Abstract
We study the epidemiology, molecular basis, clinical risk factors, and outcome involved in the clonal dissemination of VIM-1-producing Klebsiella pneumoniae isolates in the hospital setting. All patients infected/colonized by carbapenem-nonsusceptible K. pneumoniae (CNSKP) in 2009 were included. Molecular epidemiology was studied by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Antibiotic resistance genes were analyzed by PCR and sequencing. Plasmids were studied by PFGE with S1 nuclease digestion and for incompatibility group by a PCR-based replicon typing scheme. Risk factors associated with CNSKP colonization/infection were assessed by an observational case-control study. All 55 patients studied were infected (n = 28) or colonized (n = 27) by VIM-1-producing K. pneumoniae. All but one acquired isolates of a single clone (PFGE cluster 1 [C1], sequence type 15 [ST15]), while another clone (PFGE C2, ST340) was detected in four patients. C1 isolates also produced the new extended-spectrum β-lactamase SHV-134. blaVIM-1 was carried in a class 1 integron and an untypeable plasmid of ∼50 bp. The number of days that the patient received mechanical ventilation, the use of parenteral nutrition, previous treatment with linezolid, and treatment with extended-spectrum cephalosporins for more than 7 days were detected to be independent risk factors for CNSKP acquisition. The VIM-1-producing K. pneumoniae ST15 clone has a high capacity to spread among intensive care unit patients with severe underlying conditions. A high rate of associated mortality and great difficulty in controlling the spread of this clone, without permanent behavioral changes in the personnel, were observed.
INTRODUCTION
Carbapenemases hydrolyze all beta-lactam antibiotics, including carbapenems (5, 19, 33, 37), and their high potential for rapid, wide dissemination constitutes a major clinical and public health threat (5, 18, 19, 24, 26–28, 37). Class B carbapenemases include metallo-β-lactamases (MBLs), such as VIM, IMP, and NDM-1 (19, 24). Acquired MBLs are increasingly reported from Enterobacteriaceae (5, 9, 14, 10, 24, 35, 37).
Klebsiella pneumoniae is a major cause of nosocomial infections, particularly in intensive care units (ICUs), representing a relevant potential clinical risk. During the last decade, VIM-type MBLs have spread in K. pneumoniae (2, 21, 31, 32, 35, 38) and outbreaks of such strains have been reported (2, 21, 32).
In Spain, VIM-1-producing Enterobacteriaceae have been described to be associated with single cases, small outbreaks (6, 31, 36), or polyclonal spread affecting different bacterial species (25, 35).
This study describes an extensive nosocomial outbreak caused by a clonal multidrug-resistant strain of K. pneumoniae producing VIM-1. Our aims were to assess the epidemiology, molecular basis, clinical risk factors, and outcomes involved in the acquisition and dissemination of VIM-1-producing K. pneumoniae in the hospital setting.
MATERIALS AND METHODS
Study design and bacterial isolates.
During 2009, an increase in the isolation of carbapenem-nonsusceptible K. pneumoniae (CNSKP) isolates was noted in the University Hospital Puerta de Hierro Majadahonda (UHPHM) in Madrid, Spain. This observation prompted the present investigation. UHPHM is a tertiary care hospital with 613 inpatient beds, 52 intensive care beds, and over 17,000 hospital admissions per year.
All the patients infected and/or colonized by CNSKP isolates between January 2009 and December 2009 were included in this study. An infected case caused by CNSKP was defined according to CDC criteria (17); a colonized case was defined as a patient carrying CNSKP without clinical evidence of infection.
Bacterial identification and susceptibility testing.
Species identification and antibiotic susceptibility testing were performed by broth microdilution (MicroScan; Siemens Healthcare Diagnostics, Deerfield, IL) and by Etest (AB Biodisk, Solna, Sweden). Susceptibility results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (8). Antibiotic-susceptible Escherichia coli ATCC 25922 and carbapenem-resistant E. coli MN4 (30) were used as quality control strains. Tigecycline susceptibility was determined by the Etest and interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (13).
Carbapenem nonsusceptibility was considered to be resistance or intermediate susceptibility to one of more of the three carbapenem antibiotics tested: imipenem, meropenem, and ertapenem.
The modified Hodge test was carried out using an imipenem disk; imipenem susceptibility alone and in combination with EDTA was also determined (Etest; AB Biodisk, Solna, Sweden).
Molecular epidemiology.
The genetic relatedness between the K. pneumoniae isolates was determined by pulsed-field gel electrophoresis (PFGE) after total chromosomal DNA digestion with XbaI (30).
K. pneumoniae isolates representing the different PFGE clusters were further studied by multilocus sequence typing (MLST) according to the Institut Pasteur scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html; last accessed December 2010).
Antibiotic resistance mechanisms and class 1 integron characterization.
Standard PCR conditions were used to amplify genes coding for carbapenemases (blaKPC, blaVIM, blaIMP, blaNDM), extended-spectrum β-lactamases (ESBLs; blaTEM, blaSHV, and blaCTX-M), and plasmid-mediated AmpC (blaCMY, blaFOX, blaMOX, blaDHA, blaEBC blaACC). Aminoglycoside acetylase resistance genes [aac(3)-IIa, aac(6′)-Ib] and 16S RNA methylases (armA, rmtA, rmtB) were also screened (15, 30).
Class 1 integron structures, the integrase gene intI1, and the variable regions, as well as the linkage of the blaSHV-134 allele with IS26, were screened by PCR amplification and DNA sequencing (12, 34).
In addition, the entire sequences of the ompK35 and ompK36 genes in four isolates representing the two CNSKP clones detected were analyzed by PCR and DNA sequencing (20).
Conjugation assay and plasmid characterization.
Conjugation experiments were performed with four donor K. pneumoniae clinical isolates, previously selected according to PFGE results, using the kanamycin-azide resistant E. coli HB101 as a recipient. Putative transconjugants were selected on Mueller-Hinton agar plates containing kanamycin (100 μg/ml), azide (160 μg/ml), and cefotaxime (4 μg/ml).
Plasmids were classified according to their incompatibility group by a PCR-based replicon-typing scheme (3, 4). PFGE with S1 nuclease digestion of whole genomic DNA (S1-PFGE) was used to detect plasmids as previously described (16).
Case-control clinical study.
A retrospective observational case-control study was conducted among patients admitted to the medical ICU (MICU) or surgical ICU (SICU) between January and December 2009.
All the ICU patients were screened by culture of rectal swab specimens at least once a week until hospital discharge. A case was defined as a patient admitted to either of the two ICUs during the study period who presented infection or colonization by VIM-1-producing K. pneumoniae after 48 h of admission in the ICU. Control patients were randomly selected from monthly lists of patients admitted to the ICUs during the same period as the case patient with an ICU stay greater than 48 h and with no evidence of CNSKP infection or colonization in any of the clinical or surveillance samples taken.
Death associated with the infection was defined as persistence of signs and symptoms of infection by VIM-1-producing K. pneumoniae at the time of death or death occurring within 14 days after the diagnosis of VIM-1-producing K. pneumoniae infection without evidence of another obvious cause.
Information about the potential clinical risk factors (underlying medical conditions, as well as surgical procedures, including use and duration of invasive devices and type and duration of antimicrobial treatment) were recorded. In addition, Charlson modified index (1, 7) and Apache II scores were also recorded on admission to the ICU (22).
The periods used for the determination of risk factors were from admission until the date of colonization or infection by VIM-1-producing K. pneumoniae (for cases) and from admission to the time of discharge from the ICU (for controls).
Statistical analysis.
Comparison of discrete variables was performed by univariate logistic regression. Continuous variables were compared by Student's t test or the Wilcoxon rank sum (Mann-Whitney) test, as appropriate. Two multivariate logistic regression models were developed with the purpose of determining the potential independent risk factors associated with VIM-1-producing K. pneumoniae infection/colonization, adjusted by Apache II score: a general model and a model that included only the use of antibiotics as a risk factor. On the basis of a complete model, which included all the risk factors that showed a P value of <0.20 in the univariate analysis, the final model was built using the maximum likelihood method using a backward strategy of selecting variables. Adjusted odds ratios (ORs), including their 95% confidence intervals (CIs), were calculated. Two-tailed tests were used for all analyses. P values of <0.05 were considered statistically significant. Statistical procedures were performed using the Stata (version 9.0 SE) statistical package.
RESULTS AND DISCUSSION
Patients and infections.
During 2009, CNSKP isolates were obtained from clinical samples from 55 patients admitted to either the SICU (n = 36, 65.5%) or the MICU (n = 19, 34.5%). Of the 55 patients, 28 (50.9%) were infected by CNSKP, while 27 (49.1%) had no clinical evidence of infection and were considered colonized. The incidence of new cases in the SICU was significantly higher than that in the MICU (16.7 versus 6.1 cases/1,000 patient-days) (relative risk, 2.7; 95% CI, 1.6 to 4.7; P < 0.001). Thirty-five (63.6%) patients were male. Twenty-one (38.2%) were >65 years old, and 34 (61.8%) were >18 and ≤65 years old. The median MICU/SICU stay was 36 days (range, 3 to 101 days).
The clinical characteristics, treatments, and outcomes of the patients infected by CNSKP are detailed in Table 1. The most prevalent infections diagnosed were pneumonia and catheter-associated bacteremia, with seven cases (25%) each. Of the 27 patients colonized, CNSKP was recovered from exudates of the rectum (22 cases; 78.6%), trachea (3 cases; 11.1%), urethra (1 case; 3.7%), and bladder catheter (1 case; 3.7%).
Table 1.
Clinical characteristics of patients infected with VIM-1-producing Klebsiella pneumoniaea
| Gender | Age (yr) | Unit | Charlson index score | Apache II index | Type of infection | Antimicrobial therapy | Total no. of antimicrobial days | Adjunctive therapy | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| M | 60 | SICU | 3 | 28 | Pneumonia | TGC + CST | 50 | None | Death |
| F | 66 | MICU | 3 | 12 | UTI | AMK | 8 | BC change | Cure |
| M | 67 | MICU | 4 | 25 | Pneumonia | Meropenem | 12 | CVC change | Cure |
| M | 53 | SICU | 2 | 8 | Intra-abdominal | TGC | 9 | None | Cure |
| M | 46 | MICU | 1 | 21 | CAB | TGC | 4 | CVC removal | Cure |
| M | 35 | SICU | 3 | 17 | CAB | TGC | 34 | CVC change | Cure |
| F | 65 | MICU | 0 | 26 | Meningitis | TGC + CST | 26 | CVC change | Cure |
| F | 64 | MICU | 7 | 23 | UTI | TGC | 11 | None | Death |
| F | 38 | SICU | 0 | 20 | CAB | None | 0 | PVC change | Cure |
| M | 61 | MICU | 4 | 15 | LRTI | TGC + CST | 10 | CVC change | Death |
| F | 54 | SICU | 4 | 23 | Pneumonia, CAB | TGC + CST | 10 | CVC and ET change | Death |
| M | 68 | SICU | 6 | 17 | Meningitis | TGC | 7 | CVC and VS change | Death |
| M | 61 | MICU | 3 | 28 | Pneumonia | TGC + CST | 25 | None | Cure |
| M | 43 | MICU | 2 | 31 | Pneumonia | TGC + CST | 43 | None | Cure |
| F | 72 | SICU | 3 | 23 | UTI | None | 0 | CVC change | Death |
| M | 46 | SICU | 3 | 15 | Meningitis | None | 0 | PVC change | Death |
| M | 34 | MICU | 0 | 11 | UTI | None | 0 | BC change | Cure |
| M | 43 | SICU | 3 | 18 | Pneumonia, CAB | TGC + CST | 7 | CVC and PVC change | Death |
| M | 51 | SICU | 4 | 26 | LRTI | Ertapenem | 12 | PVC removal | Cure |
| M | 39 | SICU | 1 | 24 | LRTI | AMK + TGC + CST | 28 | None | Cure |
| M | 54 | SICU | 2 | 21 | LRTI | TGC + CST | 28 | None | Death |
| M | 80 | SICU | 2 | 16 | Intra-abdominal | TGC | 3 | CVC change | Death |
| F | 58 | MICU | 2 | 23 | UTI | TGC | 2 | BC change | Cure |
| M | 69 | SICU | 7 | 19 | CAB | TGC + CST | 38 | CVC change | Death |
| M | 59 | SICU | 6 | 29 | Soft tissue | TGC | 31 | None | Death |
| F | 63 | SICU | 4 | 21 | LRTI | TGC + CST | 21 | CVC removal | Cure |
| M | 35 | SICU | 0 | 17 | CAB | TGC | 2 | None | Cure |
| F | 66 | SICU | 3 | 29 | Pneumonia | TGC + CST | 38 | CVC and PD change | Death |
M, male; F, female; MICU, medical intensive care unit; SICU, surgical intensive care unit; AMK, amikacin; TGC, tigecycline; CST, colistin; CAB, catheter-associated bacteremia; UTI, urinary tract infection; LRTI, lower respiratory tract infection; CVC, central venous catheter; PVC, peripheral venous catheter; BC, bladder catheter; ET, endotracheal tube; VS, ventricular shunt; PD, percutaneous drainage.
Of the 55 patients studied, 45 (81.8%) had previous underlying conditions, and 25 of them (55.6%) had more than one. The most prevalent underlying diseases were neoplasia (13 cases; 25.5%), immunosuppression due to solid organ transplantation (12 cases; 21.8%), and heart diseases (21 cases; 38.2%).
Thirteen (46.4%) infected patients and 11 (40.7%) colonized patients, all of them with severe underlying diseases, died.
Twenty-two of the 28 (78.6%) infected patients were treated with antibiotics active against CNSKP, mainly tigecycline in monotherapy (9 cases; 32.1%) or in combination with colistin (11 cases; 39.3%) (Table 1). Of the remaining 6 infected patients, 2 were treated with meropenem or ertapenem because their isolates' MICs for both antibiotics were 1 μg/ml and interpreted to be susceptible according to previous CLSI criteria (ertapenem MIC ≤ 2 μg/ml and meropenem MIC ≤ 4 μg/ml; document M100-S20 [7a]), 2 did not receive treatment due to early death, and the clinical condition of 2 patients improved after central venous catheter or bladder catheter removal (Table 1).
Nineteen of the 28 patients infected (67.9%) received adjuvant treatment, mainly change or removal of the central venous catheter (14 patients) (Table 1).
Susceptibility testing of CNSKP isolates.
During the study period, 335 CNSKP isolates were recovered from different samples from the 55 patients studied. All of them were resistant to ampicillin, amoxicillin-clavulanic acid, cefoxitin, cefotaxime, ceftazidime, piperacillin-tazobactam, gentamicin, tobramycin, ciprofloxacin, and co-trimoxazole; 36% were susceptible to amikacin. The MICs for tigecycline ranged from 0.25 μg/ml to 4 μg/ml (MIC50 = 0.5 μg/ml, MIC90 = 1 μg/ml). All isolates were susceptible to colistin (MICs < 2 μg/ml).
Imipenem, meropenem, and ertapenem MICs ranged from 1 to >8 μg/ml (MIC50 and MIC90, >8 μg/ml in all cases). The modified Hodge test with imipenem or imipenem-EDTA Etest strips was positive for all CNSKP isolates.
Molecular epidemiology.
In total, 99 representative CNSKP isolates were subjected to further molecular epidemiology studies. PFGE results revealed two well-defined clusters. Cluster 1 (C1) was predominant in both infected (n = 27) and colonized (n = 27) patients, while isolates of cluster 2 (C2) were isolated from only four patients, although only one was infected; the three patients colonized by isolates of C2 were also infected (n = 1) or colonized (n = 2) by isolates of C1.
According to the minor banding pattern differences found, C1 could be subdivided into four subtypes, C1A to C1D; two subtypes were also identified in C2, C2A and C2B (Fig. 1). By MLST analysis, C1 and C2 were identified to be sequence type 15 (ST15) (six isolates tested, three of subtype C1A and one each of C1B, C1C, and C1D) and ST340 (three isolates tested, two of subtype C2A and one of C2B), respectively (Fig. 1). This is the first description of VIM-1-producing K. pneumoniae ST15, an MLST type previously associated with CTX-M-15-producing epidemic clones in Hungary and Denmark (11, 29). ST340 is a single-locus variant of the widely disseminated KPC-producing ST258 strain (18).
Fig 1.
Dendrogram illustrating the PFGE profiles and MLST types of the two different clones of VIM-1-producing Klebsiella pneumoniae isolates and the number of patients infected and/or colonized by each. One patient was infected by clone C1/ST15 and colonized by clone C2/ST340. Two patients were colonized by both C1/ST15 and C2/ST340.
Antibiotic resistance genes and class 1 integron characterization.
blaVIM-1 was identified in both the C1/ST15 and C2/ST340 clones. blaVIM-1 was carried in a class 1 integron in the following cassette combination: intI1 (integrase gene)–blaVIM-1–aac(6′)-Ib (tobramycin resistance gene, also called aacA4)–dhfrII (trimethoprim resistance gene)–aadA1 (streptomycin resistance gene)–catB2 (chloramphenicol resistance gene)–qacEδ1/sul-1 (quaternary ammonium compound resistance gene/sulfonamide resistance gene).
C1/ST15 isolates also had the blaSHV-134 gene encoding the new ESBL SHV-134 (http://www.lahey.org/studies/webt.asp). blaSHV-134 has a single nucleotide change at position 448 (C → G), in comparison with the sequence of blaSHV-12, which codes for an amino acid substitution at position 154 (Q → E) (GenBank accession number HM559945). The insertion sequence IS26 was detected linked 73 bp upstream of blaSHV-134, as previously described in the blaSHV-12 gene (12). This fact supports the idea of genetic evolution of blaSHV-134 from the blaSHV-12 gene.
The blaTEM-1 gene and the aminoglycoside resistance gene aac(3′)-IIa were also identified in K. pneumoniae isolates representative of both clones.
A selected sample of four K. pneumoniae isolates representing the different clones (two C1/ST15 isolates and two C2/ST340 isolates) with MICs of >8 μg/ml to imipenem, meropenem, and ertapenem was studied further to determine ompK35 and ompK36 sequences. In the two C1/ST15 isolates, the DNA sequences of ompK35 and ompK36 showed point mutations at positions 690 (G → A) and 360 (C → A), generating TGA and TAA premature stop codons, respectively. In the two C2/ST340 isolates, the sequence of ompK35 had a point mutation at position 520 (C → T), generating a TAG premature stop codon, whereas no changes were detected in the ompK36 sequence.
Conjugation assay and plasmid characterization.
Carbapenem-nonsusceptible E. coli transconjugants were obtained from isolates of the C1/ST15 and C2/340 clones (Table 2). All transconjugants carried a plasmid of ∼50 kb that was untypeable by PCR and from which positive identification of the blaVIM-1 gene and a class 1 integron was obtained (Table 3). Untypeable plasmids of a similar size were found in a recent study by Miró et al. (25). Previous European studies had detected blaVIM-1 in association with plasmids of incompatibility groups N (32), I (35), and HI2 (25, 35).
Table 2.
Antibiotic susceptibilities of VIM-1-producing Klebsiella pneumoniae isolates K534 of clone C1/ST15 and K535 of clone C2/ST340 and their transconjugants, TC1K534, TC2K534 and TC1K535
| Antibiotica | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|
| K534 | TC1K534 | TC2K534 | K535 | TCK535 | E. coli HB101 | |
| Ampicillin | >16 | >16 | >16 | >16 | >16 | ≤4 |
| Amox/clav | >16 | >16 | 8 | >16 | >16 | ≤4 |
| Cefazolin | >16 | >16 | >16 | >16 | >16 | ≤4 |
| Cefoxitin | >16 | >16 | ≤4 | >16 | 16 | ≤4 |
| Cefotaxime | >64 | 32 | 4 | >64 | 64 | ≤1 |
| Ceftazidime | >128 | 128 | 32 | >128 | 64 | ≤1 |
| Cefepime | >16 | 8 | 2 | >16 | 4 | ≤1 |
| Aztreonam | >16 | ≤1 | >16 | ≤1 | ≤1 | ≤1 |
| Imipenem | >32 | 4 | 0.12 | 16 | 4 | 0.12 |
| Meropenem | >32 | 1 | 0.03 | 4 | 1 | 0.03 |
| Ertapenem | >32 | 1 | 0.015 | 4 | 1 | 0.015 |
| Piper/taz | >64 | >64 | ≤8 | >64 | >64 | ≤8 |
| Ciprofloxacin | >2 | ≤0.12 | ≤0.12 | >2 | ≤0.12 | ≤0.12 |
| Gentamicin | >8 | ≤ 2 | >8 | 4 | ≤2 | ≤2 |
| Tobramycin | >8 | 4 | >8 | 8 | 4 | ≤2 |
| Amikacin | 32 | ≤8 | 16 | ≤8 | ≤8 | ≤8 |
Amox/clav, amoxicillin-clavulanic acid; Piper/taz, piperacillin-tazobactam.
Table 3.
Molecular characteristics of VIM-1-producing Klebsiella pneumoniae isolates K534 of clone C1/ST15 and K535 of clone C2/ST340 and their transconjugants, TC1K534, TC2K534 and TC1K535
| Characteristic | Resulta |
|||||
|---|---|---|---|---|---|---|
| K534 | TC1K534 | TC2K534 | K535 | TCK535 | E. coli HB101 | |
| Modified Hodge test | Pos | Pos | Neg | Pos | Pos | Neg |
| Imipenem-EDTA synergy test | Pos | Pos | Neg | Pos | Pos | Neg |
| blaVIM-1 | Pos | Pos | Neg | Pos | Pos | |
| blaSHV-134 | Pos | Neg | Pos | Neg | Neg | |
| blaTEM-1 | Pos | Neg | Pos | Pos | Neg | |
| aac(6′)-Ib | Pos | Pos | Neg | Pos | Pos | |
| aac(3′)-IIa | Pos | Neg | Pos | Pos | Neg | |
| Class 1 integron | Pos | Pos | Neg | Pos | Pos | |
| PFGE profile | C1 | C2 | ||||
| MLST | ST15 | ST340 | ||||
| Plasmid Inc group | Untypeable | FIIA | Untypeable | |||
| PFGE S1 fragment size (kb) | ∼170, ∼75, ∼50 | ∼50 | ∼75 | ∼250, ∼170, ∼50 | ∼50 | |
Pos, positive; Neg, negative.
In addition, cefotaxime-resistant but carbapenem-susceptible transconjugants were also obtained from isolates of the C1/ST15 clone that were TEM-1 and SHV-134 producers and carried a single IncFIIA plasmid of ∼75 kb (Tables 2 and 3). Simultaneous production of VIM-1 and ESBLs of the SHV family has previously been described in K. pneumoniae (2, 32, 35), but this is the first report of the novel ESBL SHV-134 and VIM-1. blaSHV-134 is located in a conjugative IncFII plasmid other than the plasmid harboring the blaVIM-1 gene. Previous reports described the association of blaSHV-12 with IncFII plasmids (12, 29).
Case-control clinical study.
All the 55 patients infected and/or colonized by VIM-1-producing K. pneumoniae isolates and 55 control patients were included in the study. No differences between case and control patients regarding age, gender, and frequency of surgery could be found (Table 4).
Table 4.
Univariate analysis of risk factors linked to infection or colonization by VIM-1-producing Klebsiella pneumoniae
| Characteristica | Case patients (n = 55) | Control patients (n = 55) | OR | 95% CI | P |
|---|---|---|---|---|---|
| Median (range) no. of days at risk | 18 (5–63) | 7 (3–110) | 0.001b | ||
| Mean (SD) Apache II risk index per 10 units of increment | 41.0 (22.9) | 18.8 (8.3) | 1.5 | 0.9–2.5 | 0.08 |
| Mean (SD) Charlson index | 3.2 (2.2) | 3.5 (2.2) | 0.9 | 0.8–1.1 | 0.5 |
| No. (%) of patients with the following specific risk factors: | |||||
| Chronic lung disease | 1 (1.8) | 5 (9.1) | 0.2 | 0.03–2.2 | 0.1 |
| Diabetes mellitus | 7 (12.8) | 11 (20.0) | 0.6 | 0.2–1.6 | 0.3 |
| Chronic renal insufficiency | 6 (10.9) | 6 (10.9) | 1 | 0.3–3.3 | 1 |
| Malignancy | 14 (25.5) | 13 (23.6) | 1.1 | 0.5–2.6 | 0.8 |
| Cardiovascular disease | 21 (38.2) | 24 (43.6) | 0.8 | 0.4–1.7 | 0.6 |
| Liver disease | 3 (5.6) | 6 (10.9) | 0.5 | 0.1–2 | 0.3 |
| Blood disease | 3 (5.5) | 4 (7.3) | 0.7 | 0.2–3.5 | 0.7 |
| Neurologic disease | 10 (18.2) | 5 (9.9) | 2.2 | 0.7–7.0 | 0.1 |
| Immunosuppression | 12 (21.8) | 16 (29.1) | 0.7 | 0.3–1.7 | 0.3 |
| No. (%) of patients with the following health care-associated factors: | |||||
| Presence of CVC (days) | |||||
| 0–7 | 6 (10.9) | 34 (61.8) | 1 | ||
| 8–21 | 29 (52.7) | 16 (29.1) | 10.3 | 3.6–29.7 | <0.001 |
| >21 | 20 (36.4) | 5 (9.1) | 22.7 | 6.1–83.9 | <0.001 |
| Presence of PVC (days) | |||||
| 0–7 | 14 (25.5) | 32 (58.2) | 1 | ||
| >7 | 41 (74.6) | 23 (41.8) | 4.1 | 1.8–9.2 | 0.001 |
| Presence of mechanical ventilation (days) | |||||
| 0–7 | 7 (14.9) | 34 (72.3) | 1 | ||
| 8–14 | 14 (29.8) | 8 (17) | 6.7 | 2.4–18.6 | <0.001 |
| >14 | 26 (55.3) | 5 (10.6) | 5.0 | 7.0–84.2 | <0.001 |
| Presence of nasogastric tube (days) | |||||
| 0–7 | 9 (19.2) | 25 (74.5) | 1 | ||
| 8–14 | 12 (25.5) | 9 (19.2) | 4.4 | 1.6–12.3 | 0.004 |
| >14 | 26 (55.3) | 3 (6.4) | 5.0 | 6.5–74.2 | <0.001 |
| Tracheostomy | 31 (66) | 9 (19.2) | 7.0 | 3.0–16.5 | <0.001 |
| Parenteral nutrition | 33 (70.2) | 16 (34) | 4.1 | 2.4–12.6 | <0.001 |
| Mean total with invasive devices | 5.2 (1) | 3.6 (1.4) | 2.8 | 1.9–4.1 | <0.001 |
| Median (range) total hospital LOS (days) | 36 (3–101) | 7 (3–110) | <0.001b | ||
| Mean (SD) total no. of antimicrobials | 5.0 (2.3) | 2.9 (2.4) | 1.4 | 1.2–1.7 | <0.001 |
| No. (%) of patients with the following antimicrobial use characteristics: | |||||
| Total length of antimicrobial use (days) | |||||
| 0–14 | 20 (36.4) | 40 (72.7) | 1 | ||
| 15–21 | 15 (27.3) | 8 (14.6) | 3.8 | 1.4–10.3 | 0.01 |
| >21 | 20 (36.4) | 7 (12.7) | 5.7 | 2.1–15.8 | 0.001 |
| Extended-spectrum cephalosporin use for >7 days | 15 (27.8) | 4 (7.3) | 4.8 | 1.5–15.5 | 0.009 |
| Antimicrobial(s) used | |||||
| Quinolones | 46 (83.6) | 30 (54.6) | 4.6 | 1.8–10.4 | 0.001 |
| Carbapenems | 32 (58.2) | 15 (27.3) | 3.7 | 1.7–8.3 | 0.001 |
| Piperacillin-tazobactam | 24 (43.6) | 12 (21.8) | 2.8 | 1.2–6.4 | 0.016 |
| Aminoglycosides | 5 (9.1) | 6 (10.9) | 1.2 | 0.4–4.3 | 0.751 |
| Linezolid | 38 (69.1) | 12 (21.8) | 8.0 | 3.4.18.9 | <0.001 |
CVC, central venous catheter; PVC, peripheral venous catheter; LOS, length of stay.
Mann-Whitney U test.
The total hospital stay was longer for cases than for controls (median stay, 36 versus 7 days; P < 0.001), as was the Apache II index (21.4 versus 18.8; P = 0.08). No significant differences in frequency of underlying diseases were observed between the two groups; however, the case-fatality ratio was significantly higher for case patients than for control patients (45.5% and 30.9%, respectively; P = 0.003).
In general, infection or colonization by VIM-1-producing K. pneumoniae was significantly associated with total previous exposure to invasive devices and a lengthened stay in the ICU (P < 0.001). Previous exposures included, in particular, central and peripheral venous catheters, mechanical ventilation, a nasogastric tube, parenteral nutrition, and a tracheotomy. Cases also received more antibiotics and received antibiotics for longer periods of time than their control counterparts (Table 4). Furthermore, use of quinolones, carbapenems, piperacillin-tazobactam, linezolid, and extended-spectrum cephalosporins for more than 7 days was associated with a greater risk of acquiring CNCKP (Table 4). The main independent risk factors identified by the general multivariate model were the number of days that the patient received mechanical ventilation and the use of parenteral nutrition; previous treatment with linezolid and extended-spectrum cephalosporins for more than 7 days was identified as a risk factor by the antimicrobial use model (Table 5).
Table 5.
Potential risk factors associated with VIM-1-producing Klebsiella pneumoniae colonization or infection as determined by multivariate analysisa
| Model | OR | 95% CI | P |
|---|---|---|---|
| General model | |||
| Presence of mechanical ventilation (days) | 1.2 | 1.1–1.3 | <0.001 |
| Use of parenteral nutrition | 3.6 | 1.4–9.4 | 0.009 |
| Model with specific antimicrobialsb | |||
| Extended-spectrum cephalosporins (>7 days) | 4.2 | 1.1–15.3 | 0.032 |
| Linezolid | 7.5 | 3.1–18.3 | <0.001 |
Multivariate analyses were performed by logistic regression adjusted for Apache II index.
The remaining risk factors included in the general model are not shown because the ORs, 95% CIs, and P values did not change significantly.
Taking into account the fact that the size of this observational study precludes the identification of several independent factors, which would otherwise be interrelated, the factors that were identified indicated that very sick patients who required intensive care for long periods of time was at greater risk for infection or colonization by VIM-1-producing K. pneumoniae. These factors give greater opportunities to acquire bacteria, especially in an environment where intensive antibiotic treatments select those species resistant to many antibiotics. One of the interesting findings in this observational study is the independent association of a previous exposure of cases to linezolid. On the one hand, it could be signaling those patients who developed more severe infections and were empirically treated for infections with Gram-positive microorganisms, but, on the other hand, linezolid could be helping to eradicate patients' Gram-positive bacterial flora, thereby allowing colonization by resistant Gram-negative bacteria.
Epidemic curve and infection control measures.
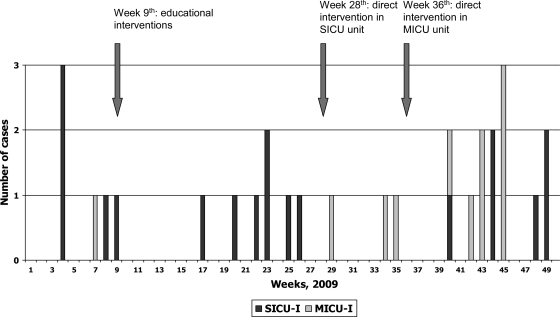
The epidemic curve of patients infected by CNSKP in the two ICU units is displayed in Fig. 2. Almost all the MICU cases appeared in the second half of the year, from week 27 to year end. The dissemination of CNSKP from the SICU to the MICU during the summer period was linked to the fact that the MICU was closed; therefore, MICU patients were allocated to one of the SICU modules and nurses, registered nurses, warden, and cleaning personnel were shared by MICU and SICU patients. This circumstance allowed dissemination of the bacteria between the two populations, indicating that contact transmission is a key factor for the spread of the epidemic clone.
Fig 2.
Epidemic curve of cases of infection by VIM-producing Klebsiella pneumoniae in two intensive care units. SICU-I, infected patients in surgical intensive care unit; MICU-I, infected patients in medical intensive care unit.
Infection control personnel identified all patients with VIM-1-producing K. pneumoniae isolates recovered from any clinical specimen according to the microbiology laboratory reports. In addition, active surveillance of all patients admitted to the ICUs was performed once a week. Patients identified to be harboring VIM-1-producing K. pneumoniae were assigned to contact precautions, including a single room, when available, or patient cohorting and the use of gowns and gloves that were discarded after caring for a patient. In addition, standard precautions were reinforced for all patients admitted to the ICUs, including improvement of hand hygiene compliance through the use of alcohol rubs before and after caring for patients. The outbreak described here did not stop during 2010 (data not shown) but remained at a lower incidence toward endemicity (incidence rate, 4.4 cases/1,000 patient-days in 2010). Eradication of these bacteria in this vulnerable population can be very difficult to achieve, in spite of drastic measures, such as an aggressive infection control strategy.
In this outbreak, we observed three periods without cases of infection: between weeks 10 and 16, 31 and 33, and 36 and 39 (Fig. 2). These periods followed both the educational series in the SICU on week 9 and the direct observation of the personnel and the requirement to comply with basic hand hygiene and contact precaution measures on weeks 28 to 29 and 36 to 37 for the SICU and MICU, respectively (Fig. 2). The latter intervention is costly and could not be continued due to economic constraints. Other potentially effective but costly interventions, which include the cohorting of carriers and staff or even closing of the units and the total removal of all environmental as well as patient reservoirs, were not undertaken (23).
ACKNOWLEDGMENTS
This study was supported by the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III; the Spanish Network for Research in Infectious Diseases (REIPI C03/14 and REIPI RD06/0008); a research grant from the Fondo de Investigaciones Sanitarias (FIS PI09/917); and research intramural grants from the Instituto de Salud Carlos III (MPY 022/09) and from the the Dirección General de Salud Pública, Ministry of Health, Spain (reference DGVI 1409/10-TS-15).
J. Oteo, A. Vindel, O. Cuevas, and J. Campos are members of the Spanish Network in infectious Pathology (REIPI).
We also thank Martin Hadley-Adams for assisting with the English language and preparation of the manuscript.
Footnotes
Published ahead of print 17 October 2011
REFERENCES
- 1. Beddhu S, Bruns FJ, Saul M, Seddon P, Zeidel ML. 2000. A simple comorbidity scale predicts clinical outcomes and costs in dialysis patients. Am. J. Med. 108:609–613 [DOI] [PubMed] [Google Scholar]
- 2. Cagnacci S, et al. 2008. Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem hydrolysing VIM-1 metallo-β-lactamase: first Italian outbreak. J. Antimicrob. Chemother. 61:296–300 [DOI] [PubMed] [Google Scholar]
- 3. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 4. Carattoli A, et al. 2006. Replicon typing of plasmids encoding resistance to newer β-lactams. Emerg. Infect. Dis. 12:1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carmeli Y, et al. 2010. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin. Microbiol. Infect. 16:102–111 [DOI] [PubMed] [Google Scholar]
- 6. Cendejas E, Gómez-Gil R, Gómez-Sánchez P, Mingorance J. 2010. Detection and characterization of Enterobacteriaceae producing metallo-β-lactamases in a tertiary-care hospital in Spain. Clin. Microbiol. Infect. 16:181–183 [DOI] [PubMed] [Google Scholar]
- 7. Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40:373–383 [DOI] [PubMed] [Google Scholar]
- 7a. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing, 20th informational supplement, M100-S20, vol. 30, no. 1 Clinical and laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing, 20th informational supplement, M100-S21, vol. 31, no. 1 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Cornaglia G, et al. 2007. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 10. Crowder MW, Spencer J, Vila AJ. 2006. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39:721–728 [DOI] [PubMed] [Google Scholar]
- 11. Damjanova I, et al. 2008. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new “MRSAs”? J. Antimicrob. Chemother. 68:978–985 [DOI] [PubMed] [Google Scholar]
- 12. Diestra K, et al. 2009. Characterization of plasmids encoding blaESBL surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 63:60–66 [DOI] [PubMed] [Google Scholar]
- 13. European Committee on Antimicrobial Susceptibility Testing 2011. Clinical breakpoints. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/clinical_breakpoints/ Last accessed 15 January 2011 [Google Scholar]
- 14. Falcone M., et al. 2009. Infections with VIM-1 metallo-β-lactamase-producing Enterobacter cloacae and their correlation with clinical outcome. J. Clin. Microbiol. 47:3514–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fritsche TR, Castanheira M, Miller GH, Jones RN, Armstrong ES. 2008. Detection of methyltransferases conferring high-level resistance to aminoglycosides in Enterobacteriaceae from Europe, North America, and Latin America. Antimicrob. Agents Chemother. 52:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. GarcíA A, et al. 2007. Acquisition and diffusion of blaCTX-M-9 gene by R478-IncHI2 derivative plasmids. FEMS Microbiol. Lett. 271:71–77 [DOI] [PubMed] [Google Scholar]
- 17. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. 1988. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128–140 [DOI] [PubMed] [Google Scholar]
- 18. Giakoupi P, et al. 2009. KPC-2-producing Klebsiella pneumoniae infections in Greek hospitals are mainly due to a hyperepidemic clone. Euro Surveill. 14(21):pii=19218. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19218 [DOI] [PubMed] [Google Scholar]
- 19. Grundmann H, et al. 2010. Carbapenem-nonsusceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 15(46):pii=19711. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19711 [DOI] [PubMed] [Google Scholar]
- 20. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kassis-Chikhani N, et al. 2010. Extended measures for controlling an outbreak of VIM-1 producing imipenem-resistant Klebsiella pneumoniae in a liver transplant centre in France, 2003-2004. Euro Surveill. 15(46):pii=19713. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19713 [DOI] [PubMed] [Google Scholar]
- 22. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. 1985. APACHE II: a severity of disease classification system. Crit. Care Med. 13:818–829 [PubMed] [Google Scholar]
- 23. Laurent C, et al. 2008. Intensive care unit outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae controlled by cohorting patients and reinforcing infection control measures. Infect. Control Hosp. Epidemiol. 29:517–524 [DOI] [PubMed] [Google Scholar]
- 24. Miriagou V, et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 25. Miró E, et al. 2010. Spread of plasmids containing the blaVIM-1 and blaCTX-M genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J. Antimicrob. Chemother. 65:661–665 [DOI] [PubMed] [Google Scholar]
- 26. Munoz-Price LS, et al. 2010. Successful eradication of a monoclonal strain of Klebsiella pneumoniae during a K. pneumoniae carbapenemase-producing K. pneumoniae outbreak in a surgical intensive care unit in Miami, Florida. Infect. Control Hosp. Epidemiol. 31:1074–1077 [DOI] [PubMed] [Google Scholar]
- 27. Nadkarni AS, Schliep T, Khan L, Zeana CB. 2009. Cluster of bloodstream infections caused by KPC-2 carbapenemase-producing Klebsiella pneumoniae in Manhattan. Am. J. Infect. Control 37:121–126 [DOI] [PubMed] [Google Scholar]
- 28. Navon-Venezia S, et al. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nielsen JB, Skov MN, RLJørgensen Heltberg O, Hansen DS, Schønning K. 2011. Identification of CTX-M-15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur. J. Clin. Microbiol. Infect. Dis. 30:773–778 [DOI] [PubMed] [Google Scholar]
- 30. Oteo J, et al. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32:534–537 [DOI] [PubMed] [Google Scholar]
- 31. Oteo J, et al. 2010. Outbreak of VIM-1-carbapenemase-producing Enterobacter cloacae in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 29:1144–1146 [DOI] [PubMed] [Google Scholar]
- 32. Psichogiou M, et al. 2008. Ongoing epidemic of blaVIM-1-positive Klebsiella pneumoniae in Athens, Greece: a prospective survey. J. Antimicrob. Chemother. 61:59–63 [DOI] [PubMed] [Google Scholar]
- 33. Rossolini GM, Mantengoli E, Docquier JD, Musmanno RA, Coratza G. 2007. Epidemiology of infections caused by multiresistant gram-negatives: ESBLs, MBLs, panresistant strains. New Microbiol. 30:332–339 [PubMed] [Google Scholar]
- 34. Sunde M. 2005. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J. Antimicrob. Chemother. 56:1019–1024 [DOI] [PubMed] [Google Scholar]
- 35. Tato M, et al. 2007. Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45:1171–1178 [DOI] [PubMed] [Google Scholar]
- 36. Treviño M, Moldes L, Martínez-Lamas L, Varón C, Regueiro BJ. 2009. Carbapenem-resistant Enterobacter cloacae and the emergence of metallo-β-lactamase-producing strains in a third-level hospital (Santiago de Compostela, NW Spain) Eur. J. Clin. Microbiol. Infect. Dis. 28:1253–1258 [DOI] [PubMed] [Google Scholar]
- 37. Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yildirim I, Ceyhan M, Gur D, Mugnaioli C, Rossolini GM. 2007. First detection of VIM-1 type metallo-β-lactamase in a multidrug-resistant Klebsiella pneumoniae clinical isolate from Turkey also producing the CTX-M-15 extended-spectrum β-lactamase. J. Chemother. 19:467–468 [DOI] [PubMed] [Google Scholar]