Abstract
CYP164 family P450 enzymes are found in only a subset of mycobacteria and include CYP164A1, which is the sole P450 found in Mycobacterium leprae, the causative agent of leprosy. This has previously led to interest in this enzyme as a potential drug target. Here we describe the first crystal structure of a CYP164 enzyme, CYP164A2 from Mycobacterium smegmatis. CYP164A2 has a distinctive, enlarged hydrophobic active site that extends above the porphyrin ring toward the access channels. Unusually, we find that CYP164A2 can simultaneously bind two econazole molecules in different regions of the enlarged active site and is accompanied by the rearrangement and ordering of the BC loop. The primary location is through a classic interaction of the azole group with the porphyrin iron. The second econazole molecule is bound to a unique site and is linked to a tetracoordinated metal ion complexed to one of the heme carboxylates and to the side chains of His 105 and His 364. All of these features are preserved in the closely homologous M. leprae CYP164A1. The computational docking of azole compounds to a homology model of CYP164A1 suggests that these compounds will form effective inhibitors and is supported by the correlation of parallel docking with experimental binding studies of CYP164A2. The binding of econazole to CYP164A2 occurs primarily through the high-spin “open” conformation of the enzyme (Kd [dissociation constant] of 0.1 μM), with binding to the low-spin “closed” form being significantly hindered (Kd of 338 μM). These studies support previous suggestions that azole derivatives may provide an effective strategy to improve the treatment of leprosy.
INTRODUCTION
Cytochrome P450 enzymes (CYPs) are prevalent across many organisms in which they perform a wide range of monooxygenation reactions essential for metabolism and detoxification (reviewed in reference 45). The criticality of many of these reactions has resulted in CYPs being targeted in drug development, for example, as antifungals (20, 29, 48). Most eukaryotes contain multiple forms of CYPs (grouped into families on the basis of substrate specificity [42]); in contrast, CYPs are less common in prokaryotes, although genome studies have now identified CYPs in actinomycetes, such as mycobacteria, and streptomycetes (9, 30). This has spurred an interest in developing antibacterials to target these enzymes (1, 6, 23). Such studies could build on existing expertise; for example, azole-based antifungals have been used to target CYP51 in eukaryotes, for which a homolog has been identified in Mycobacterium tuberculosis (5). This is one of 20 CYPs identified in the M. tuberculosis genome (42). In contrast, unlike other mycobacterial species, Mycobacterium leprae has retained only one CYP (9, 10), although many pseudogenes are evident. This suggests that it has an important function, and as mycobacteria exhibit sensitivity to azole compounds, this could have relevance in the treatment of leprosy, one of the oldest human diseases.
Studies of the M. leprae genome show that it has undergone severe gene degradation (10), with 60% fewer active genes than in M. tuberculosis. There are many CYP pseudogenes whose functions appear to have been lost as the bacterium has shifted to be an obligate pathogen, where many of the critical functions are dependent on the intracellular reactions of the host. This may partly explain the long doubling time of M. leprae, approximately 14 days (10), which complicates laboratory studies of M. leprae. In addition, there is no readily accessible host model system: the nine-banded armadillo and nude mice are capable of hosting the bacteria but lack clinically relevant responses (59).
The single remaining M. leprae P450 enzyme belongs to a new and unusual family of CYPs of unknown function, termed CYP164 (10, 42). The M. leprae enzyme CYP164A1 has a close homolog (59% identical sequence) in Mycobacterium smegmatis, CYP164A2 (24, 28). The closest CYPs for which structures and functions are known are P450eryF (12) and CYP142 (16), involved in erythromycin biosynthesis and cholesterol hydroxylation, respectively. The importance of these two processes to microbial viability, and the retention of CYP164A1 as the single remaining P450 gene in M. leprae, strongly suggests a critical role for CYP164A1 in M. leprae survival. As such, CYP164A1 was previously proposed to be a viable target for the development of effective leprosy therapeutics (62).
CYP164A2 is also of interest for its unusual biochemical property of readily altering the spin state in response to changes in ionic strength, pH, and cosolvents (13, 14, 62–64). Changes from the low-spin state to the high-spin state in P450 enzymes are an essential part of both substrate binding and inhibitor binding and often correlate with structural changes in both the active-site arrangement and access to this site (45). Unlike M. tuberculosis CYPs, for which the structures of several have recently been characterized (16, 34, 39, 46), no direct structural information has previously been reported for CYP164 enzymes. In addition to the experimental difficulty in culturing M. leprae effectively, a further obstacle has been that the recombinant expression of CYP164A1 has also proven problematic (62; our unpublished data). In order to progress our understanding of CYP164 family enzymes, in this study we report a crystal structure of the very close homolog CYP164A2 from Mycobacterium smegmatis. This protein tightly binds azole inhibitors (Kd [dissociation constant] = 1 to 6 μM [62]), and a crystal structure of a CYP164A2-econazole complex is also described. This closely homologous structure was then used to construct a model for CYP164A1 from M. leprae. This approach provides important molecular details on the CYP164 family of P450s, which could prove to be a novel route to potential new leprosy therapeutics.
MATERIALS AND METHODS
Expression, purification, and crystallization of recombinant CYP164A2.
CYP164A2 was expressed and purified as described previously (62). In brief, after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were incubated at 25°C for 18 h and then lysed and centrifuged, and the supernatant was subjected to Ni2+-nitrilotriacetic acid (NTA) agarose affinity chromatography. The purified protein was dialyzed in a final buffer containing 20 mM Tris (pH 7.4). The protein concentration was determined by using reduced carbon monoxide difference spectra (43).
Crystals of CYP164A2 were obtained by vapor diffusion, mixing equal volumes of the protein (40 mg/ml) and reservoir (0.1 M HEPES [pH 7.5]–0.8 M sodium dihydrogen phosphate and 0.8 M potassium dihydrogen phosphate–3% [wt/vol] 6-aminohexanoic acid [crystallization buffer 1 {CB1}]) solutions. Crystals of the inhibitor complex were obtained by combining CYP164A2 (11 mg/ml in Tris-HCl [pH 7.4]–100 mM NaCl) with a 5-fold molar excess of the azole inhibitor econazole in dimethyl sulfoxide (DMSO) and equilibrating the mixture against 0.1 M potassium thiocyanate and 30% (wt/vol) polyethylene glycol 2000 monomethyl ether (PEG 2000 MME) (CB2). Prior to data collection, crystals were progressively transferred into 30% ethylene glycol for cryopreservation.
Crystal structure determination.
The structure of CYP164A2 was solved by multiwavelength anomalous dispersion (MAD) using the heme Fe anomalous signal from diffraction data collected at the Diamond Light Source (Oxford, United Kingdom). Three data sets were collected from a single crystal and corresponded to peak, inflection, and high-resolution remote. These were indexed separately by using MOSFLM (33) and scaled with SCALA (19). Heavy-atom position location, phasing, and density modification were performed by using the SHELXC/D/E package (60), and further density modification, solvent flattening, and histogram mapping were performed with the CCP4 program DM (11). The computed phases were combined with a high-resolution native data set to improve the resolution of the final structure. An initial model was built into the electron density map ARP/wARP (50); after 10 cycles of autobuilding, 98% of the sequence could be traced into the electron density. Further iterative rounds of refinement and manual model building of the CYP164A2 structure with REFMAC (41) and COOT (18) produced the final model.
The structure of the econazole-CYP164A2 inhibitor complex was solved by molecular replacement with PHASER (37), using the native structure as the search model. COOT was used for model building, and two molecules of econazole were placed into the electron density and refined with REFMAC. Refinement and structural statistics are shown in Table 1. The final crystal structures were checked with MOLPROBITY (8) for good stereochemistry and to ensure that there were no residues in the disallowed regions of the Ramachandran plot (Table 1).
Table 1.
Crystallographic and refinement statistics of CYP164A2 and the econazole-CYP164A2 complex
| Parameterb | Value (value forhighest-resolution shell)a |
||||
|---|---|---|---|---|---|
| CYP164A2 high resolution | CYP164A2 peak | CYP164A2 inflection | CYP164A2 remote | CYP164A2-econazole complex | |
| Wavelength (Å) | 0.911 | 1.739 | 1.742 | 1.704 | 0.979 |
| Resolution range (Å) | 92.45–1.9 (2.0–1.9) | 50–2.5 (2.6–2.5) | 50–2.5 (2.6–2.5) | 50–2.5 (2.6–2.5) | 70.2–2.1 (2.2–2.1) |
| Space group | C2 | P22121 | |||
| Unit cell | a = 178.4 Å, b = 102.8 Å, c = 92.4 Å, α = γ = 90°, β = 90.1° | a = 38.8 Å, b = 85.8 Å, c = 121.9 Å, α = β = γ = 90° | |||
| Completeness (%) | 99.5 (95.1) | 98.6 (85.4) | 98.8 (86.5) | 98.7 (86.3) | 97.1 (92.1) |
| No. of reflections | 131,656 (10,886) | 57,147 (4,139) | 56,848 (4,112) | 57,059 (4,163) | 22,538 (3,041) |
| Redundancy | 10.3 (7.3) | 7.3 (6) | 7.3 (6) | 7.3 (5.9) | 11.7 (11.7) |
| I/σI | 32.1 (1.9) | 13.1 (3.0) | 22.2 (3.1) | 26.1 (4.0) | 17.7 (5.7) |
| Rsym | 0.065 (0.676) | 0.093 (0.347) | 0.09 (0.337) | 0.079 (0.266) | 0.109 (0.501) |
| Refinement statistics | |||||
| Total no. of reflections | 124,989 (8,826) | 22,446 (1,448) | |||
| Rwork | 0.183 | 0.200 | |||
| Rfree | 0.202 | 0.232 | |||
| RMSD of bond lengths (Å) | 0.01 | 0.006 | |||
| RMSD of bond angles (°) | 1.25 | 0.931 | |||
| No. of protein atoms | 8,961 | 3,055 | |||
| No. of solvent molecules | 718 | 115 | |||
Values in parentheses refer to data from the highest-resolution shell.
Rsym=ΣhklΣl|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl). Rwork=Σhkl|F0(hkl)− FC(hkl)|/Σh|F0(hkl)| for main set (95%) of reflections used for refinement. Rfree=Σhkl|F0(hkl)− FC(hkl)|/Σh∣F0(hkl)∣ calculated using subset (5%) of reflections not used in refinement.
Molecular modeling of M. leprae CYP164A1 and inhibitor docking.
Molecular models for CYP164A1 with and without bound inhibitor were generated by using the program MODELLER (56). The templates in each case were the experimentally derived molecular coordinates of CYP164A2. Sequence alignment indicated an overall identity of 59%; amino acids for which there was no electron density were omitted and computationally generated. One hundred models were constructed and scored, and the top five candidates from each population were compared with MOLPROBITY (9). The model with the best geometry was subjected to energy minimization using the program NAMD (51), with internal heme restraints selected. The final models can be downloaded from www.bristol.ac.uk/biochemistry/brady/coordinates.
The binding of the azole inhibitors to the final CYP164A1 structures generated with and without the inhibitor bound was tested computationally by using the docking program GOLD (27). In each case, the inhibitor was first placed within the active-site cavity by the program, which was defined as atoms within a 12-Å radius from the heme Fe. The binding of a range of inhibitor conformations was then explored, each for a minimum of 10 runs to identify the conformation with the most favorable binding energy. The generated models were all visually inspected, and a final selection criterion was used so that the azole group had to be within 2 Å of the primary econazole position.
Experimental determinations of econazole and fatty acid binding.
Assays of binding of econazole to CYP164A2 were performed as previously described (62). The absolute spectra of 3 μM M. smegmatis CYP164A2 were determined in the absence and presence of 16 to 225 μM econazole nitrate (molecular weight [MW], 445; Sigma) under low-spin-state (LS buffer [50 mM Tris-HCl {pH 8.1}, 60% {vol/vol} ethylene glycol]) and high-spin-state (HS buffer [5 M NaCl]) solution conditions. Determinations of spectra were then repeated under solution conditions similar to those used for the crystallization of ligand-free CYP164A2 (CB1 [0.1 M HEPES {pH 7.4}, 0.8 M NaH2PO4, 0.8 M KH2PO4, 6% {wt/vol} 6-aminohexanoic acid]) and the CYP164A2-econazole complex (CB2 [0.1 M potassium thiocyanate, 30% {wt/vol} PEG 2000-MME, 0.1 M NaCl]) and with a control buffer (Cont [0.1 M Tris-HCl {pH 8.1}, 25% {wt/vol} glycerol]). Spin-state equilibria were determined as previously described (31). UV-visible light (UV-Vis) type II binding spectra were determined for the interaction of econazole nitrate with 3 μM M. smegmatis CYP164A2 under each buffer condition. Econazole nitrate binding was characterized by using both direct azole saturation plots and Hill plots {log[ΔA/(ΔAmax − ΔA)] versus log[azole]}. In addition, Job plots {ΔA versus [CYP]/([CYP] + [azole])} (22) were performed to determine binding stoichiometry, with the total concentration of CYP164A2 and econazole kept constant at 9 μM and with the CYP164A2/econazole ratio varied in 0.5 μM steps using HS and control buffers. Fatty acid binding spectra for 3 μM CYP164A2 in 50 mM Tris-HCl (pH 8.6)–25% (wt/vol) glycerol were determined by progressive titration using 5-mg/ml solutions of a range of fatty acids in dimethylformamide (DMF) with an equivalent addition of DMF to the reference cuvette. Absorbance difference spectra were measured between 500 and 350 nm after each addition of fatty acid. Binding determinations were made by using acetic acid, 6-aminohexanoic acid, lauric acid (C12:0), myristic acid (C14:0), linoleic acid (C18:2), and arachidic acid (C20:0). Binding determinations using 5-mg/ml solutions of n-dodecane and n-hexadecan-1-ol in DMF were also performed. Curve fitting of data was performed by using ProFit 6.1.9 (QuantumSoft, Zurich, Switzerland).
Protein structure accession numbers.
The atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession numbers 3R9B for CYP164A2 and 3R9C for the econazole-CYP164A2 complex.
RESULTS
Structure of CYP164A2.
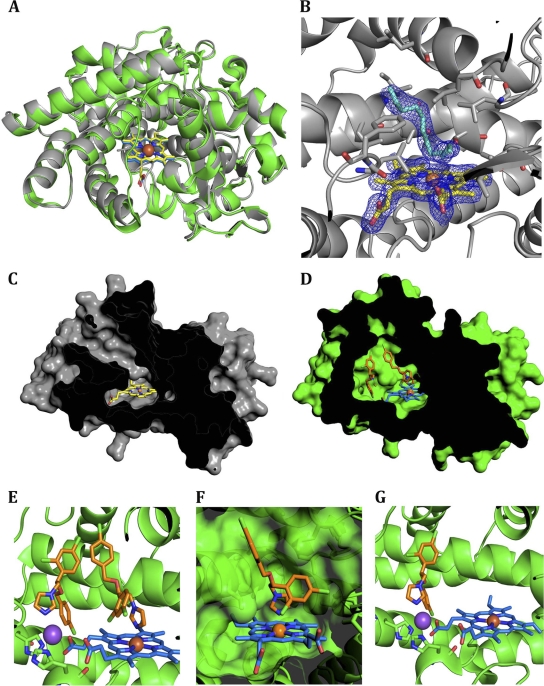
The crystal structure of CYP164A2 contains three independent views of the enzyme in the asymmetric unit in all of which the features described below are conserved. CYP164A2 adopts a classic, mainly α-helical P450 structure (Fig. 1A). A comparison of the native structure to the structure of P450cam (PDB accession number 2CPP [55]) shows that the overall fold of the P450 enzyme is well conserved (core alpha carbon [Cα] root mean square deviation [RMSD] of 1.9 Å) despite a low level of sequence identity (24.6%). There are three sections of the structure for which there is no corresponding electron density: residues 1 to 14, BC loop residues 85 to 93, and the C-terminal His6 tag. The conventional FG and BC loops (nomenclature as defined in reference 55) are both extended in CYP164A2, increasing the size of the substrate channel and volume of the active site (relative to those of P450cam). In the absence of a substrate, the stub of the BC loop partially occludes the active-site pocket, with the Leu 98 and Phe 99 side chains projecting toward the heme group. This forms part of a highly hydrophobic active site that is also lined by Leu 180 and Leu 184 (F helix); Leu 252, Ile 255, Ala 256, and Thr 260 (I helix); Val 303 and Val 306 (KL loop); and Leu 404 (Fig. 1B). Due to the disorder of the BC loop, the active-site heme is found to be solvent accessible with the enzyme in an “open” conformation. The heme iron has five ligands: the four pyrrole nitrogens plus the sulfhydryl side chain of Cys 355. There is no obvious water molecule bound in the sixth axial position. However, although the crystals were grown in the absence of an added inhibitor or substrate analog, a close inspection of the active site revealed that it contained an ambiguous V-shaped electron density with its apex ∼3.6 Å from the axial heme position and extending along the access channel (Fig. 1B). This contiguous piece of density has been modeled as an alkyl chain similar to those of previously reported crystal structures of P450 complexes with long-chain fatty acids (e.g., PDB accession number 1JPZ, for P450BM-3 complexed with N-palmitoylglycine [21]), although the density becomes broken toward the entrance of the solvent channel and here has been modeled as water molecules. In this region, the Tyr 309 and Arg 73 side chains could potentially provide anchorage for fatty acid head groups. Attempts to characterize the identity of the bound compounds using mass spectrometry were inconclusive, although preliminary binding studies indicated that fatty acids (C12 to C18) bound to CYP164A2 produce characteristic type I UV-Vis difference spectra (see Fig. 4). It is possible that there is a mixed population of alkyl chains bound from a variety of fatty acids, presumably isolated along with the enzyme.
Fig 1.
Crystal structure of Mycobacterium smegmatis CYP164A2. (A) Molecular fold and overlay of CYP164A2 structures. The enzyme structure is shown in gray, and the heme moiety is shown in yellow. The econazole-enzyme complex is shown in green, with the heme shown in blue. The enzyme is viewed through the BC loop, and the central iron is shown as an orange sphere. (B) Active site of CYP164A2. The residues that form the active-site cavity are represented as sticks. The heme is displayed with the electron density contoured at 1 sigma, and the ambiguous “V-shaped” electron density in the substrate binding region is also shown. (C) Surface representation of the CYP164A2 central cavity in the open state. (D) Depiction of the closed cavity of econazole-bound CYP164A2. (E) Occupation of two econazoles in the active site of CYP164A2. The econazoles are represented as sticks, in orange, and the sodium is pictured as a purple sphere. His 105 and His 364 are shown behind and in front of the sodium, respectively. (F) Transparent surface representation of the primary econazole binding site. (G) The secondary econazole bound to the tetracoordinated sodium cation. All images were created with PyMOL (version 1.3; Schrödinger).
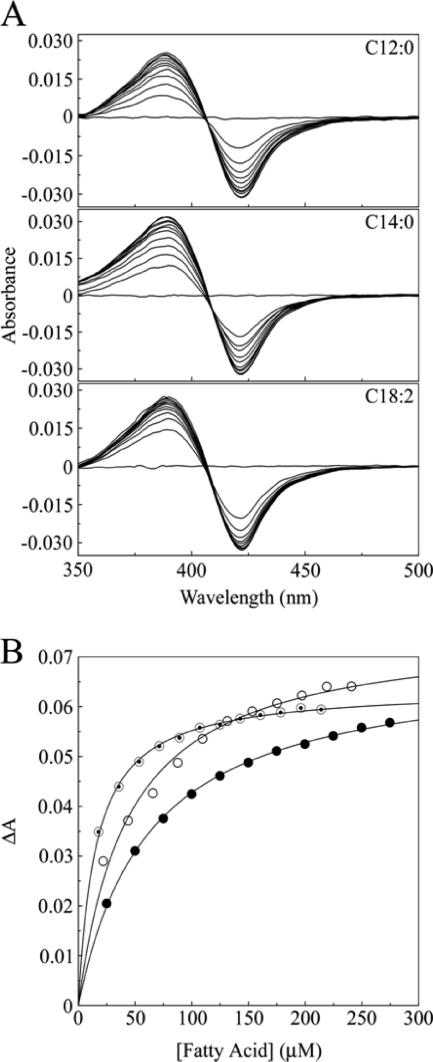
Fig 4.
Fatty acid binding to CYP164A2. (A) UV-Vis difference spectra were determined by the progressive titration of lauric (C12:0), myristic (C14:0), and linoleic (C18:2) acids against 3 μM CYP164A2 in 50 mM Tris-HCl (pH 8.6)–25% (wt/vol) glycerol. (B) Fatty acid binding saturation curves for lauric (filled circles), myristic (hollow circles), and linoleic (bullets) acids were constructed from the UV-Vis difference spectra. Data sets were fitted by using the Michaelis-Menten equation to derive Ks values for each fatty acid.
Structure of the econazole-CYP164A2 inhibitor complex.
Cocrystallization of CYP164A2 was attempted with a wide range of azole derivatives (miconazole, clotrimazole, fluconazole, voriconazole, itraconazole, ketoconazole, and econazole) at various concentrations and under >400 conditions. Crystals were obtained for several of the inhibitor complexes, but only the econazole complex diffracted to a resolution suitable for structure determinations. The crystals contained one CYP164A2 molecule in the asymmetric unit, with density absent for residues 1 to 14 and the His6 tag. An overlay of the structures of the enzyme and econazole-enzyme complex (Fig. 1A) shows them to be closely similar, with a Cα RMSD of 0.58 Å.
Econazole-bound CYP164A2 shows one major difference from the open-conformation structure. In the complex, the BC loop, which was disordered in the absence of an inhibitor, is clearly visible in the electron density. The loop forms a cap to the channel leading to the heme, producing a “closed” conformation, which restricts solvent access (Fig. 1C and D). At the end of the loop, residues 97 to 100, with the sequence FLFL, are displaced up to 11 Å away from their location in the open-conformation structure. This leads to the solvent exposure of several of these hydrophobic residues, the entropic cost of which is presumably overcome by the tight network of electrostatic interactions in the Glu 86-Ser 96 loop. This region, which is completely disordered in the absence of an inhibitor, now forms a tightly restrained loop with a plethora of electrostatic and charged hydrogen bonds: on the inside the loop, between side-chain and main-chain atoms of Glu 86, Arg 90, and Ser 96, and on the exterior of the loop, the Gln 92 main-chain carbonyl links to the neighboring side chains of Arg 209 and Asp 210. The loop interfaces with the G helix primarily via the side chains of Phe 198 and Met 202, leading to a displacement of the helix by about 1 Å relative to its position in the absence of the inhibitor. Although some of these changes within the complex may be partially stabilized by fortuitous crystal contacts, the BC loop is clearly dynamic and may enable this enzyme to accommodate a range of large substrates within the active site.
Buried within the enzyme core by these changes are, unusually, two molecules of econazole (Fig. 1C). One of these coordinates directly with the heme iron at the sixth axial position via the nitrogen in the 3 position of the azole ring in the “conventional” arrangement (2.1 Å) (Fig. 1D), as previously described for other CYP-econazole complexes (PDB accession numbers 3JUS and 2UVN [47]). The complementary shape of the active-site pocket induces minor differences: the 4-chlorophenyl ring is positioned away from the heme as Leu 252 protrudes into the active site, whereas the 2,4-dichlorophenyl substituent fits into a hydrophobic pocket lined by Leu 184 (F helix), Val 303 plus Val 306 (KL loop), and Leu 404. The active-site residues close to the heme are not noticeably perturbed by the binding of econazole, which concurs with the previously reported high-affinity binding (Kd = 2.52 μM [62]).
Unexpectedly, a second econazole molecule was observed to be bound within the space occupied by residues 94 to 101 in the open form and vacated by the loop arrangement described above. This site is approximately 12 Å from the heme iron. The two econazole molecules cooperate through a parallel, displaced π-stacking interaction similar to that identified previously in aromatic-amino-acid side-chain pairs (38). In this second econazole molecule, the 4-chlorophenyl ring binds in a pocket formed by Leu 108 plus Val 112 (C helix) and Leu 367 in the position previously occupied by Phe 97, whereas the 2,4-dichlorophenyl group is oriented toward the substrate access channel and interacts with the main-chain carbonyl groups of Pro 94 and Leu 98. The econazole molecule forms a close fit to a fortuitously highly complementary pocket. This conformation appears to also be stabilized by a metal ion that binds the nitrogen in the 3 position of the imidazole ring and is also coordinated by one of the carboxylates of the heme ring and the imidazole side chains of His 105 and His 364 (Fig. 1E). His 105 is at the base of the C helix and undergoes a small shift (1 Å) between the open and closed structures. Modeling of the metal ion was attempted with a range of possible candidates. Although potassium was present in the crystallization buffer, the tetrahedral geometry and bonding distances suggested that the ligand was unlikely to be potassium. Refinement of the model proceeded most successfully with sodium at this site, which is present at high concentrations in the crystallization buffer. No obvious contaminating cations could be found by mass spectrometry analysis of the inhibitor solution.
Spectroscopic analysis of the CYP164A2-econazole complex.
The heme iron in cytochrome P450 proteins is known to undergo a transition from a low-spin (hexacoordinated) to a high-spin (pentacoordinated) state during catalysis, attributable to the displacement of an axially bound water molecule from the heme iron when the substrate binds to the CYP enzyme (45). This transition can be readily observed spectroscopically by the blue shift of the Soret band from 418 to 392 nm. CYP164A2 was previously noted to be unusual for a P450 in that the spin state can be readily modulated in the absence of a substrate by the addition of metal cations (spin equilibrium shifted toward the high-spin state) and by the addition of cosolvents such as ethylene glycol (spin-state equilibrium shifted toward the low-spin state) at room temperature and pressure (60). The spin states of the low- and high-spin CYP164A2 standards (Fig. 2A) were nearly 100% low spin and >95% high spin, respectively, compared to previously reported low-spin and high-spin UV-Vis spectra of P450cam (32, 61) determined by comparisons of the A418/A392 and A392/A418 ratios for the low- and high-spin forms. In contrast, azole antifungal agents bind to CYP enzymes through the direct coordination of the azole nitrogen atom with the heme iron to form a hexacoordinated low-spin complex characterized by a red shift of the heme Soret peak to 425 to 430 nm (25), with additional hydrophobic interactions between the side chains of the azole antifungal molecule and the amino acid side-chain residues lining the substrate binding pocket/access channel of the CYP that are not manifest in the UV-Vis spectrum.
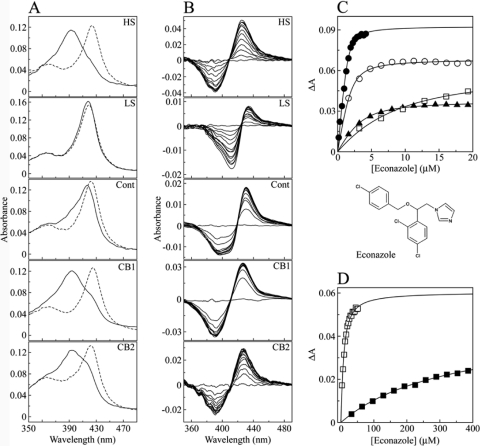
Fig 2.
Spectral titration of econazole against CYP164A2. (A) Absolute oxidized spectra (solid lines) of 3 μM CYP164A2 were measured under >95% high-spin (5 M NaCl) (HS), near-100% low-spin (50 mM Tris-HCl [pH 8.1], 60% [vol/vol] ethylene glycol) (LS), control buffer (0.1 M Tris-HCl [pH 8.1], 25% [wt/vol] glycerol) (Cont), crystallization buffer 1 (CB1) (0.1 M HEPES [pH 7.4], 0.8 M NaH2PO4, 0.8 M KH2PO4, 6% [wt/vol] 6-aminohexanoic acid), and crystallization buffer 2 (CB2) (0.1 M KSCN, 30% [wt/vol] PEG 2000-MME, 0.1 M NaCl) conditions. The heme Soret region (330 to 490 nm) is shown for clarity. The absolute spectra were also determined in the presence of econazole (dashed lines) at 16, 225, 32, 16, and 115 μM in HS, LS, control, CB1, and CB2 buffers, respectively. (B) Econazole was progressively titrated against 3 μM CYP164A2 in each of the five buffers until saturation was reached, with the type II difference spectrum recorded after each addition of azole. (C) Binding saturation curves were constructed from the spectral difference (ΔApeak-trough) against the econazole concentration for HS (filled circles), control (filled triangles), CB1 (hollow circles), and CB2 (hollow squares) buffers. (D) Binding saturation in CB2 (hollow squares) and LS (filled squares) buffers occurred at higher econazole concentrations. The modified Morrison equation (35) for tight ligand binding was used to fit the saturation curves, except for LS buffer, where the Michaelis-Menten equation was used.
The unexpected observation of two econazole binding sites in CYP164A2 led us to explore the mechanism of inhibitor binding to this enzyme. To probe the correlation of econazole binding to CYP164A2 under crystallization conditions, reference low- and high-spin-state spectra were initially obtained under conditions previously derived (Fig. 2A) (62). Solution spectra were then also obtained for CYP164A2 under conditions similar to those of the crystallization buffers CB1 and CB2, which were used to grow free CYP164A2 and CYP164A2-econazole complex crystals, respectively, in addition to a control Tris-HCl buffer. Reference spectra indicated that when CYP164A2 resides in the high-spin conformation at a level of >95%, the Soret peak maximum is at 392 nm, in comparison to the Soret peak maximum at 418 nm when nearly 100% in the low-spin state (Fig. 2A). This spin-state-induced blue shift of the Soret peak is also characteristic of cytochrome P450 enzymes during substrate binding (25). The low-spin components of CYP164A2 in the control, CB1, and CB2 buffers were 66, 17, and 34%, respectively. Therefore, the crystals of both CYP164A2 and its complex with econazole are grown from solutions in which the high-spin conformation of the free enzyme predominates; however, once econazole is bound to CYP164A2, the resultant CYP164A2-econazole complex adopts the low-spin conformation.
The addition of econazole caused a red shift of the Soret peak in all five buffers (Fig. 2A). The largest shift was observed for HS buffer, from 392 to 424 nm, closely followed by CB1 (393 to 425 nm) and CB2 (393 to 422 nm), indicating that econazole readily bound to CYP164A2 predominantly in the high-spin conformation. In contrast, there was little shift in the Soret peak of CYP164A2 in LS buffer in the presence of 225 μM econazole (418 to 419 nm), indicating that the low-spin form of CYP164A2 bound econazole poorly and not to 100% completion. The control buffer (66% low spin) gave an intermediate Soret peak shift from 418 to 422 nm, again indicating that econazole binding did not go to completion. The low-spin buffer contained 60% (vol/vol) ethylene glycol. Confirmation that 3 μM CYP164A2 protein was still native after 10 min in the low-spin buffer was obtained by carbon monoxide difference spectroscopy in the presence of sodium dithionite (44), with the rate of formation and the final concentration of the CYP164A2-CO adduct being similar to those obtained with the control buffer, with a characteristic red shift of the Soret band to 449 nm (data not shown). Consequently, the failure of econazole to bind efficiently to low-spin CYP164A2 was not due to enzyme denaturation by 60% (vol/vol) ethylene glycol and was most likely due to the heme being inaccessible to econazole in the closed low-spin CYP164A2 conformation.
Azole titration experiments (Fig. 2B to D) confirmed that low-spin CYP164A2 bound econazole poorly (Kd of 338 μM), compared with the tight binding observed with the high-spin conformation (Kd of 0.1 μM). The variation in binding affinities in the other buffers partly reflects the proportion of high-spin-state enzymes under each condition (Table 2), although the increased viscosity caused by 30% (wt/vol) PEG 2000-MME in CB2 may be a factor contributing to the relatively poor binding of econazole to CYP164A2 in this buffer.
Table 2.
Spin-state dependency of parameters of econazole binding with CYP164A2
| Buffer | Low-spin component (%) | Kd for econazole (μM) | Hill plot slope (SD) |
|---|---|---|---|
| HS | <5 | 0.098 (±0.016) | 2.34 (±0.05) |
| LS | ∼100 | 338 (±17) | 1.01 (±0.04) |
| Control | 66 | 1.07 (±0.15) | 1.41 (±0.09) |
| CB1 | 17 | 0.37 (±0.10) | 1.79 (±0.11) |
| CB2 | 34 | 4.05 (±0.56) | 1.04 (±0.04) |
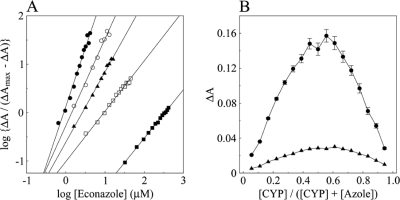
Fitting of the econazole binding saturation curves both directly and to a linear form of the Hill equation (Fig. 3A) gave apparent Hill constants of 1.9 to 2.3 for the high-spin form of the enzyme but a value of 1.0 under low-spin conditions, suggesting possible allosterism (positive cooperativity) for econazole binding to the high-spin form of CYP164A2 (Table 2). However, fitting of the data to Job plots (Fig. 3B) (22), which may be a more reliable method for the determination of binding stoichiometry with tight-binding ligands, indicated mole fractions in the complex of 0.53 and 0.56, respectively, indicating a virtual 1:1 enzyme/inhibitor ratio. We note from the crystal structure that the spectroscopic analyses are sensitive primarily only to the binding of the first econazole molecule and do not provide a direct method of measuring binding to the second econazole site.
Fig 3.
Hill and Job plots for econazole binding to CYP164A2. (A) Hill plots were constructed from the saturation curves shown in Fig. 2 for HS (filled circles), LS (filled squares), control (filled diamonds), CB1 (hollow circles), and CB2 (hollow squares) buffers. (B) Job plot titrations (22) were performed with HS (filled circles) and control (filled triangles) buffers. The combined CYP164A2 and econazole concentrations were maintained at 9 μM, with the CYP164A2 concentration being increased in 0.5 μM steps up to 8.5 μM while the econazole concentration was decreased accordingly. The CYP164A2 solution was divided between sample and reference cuvettes, and the background absorbance was measured. Econazole was then added to the sample cuvette, and an equivalent volume of DMSO was added to the reference cuvette, followed by thorough mixing. After 5 min, the type II difference spectrum was measured at between 500 and 350 nm to obtain the ΔA values.
In summary, combining both the structural and spectroscopic data, the closed conformation of CYP164A2 observed in its complex with econazole exhibits obvious hexacoordination for the heme iron and spectra consistent with type II inhibitor binding. We concluded that the crystals contained low-spin-state CYP164A2. This is formed when econazole binds preferentially to the high-spin open conformation of CYP164A2 to form a low-spin closed CYP164A2-econazole complex in which the econazole is directly coordinated with the heme iron as the sixth axial ligand. The binding of econazole directly to the low-spin closed form of CYP164A2 was severely hindered, reflected in the 3,000-fold-higher apparent Kd value than that obtained with the high-spin form. In contrast, crystals of the open-conformation enzyme were obtained under high-salt buffer conditions, in which the high-spin form predominates. In this structure, the heme iron has five ligands, with the density ascribed to the unidentified alkyl chain lying too distant for regular coordination. There is no density for a bound water molecule at the axial position. All of these features are consistent with the open structure observed in the crystal representing a high-spin form of the enzyme. These correlations match those previously described for other forms of CYPs, such as those for P450eryF (13). Finally, the high affinity of the high-spin conformation of CYP164A2 (Kd of 0.1 μM) for econazole suggests that azoles may be a more effective antibacterial agent target than previously thought (60).
Spectroscopic screening of potential CYP164A2 substrates.
Determinations of binding made by using acetic acid, 6-aminohexanoic acid, arachidonic acid, n-dodecane, or n-hexadecan-1-ol did not produce UV-Vis difference spectra in the 500- to 350-nm region, indicating that these compounds did not perturb the heme environment of CYP164A2 either by the displacement of the axial ligated water molecule or by direct coordination with the heme iron. However, lauric acid, myristic acid, and linoleic acid produced strong type I binding spectra with 3 μM CYP164A2 (Fig. 4), with a peak at 389 nm and a trough at 421 nm, indicating the displacement of the axially ligated water molecule from the heme iron and a low- to high-spin-state change. Binding affinities (Ks) were determined to be 62 (±3), 44 (±8), and 16 (±1) μM for lauric, myristic, and linoleic acids, respectively, using the Michaelis-Menten equation. The slight fall in pH incurred during the fatty acid titration was not the cause of the observed type I difference spectra (by the displacement of the spin-state equilibrium), as titration with acetic acid produced no discernible UV-Vis difference spectrum. Therefore, the CYP164A2 in vivo substrate is likely to be either a fatty acid or a fatty acid derivative retaining a functional carboxyl group, as n-dodecane and n-hexadecan-1-ol did not produce a UV-Vis difference spectrum with CYP164A2.
Molecular model of CYP164A1.
Two models of CYP164A1, with (closed) and without (open) econazole, were generated by homology modeling and energy minimization using the two crystal structures of CYP164A2. The open model of CYP164A1 (Fig. 5A) retains the hydrophobic nature of the active site and the enlarged access channel. The most noticeable difference is the incorporation of Met 117 and Met 424 within the active site in place of Leu 98 and Leu 404, respectively. Met 424 projects above the heme ring; the Cε is approximately 3 Å from the primary econazole position. The second methionine, Met 117, covers the secondary econazole binding site, occupied by Leu 98 in the open-conformation CYP164A2 crystal structure. Modeling of the structure based on the econazole-bound form (Fig. 5B) shows that the BC loop of CYP164A1 is capable of adopting a conformation that creates the enlarged active site including the second econazole binding site.
Fig 5.
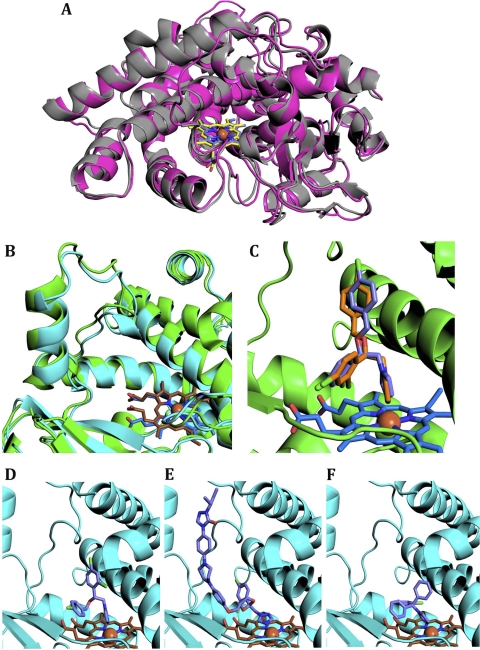
Structural modeling of M. leprae CYP164A1 complexes with azole inhibitors. (A) Cartoon overlay of CYP164A2 (colored as described in the legend of Fig. 1) and CYP164A1 in open states. The CYP164A1 model is shown in magenta, and the heme is shown in purple. (B) Overlay of the BC loop of the CYP164A1 inhibitor complex model (protein in cyan and heme in brown) on the crystal structure of the econazole-CYP164A2 complex (as described in the legend of Fig. 1). (C) Overlay of econazole docked into the CYP164A1 complex model onto the crystal structure of the econazole-CYP164A2 (closed) complex. The crystal structure is colored as described above. The top docking simulation conformation is represented in purple. (D, E, and F) The azole inhibitors bound to the active site of the closed-conformation CYP164A1 model. The inhibitors depicted are econazole (D), itraconazole (E), and clotrimazole (F).
In silico binding of azoles to CYP164A1.
In order to assess the likelihood of drug effectiveness for the treatment of leprosy, we wished to determine if the sole M. leprae cytochrome P450 is capable of binding a range of azole inhibitors. For this task, we used the model of CYP164A1 and the docking program GOLD. As a positive confirmation of the results, the azole inhibitors were also docked to the CYP164A2 crystal structures, and their binding scores were compared to previously reported experimentally determined binding affinities. Inhibitor docking to CYP164A2, using the Chemscore scoring function, was found to correlate with the CYP164A2 binding affinities, with the exception of ketoconazole (Table 3). This inhibitor had a higher-than-expected score; this is probably due to favorable hydrophobic contacts that are static in the simulation, or ketoconazole may bind to a different site. A comparison of the primary econazole to the docking simulation (Fig. 5C) clearly shows that the program is capable of docking this inhibitor into the CYP164A2 active site in the correct conformation. The remaining azole inhibitors were docked into the crystal structures and bound in orientations comparable to those of previously reported structures. We concluded that a reliable assessment of inhibitor binding to cytochrome P450s could be generated by this method.
Table 3.
Docking simulation scoresa
| Inhibitor | Kd (μM) | Simulation score |
|||
|---|---|---|---|---|---|
| CYP146A2 open | CYP164A2 closed | CYP164A1 open | CYP164A1 closed | ||
| Itraconazole | 0.78 | 48.1 | 44.1 | — | 36.5 |
| Clotrimazole | 1.20 | 32.2 | 45.1 | −10.3 | 37.6 |
| Miconazole | 2.48 | 43.6 | 43.0 | 19 | 36.3 |
| Econazole | 2.52 | 45.6 | 42.3 | 13.4 | 37.6 |
| Voriconazole | 3.24 | 27.4 | 30.8 | 2.4 | 29.2 |
| Ketoconazole | 5.69 | 47.2 | 44.1 | 0 | 35.3 |
| Fluconazole | — | 22.6 | 22.2 | −1.26 | 20.7 |
The azole inhibitors are ranked by their previously reported binding affinities (Kd) for CYP164A2 (60). The docking simulations were run for each inhibitor with the two crystal structure states of CYP164A2 and the two homology model structures of CYP164A1. The Chemscore function value for each of the simulations is provided. A dash indicates that no value was obtained or that no value has been reported.
The energy-minimized model for CYP164A1 in its open form replicates the open CYP164A2 crystal structure in having access to the heme sterically blocked by the side chain of residue 117, in this case a methionine. As the spectroscopic data indicated that econazole binds preferentially to the open form of the CYP164A2, the structure of the inhibited (closed) form of CYP164A1 was therefore believed to be a more suitable model for docking simulations.
Overall, all of the azole inhibitors returned a lower Chemscore for binding to CYP164A1 than to CYP164A2. Econazole was found to bind to CYP164A1 in a flipped conformation (Fig. 5D), where the two phenyl rings were exchanged, presumably due to steric clashes between the 4-chloro position and Ile 326. The occupation of the active site by the large inhibitor itraconazole (Fig. 5E) is possible, as the azole group binds to the heme with a slight distortion, and the rest of the molecule makes numerous hydrophobic contacts along the access channel. The routinely administered antifungal clotrimazole (Fig. 5F) bound the leprosy P450 structure in a conformation that is comparable to those of previously reported crystal structures of other P450-clotrimazole complexes (PDB accession numbers 2VRV, 3MDV [52], and 2XFH [57]). The docking calculations returned a very low Chemscore for the binding of fluconazole to CYP164A1, which is consistent with previous experimental determinations in which this compound was not observed to bind to CYP164A2. In combination, these results strongly suggest that azole-based inhibitors are likely to prove effective for targeting the sole cytochrome P450 from Mycobacterium leprae.
DISCUSSION
Although the precise function of CYP164 family enzymes, as with most mycobacterial P450s, remains unknown, the retention of CYP164A1 as the only cytochrome P450 gene in the M. leprae genome is highly suggestive of an important physiological role in the bacterial life cycle. The crystal structure of CYP164A2 shows that this family of CYP enzymes is characterized by an enlarged and highly hydrophobic active site, with the added feature that flexibility in the extended BC loop appears to provide a means to accommodate and adapt to a range of larger substrates. As further evidenced by the modeling studies, the site is sufficient to accommodate even the bulkiest of the azole inhibitors, all of which can make additional favorable contacts to the enzyme via their hydrophobic tail regions. Although our biochemical studies have yet to identify a clear substrate specificity for CYP164 enzymes, the size and composition of the active site are consistent with the binding of a range of fatty acids, and the shape of the ambiguous electron density observed for the active site of the enzyme suggests that a mixed population of fatty acid tails may be incorporated. The similarity of the active site with previously reported CYPs known to be active on fatty acids (e.g., correlation with the structure of P450BM-3 complexed with N-palmitoylglycine [21]), including the retention of the tyrosine and arginine residues in positions at the entrance of the channel, where they are capable of binding hydrophilic head groups, leads us to speculate that such compounds may form the natural substrates for CYP164 enzymes. Activity against a variety of such core compounds would be consistent with CYP164 enzymes playing an essential role within these mycobacteria. Preliminary screening studies indicated that CYP164A2 bound fatty acids with chain lengths of C12 to C18 (Ks, 16 to 62 μM), yielding type I difference spectra typical of CYP substrates (Fig. 4). The lack of UV-Vis binding spectra with alkanes and alkyl alcohols of similar chain lengths suggests that the CYP164A2 substrate is either a fatty acid or a substituted fatty acid that retains a functional carboxyl group.
CYP164A2 was previously noted to be unusual for a P450 in readily undergoing the transition from the low- to the high-spin state by increasing the ionic strength of the medium at room temperature (62). As this transition is normally accessed during substrate binding but is otherwise often difficult to attain, the CYP164 enzymes provide a valuable tool for the study of P450 actions. We cannot exclude the possibility that an unidentified ligand(s) tightly bound to the recombinant CYP164A2 may play a role in the apparent ease with which the spin state of CYP164A2 is modulated at room temperature, and further studies on this biophysical interest are ongoing.
It was speculated previously that the transition from the low- to the high-spin state involves the breaking of key structurally important internal salt bridges, leading to an open (relaxed) conformation, which promotes substrate binding (13). One feature of CYP164A2 seen in the crystal structure is the network of electrostatic interactions that order the BC loop at the entrance to the channel in the econazole complex. Their importance is highlighted by the solvent exposure of adjacent hydrophobic residues, in particular Phe 97 and Phe 99, for which the entropic cost of their exposure upon ordering must be exceeded by the contributions of the electrostatic network. It is plausible that high-salt conditions would disrupt this network, hence promoting the disordering of the loop and facilitating the exchange of the axial heme water, thus encouraging the formation of the high-spin state. This is consistent with our conclusion that the open structure of CYP164A2 is predominantly of the high-spin state, correlating with the solution data and disordered BC loop. As the lengthened BC loop accompanies an enlarged substrate binding site, the enhanced low- to high-spin transition may therefore reflect an inherent tendency of this region to disorder, hence altering the solvent accessibility of the heme pocket. Related cytochrome P450s with a lengthened BC loop are also known to be capable of accomplishing this change, for example, in M. tuberculosis CYP51 (32, 53, 54). P450cam (32) was also previously noted to change its conformation with ionic strength alterations, changing the solvent accessibility of the heme pocket, although this is believed to be triggered by a cluster of charged amino acids centering around Asp 251 and involving three of the central helices (15). This cluster is absent in the CYP164 family. CYP164A2 appears instead to provide a simple correlation between the salt dependence of a single loop structure and this transition.
Azole compounds have been reported to bind to a wide range of CYPs, but their selectivity can be refined to successfully target individual families, as previously seen with azole-based CYP51 antifungals (20, 48). Previously reported crystal structures of other P450 enzymes with econazole bound to the porphyrin iron (PDB accession numbers 3JUS and 2UVN [47]) are consistent with the association via the nitrogen in position 3 of the azole ring observed for the CYP164A2 complex. Small differences arise in the arrangements of the chlorinated phenyl rings due to the disparity in active-site geometries. CYP125 from Mycobacterium tuberculosis (39) was observed previously to bind a single econazole within the substrate access channel rather than at the heme, but this may be an artifact, as constriction toward the heme blocks further progression. The simultaneous binding of a cytochrome P450 by two azole compounds is unusual but not unique to CYP164A2. A crystal structure of human CYP3A4, which also has an enlarged active site, was reported previously with two ketoconazole molecules bound (17), an observation that is supported by binding analyses that confirm that two molecules of fluconazole can simultaneously bind to this CYP, as can two molecules of itraconazole (49). As with CYP164A2, the primary ketoconazole binds directly to the heme via the azole group. However, in contrast to CYP164A2, the second ketoconazole is located primarily via the antiparallel stacking of the methoxyl-phenyl piperazine rings. This mode of binding would be expected to be highly cooperative, whereas in CYP164A2, there are limited and offset contacts between the two econazoles, and hence, it is less clear that there is an interdependence of binding. A common feature of both enzymes is the rearrangement of the active-site entrance, particularly the BC loop, upon ligand binding, leading to a significant increase in the volume of the active site. Human CYP3A4 is known to be active on a wide range of substrates, correlating with its enlarged and adaptable active site. These same molecular features in CYP164 family enzymes suggest a similar level of promiscuity for substrates, perhaps reflecting the difficulty in experimentally assigning specificities for these enzymes. Multifunctional activity is also consistent with the preservation of CYP164A1 as the sole P450 in M. leprae.
The azole family of P450 inhibitors, originally used to treat fungal infections, have been reported to have considerable therapeutic potential against M. tuberculosis (2–4, 7). This family was also determined to have bactericidal activity against Mycobacterium smegmatis (40), with econazole found to be one of the most potent inhibitors. Azoles have widespread activity against many CYPs: M. smegmatis CYP164A2 was reported previously to bind econazole, miconazole, clotrimazole, voriconazole, ketoconazole, and itraconazole (62). These observations are consistent with the structural features of the enzyme and the docking studies reported in this study. The crystal structures also imply two possible avenues to increase the affinity and/or specificity of drugs for CYP164. First, the enlarged hydrophobic cavity immediately suggests that the further derivatization of existing azole inhibitors should be possible to increase selectivity for CYP164 and related cytochrome P450s. This is supported by the previous demonstration that CYP164A2 can accommodate larger azoles, such as itraconazole and ketoconazole, with the former having the highest affinity (Kd = 0.78 μM) for this enzyme. Second, the presence and location of the second bound econazole within the CYP164A2 structure are distinctive and immediately suggest the potential exploitation of this second binding site. This site provides a range of contact points that might be accessed directly or via the expansion of heme-bound azoles derivatized in this direction.
The inability to culture leprosy bacilli and the lack of a tractable clinical model have both impeded the identification of suitable molecular drug targets for this pathogen. This also prevents functional studies of individual enzymes, such as CYP164A1, through knockout studies or growth in selective media. Previous attempts to heterologously express CYP164A1 were reported to be unsuccessful (62). We have also unsuccessfully trialed a wide range of expression vectors and methods (data not shown). The intractability of the M. leprae enzyme to experimental studies has necessitated a structural modeling approach; this approach has been validated by parallel docking studies with CYP164A2, for which experimental data are available. The model constructed for CYP164A1 confirms that the characteristic features of the CYP164A2 active-site region are preserved in M. leprae P450, and therefore, this P450 should also be capable of similar interactions with azole family inhibitors. Although it is not currently possible to predict actual binding affinities from the docking simulations, trends in the scoring function values obtained provide an estimate of the likelihood of an effective association. The consistency of the binding conformations obtained with the crystal structures of both the econazole-CYP164A2 complex and other azole-P450 complexes provides a high level of confidence in the docking studies. Drug testing directly against leprosy bacilli is challenging, and to date, no trials with the azole antifungals have been reported. Two studies have recently tested a diarylquinoline (26) and nitroimidazopyran (36) against M. leprae. On the basis of the current study, similar trials with econazole and related inhibitors to target CYP164A1 may be worthwhile.
In summary, we report the first structural characterization of a CYP164 family enzyme both in the presence (closed, low-spin state) and in the absence (open, high-spin state) of the inhibitor econazole. The BC loop exhibits considerable flexibility and adaptability that suggest a broad substrate specificity, and a loop network of electrostatic interactions may explain the ready transition from the low- to the high-spin state observed for these enzymes. CYP164A2 can simultaneously accommodate two molecules of econazole within its enlarged active site. Molecular modeling of M. leprae CYP164A1 and its complexes with azoles is supportive of this enzyme providing a potential new drug target for M. leprae that can be accessed with azole-based therapeutics. The structures presented here could be used to initiate the rational design of inhibitors for evaluation as drugs. These studies therefore signpost a new strategy for leprosy drug development to alleviate this debilitating and deadly disease.
ACKNOWLEDGMENTS
This study was supported by a United Kingdom BBSRC postgraduate studentship award to C.R.J.A. and NHSB&T funding to N.M.B. Studies in the laboratory of R.L.B. are supported by grants from the Wellcome Trust (reference number 07746), the United Kingdom Biological and Biotechnology Research Council (BBSRC) (reference number BB/F007256), and NHSB&T. Studies in the laboratory of S.L.K. were supported by the Welsh Government and ERDF via the BEACON convergence project.
We thank the staff at Diamond Light Source for access to the synchrotron for data collection.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Ahmad Z, Pandey R, Sharma S, Khuller GK. 2008. Novel chemotherapy for tuberculosis: chemotherapeutic potential of econazole- and moxifloxacin-loaded PLG nanoparticles. Int. J. Antimicrob. Agents 31:142–146 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad Z, Sharma S, Khuller GK. 2005. In vitro and ex vivo antimycobacterial potential of azole drugs against Mycobacterium tuberculosis H37Rv. FEMS Microbiol. Lett. 251:19–22 [DOI] [PubMed] [Google Scholar]
- 3. Ahmad Z, Sharma S, Khuller GK. 2006. The potential of azole antifungals against latent/persistent tuberculosis. FEMS Microbiol. Lett. 258:200–203 [DOI] [PubMed] [Google Scholar]
- 4. Ahmad Z, et al. 2006. Antimycobacterial activity of econazole against multidrug-resistant strains of Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 28:543–544 [DOI] [PubMed] [Google Scholar]
- 5. Bellamine A, Mangla AT, Nes WD, Waterman MR. 1999. Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:8937–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byrne ST, Denkin SM, Gu P, Nuermberger E, Zhang Y. 2007. Activity of ketoconazole against Mycobacterium tuberculosis in vitro and in the mouse model. J. Med. Microbiol. 56:1047–1051 [DOI] [PubMed] [Google Scholar]
- 7. Castagnolo D, et al. 2009. Synthesis and biological evaluation of new enantiomerically pure azole derivatives as inhibitors of Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 19:2203–2205 [DOI] [PubMed] [Google Scholar]
- 8. Chen VB, et al. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 10. Cole ST, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011 [DOI] [PubMed] [Google Scholar]
- 11. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 12. Cuppvickery JR, Poulos TL. 1995. Structure of cytochrome P450eryf involved in erythromycin biosynthesis. Nat. Struct. Biol. 2:144–153 [DOI] [PubMed] [Google Scholar]
- 13. Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. 2004. An electrostatically driven conformational transition is involved in the mechanisms of substrate binding and cooperativity in cytochrome P450eryF. Biochemistry 43:6475–6485 [DOI] [PubMed] [Google Scholar]
- 14. Deprez E, Gill E, Helms V, Wade RC, Hui Bon Hoa G. 2002. Specific and non-specific effects of potassium cations on substrate-protein interactions in cytochromes P450cam and P450lin. J. Inorg. Biochem. 91:597–606 [DOI] [PubMed] [Google Scholar]
- 15. Di Primo C, Deprez E, Sligar SG, Hui Bon Hoa G. 1997. Origin of the photoacoustic signal in cytochrome P-450cam: role of the Arg186-Asp251-Lys178 bifurcated salt bridge. Biochemistry 36:112–118 [DOI] [PubMed] [Google Scholar]
- 16. Driscoll MD, et al. 2010. Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J. Biol. Chem. 285:38270–38282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ekroos M, Sjogren T. 2006. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U. S. A. 103:13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 [DOI] [PubMed] [Google Scholar]
- 20. Guardiola-Diaz HM, Foster LA, Mushrush D, Vaz AD. 2001. Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for treatment of tuberculosis. Biochem. Pharmacol. 61:1463–1470 [DOI] [PubMed] [Google Scholar]
- 21. Haines DC, Tomchick DR, Machius M, Peterson JA. 2001. Pivotal role of water in the mechanism of P450BM-3. Biochemistry 40:13456–13465 [DOI] [PubMed] [Google Scholar]
- 22. Huang CY. 1982. Determination of binding stoichiometry by the continuous variation method: the Job plot. Methods Enzymol. 87:509–525 [DOI] [PubMed] [Google Scholar]
- 23. Jackson CJ, Lamb DC, Kelly DE, Kelly SL. 2000. Bactericidal and inhibitory effects of azole antifungal compounds on Mycobacterium smegmatis. FEMS Microbiol. Lett. 192:159–162 [DOI] [PubMed] [Google Scholar]
- 24. Jackson CJ, et al. 2003. Conservation and cloning of CYP51: a sterol 14 alpha-demethylase from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 301:558–563 [DOI] [PubMed] [Google Scholar]
- 25. Jefcoate CR. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 52:258–279 [DOI] [PubMed] [Google Scholar]
- 26. Ji BH, Chauffour A, Andries K, Jarlier V. 2006. Bactericidal activities of R207910 and other newer antimicrobial agents against Mycobacterium leprae in mice. Antimicrob. Agents Chemother. 50:1558–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones G, Willett P, Glen RC, Leach AR, Taylor R. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267:727–748 [DOI] [PubMed] [Google Scholar]
- 28. Kelly SL, Lamb DC, Jackson CJ, Warrilow AG, Kelly DE. 2003. The biodiversity of microbial cytochromes P450. Adv. Microb. Physiol. 47:131–186 [DOI] [PubMed] [Google Scholar]
- 29. Lamb DC, Kelly DE, Baldwin BC, Kelly SL. 2000. Differential inhibition of human CYP3A4 and Candida albicans CYP51 with azole antifungal agents. Chem. Biol. Interact. 125:165–175 [DOI] [PubMed] [Google Scholar]
- 30. Lamb DC, et al. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277:24000–24005 [DOI] [PubMed] [Google Scholar]
- 31. Lange R, Bonfils C, Debey P. 1977. The low-spin/high-spin transition equilibrium of camphor-bound cytochrome P-450. Effects of medium and temperature on equilibrium data. Eur. J. Biochem. 79:623–628 [DOI] [PubMed] [Google Scholar]
- 32. Lepesheva GI, Virus C, Waterman MR. 2003. Conservation in the CYP51 family. Role of the B′ helix/BC loop and helices F and G in enzymatic function. Biochemistry 42:9091–9101 [DOI] [PubMed] [Google Scholar]
- 33. Leslie AGW, Powell HR. 2007. Processing diffraction data with MOSFLM, p 41–51 In Read RJ, Sussman JL. (ed), NATO Science Series II: Mathematics, Physics and Chemistry vol 245 Springer, New York, NY [Google Scholar]
- 34. Leys D, et al. 2003. Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450. J. Biol. Chem. 278:5141–5147 [DOI] [PubMed] [Google Scholar]
- 35. Lutz JD, et al. 2009. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem. Pharmacol. 77:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manjunatha UH, et al. 2006. Mycobacterium leprae is naturally resistant to PA-824. Antimicrob. Agents Chemother. 50:3350–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCoy AJ, et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGaughey GB, Gagne M, Rappe AK. 1998. Pi-stacking interactions. Alive and well in proteins. J. Biol. Chem. 273:15458–15463 [DOI] [PubMed] [Google Scholar]
- 39. McLean KJ, et al. 2009. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 284:35524–35533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McLean KJ, et al. 2002. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomycetes. Microbiology 148:2937–2949 [DOI] [PubMed] [Google Scholar]
- 41. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 42. Nelson DR. 2009. The cytochrome p450 homepage. Hum. Genomics 4:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239:2379–2385 [PubMed] [Google Scholar]
- 44. Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 239:2370–2378 [PubMed] [Google Scholar]
- 45. Ortiz de Montellano PR. 2004. Cytochrome P-450: structure, mechanism, and biochemistry, 3rd ed Kluwer Academics/Plenum, New York, NY [Google Scholar]
- 46. Ouellet H, Johnston JB, Ortiz de Montellano PR. 2010. The Mycobacterium tuberculosis cytochrome P450 system. Arch. Biochem. Biophys. 493:82–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ouellet H, Podust LM, de Montellano PR. 2008. Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J. Biol. Chem. 283:5069–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parker JE, et al. 2008. Differential azole antifungal efficacies contrasted using a Saccharomyces cerevisiae strain humanized for sterol 14 alpha-demethylase at the homologous locus. Antimicrob. Agents Chemother. 52:3597–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pearson JT, et al. 2006. Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry 45:6341–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57:1445–1450 [DOI] [PubMed] [Google Scholar]
- 51. Phillips JC, et al. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pikuleva IA, Mast N, Charvet C, Stout CD. 2010. Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain. J. Biol. Chem. 285:31783–31795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Podust LM, Poulos TL, Waterman MR. 2001. Crystal structure of cytochrome P450 14 alpha-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. U. S. A. 98:3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Podust LM, et al. 2004. Estriol bound and ligand-free structures of sterol 14 alpha-demethylase. Structure 12:1937–1945 [DOI] [PubMed] [Google Scholar]
- 55. Poulos TL, Finzel BC, Howard AJ. 1987. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 195:687–700 [DOI] [PubMed] [Google Scholar]
- 56. Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 57. Savino C, Montemiglio LC, Gianni S, Vallone B. 2010. Azole drugs trap cytochrome P450 EryK in alternative conformational states. Biochemistry 49:9199–9206 [DOI] [PubMed] [Google Scholar]
- 58. Reference deleted.
- 59. Scollard DM, et al. 2006. The continuing challenges of leprosy. Clin. Microbiol. Rev. 19:338–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sligar SG. 1976. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry 15:5399–5406 [DOI] [PubMed] [Google Scholar]
- 62. Warrilow AG, et al. 2009. Identification, characterization, and azole-binding properties of Mycobacterium smegmatis CYP164A2, a homolog of ML2088, the sole cytochrome P450 gene of Mycobacterium leprae. Antimicrob. Agents Chemother. 53:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yun CH, Ahn T, Guengerich FP. 1998. Conformational change and activation of cytochrome P450 2B1 induced by salt and phospholipid. Arch. Biochem. Biophys. 356:229–238 [DOI] [PubMed] [Google Scholar]
- 64. Yun CH, Song M, Ahn T, Kim H. 1996. Conformational change of cytochrome P450 1A2 induced by sodium chloride. J. Biol. Chem. 271:31312–31316 [DOI] [PubMed] [Google Scholar]