3. Belloc C, Lam DN, Pellerin JL, Beaudeau F, Laval A. 2005. Effect of quinolone treatment on selection and persistence of quinolone-resistant Escherichia coli in swine faecal flora. J. Appl. Microbiol. 99:954–959 [DOI] [PubMed] [Google Scholar]
- 4. Bibbal D, et al. 2007. Impact of three ampicillin dosage regimens on selection of ampicillin resistance in Enterobacteriaceae and excretion of blaTEM genes in swine feces. Appl. Environ. Microbiol. 73:4785–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bibbal D, Dupouy V, Prere MF, Toutain PL, Bousquet-Melou A. 2009. Relatedness of Escherichia coli strains with different susceptibility phenotypes isolated from swine feces during ampicillin treatment. Appl. Environ. Microbiol. 75:2999–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 7. CA-SFM 2009. Comité de l'Antibiogramme de la Société Française de Microbiologie—Recommandations 2009. Société Française de Microbiologie, Paris, France [Google Scholar]
- 8. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394–397 [DOI] [PubMed] [Google Scholar]
- 9. Cavaco LM, Aarestrup FM. 2009. Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6′)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J. Clin. Microbiol. 47:2751–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L. 2008. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrob. Agents Chemother. 52:3612–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chauvin C, Clement C, Bruneau M, Pommeret D. 2007. Time-patterns of antibiotic exposure in poultry production—a Markov chains exploratory study of nature and consequences. Prev. Vet. Med. 80:230–240 [DOI] [PubMed] [Google Scholar]
- 12. Chu G, Vollrath D, Davis RW. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582–1585 [DOI] [PubMed] [Google Scholar]
- 13. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. da Costa PM, Bica A, Vaz-Pires P, Bernardo F. 2008. Effects of antimicrobial treatment on selection of resistant Escherichia coli in broiler fecal flora. Microb. Drug Resist. 14:299–306 [DOI] [PubMed] [Google Scholar]
- 15. Dheilly A, Bouder A, Le Devendec L, Hellard G, Kempf I. 2011. Clinical and microbial efficacy of antimicrobial treatments of experimental avian colibacillosis. Vet. Microbiol. 149:422–429 [DOI] [PubMed] [Google Scholar]
- 16. Fairchild AS, et al. 2005. Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71:5865–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faure S, Perrin-Guyomard A, Delmas JM, Chatre P, Laurentie M. 2010. Transfer of plasmid-mediated CTX-M-9 from Salmonella enterica serotype Virchow to Enterobacteriaceae in human flora-associated rats treated with cefixime. Antimicrob. Agents Chemother. 54:164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faure S, Perrin-Guyomard A, Delmas JM, Laurentie M. 2009. Impact of therapeutic treatment with β-lactam on transfer of the blaCTX-M-9 resistance gene from Salmonella enterica serovar Virchow to Escherichia coli in gnotobiotic rats. Appl. Environ. Microbiol. 75:5523–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gismondo MR, Drago L, Lombardi A, Fassina C, Cesana M. 1995. Impact of rufloxacin and ciprofloxacin on the intestinal microflora in a germ-free mice model. Chemotherapy 41:281–288 [DOI] [PubMed] [Google Scholar]
- 20. Guillot JF, Boucaud JL. 1988. In vivo plasmid transfer between strains of Escherichia coli in the chicken digestive system. Pathol. Biol. (Paris) 36:655–659 (In French.) [PubMed] [Google Scholar]
- 21. Jacoby GA, Chow N, Waites KB. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacoby GA, et al. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Devendec L, Bouder A, Dheilly A, Hellard G, Kempf I. 2011. Persistence and spread of qnr, extended-spectrum beta-lactamase, and ampC resistance genes in the digestive tract of chickens. Microb. Drug Resist. 17:129–134 [DOI] [PubMed] [Google Scholar]
- 24. Mata C, et al. 2010. In vivo transmission of a plasmid coharbouring blaDHA-1 and qnrB genes between Escherichia coli and Serratia marcescens. FEMS Microbiol. Lett. 308:24–28 [DOI] [PubMed] [Google Scholar]
- 25. Medders WM, Wooley RE, Gibbs PS, Shotts EB, Brown J. 1998. Mutation rate of avian intestinal coliform bacteria when pressured with fluoroquinolones. Avian Dis. 42:146–153 [PubMed] [Google Scholar]
- 26. Miranda JM, et al. 2008. Evolution of resistance in poultry intestinal Escherichia coli during three commonly used antimicrobial therapeutic treatments in poultry. Poult. Sci. 87:1643–1648 [DOI] [PubMed] [Google Scholar]
- 27. Ng LK, Martin I, Alfa M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209–215 [DOI] [PubMed] [Google Scholar]
- 28. Sullivan A. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101–114 [DOI] [PubMed] [Google Scholar]
- 29. van Boven M, Veldman KT, de Jong MCM, Mevius DJ. 2003. Rapid selection of quinolone resistance in Campylobacter jejuni but not in Escherichia coli in individually housed broilers. J. Antimicrob. Chemother. 52:719–723 [DOI] [PubMed] [Google Scholar]
- 30. van Essen-Zandbergen A, Smith H, Veldman K, Mevius D. 2009. In vivo transfer of an incFIB plasmid harbouring a class 1 integron with gene cassettes dfrA1-aadA1. Vet. Microbiol. 137:402–407 [DOI] [PubMed] [Google Scholar]
- 31. Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57:154–155 [DOI] [PubMed] [Google Scholar]