Abstract
Despite the success of antiretroviral drugs in decreasing AIDS-related mortality, a substantial fraction of HIV-infected patients experience therapy failure due to the emergence of drug-resistant virus variants. For durable inhibition of HIV-1 replication, the emergence of such escape viruses must be controlled. In addition to antiretroviral drugs, RNA interference (RNAi)-based gene therapy can be used to inhibit HIV-1 replication by targeting the viral RNA genome. RNAi is an evolutionary conserved gene silencing mechanism that mediates the sequence-specific breakdown of the targeted mRNA. Here we investigated an alternative strategy combining the activity of a protease inhibitor (PI) with second-generation short hairpin RNAs (shRNAs) designed to specifically block the emergence of PI-resistant HIV-1 variants. We demonstrate that dominant viral escape routes can be effectively blocked by second-generation shRNAs and that virus evolution can be redirected toward less-fit variants. These results are of importance for a deeper understanding of HIV-1 evolution under combined drug and RNAi pressure and may be used to design future therapeutic approaches.
INTRODUCTION
Worldwide, more than 35 million individuals are infected with human immunodeficiency virus type 1 (HIV-1), and each year approximately 3 million persons are newly infected. Despite the major success of antiretroviral therapy (ART) that delays or prevents the onset of disease, HIV-1 infections remain incurable, and attempts to make a vaccine have so far proved unsuccessful (14). The emergence of drug-resistant viruses remains a major problem for some patients, especially those who exhibit suboptimal therapy adherence.
The RNA interference (RNAi) mechanism is a posttranscriptional gene silencing process that holds promise as a new antiviral strategy. RNAi is induced by double-stranded RNA (dsRNA) that is processed by the RNAi machinery into small interfering RNAs (siRNAs). The siRNAs are designed with perfect base pairing complementarity to the target RNA sequence and trigger cleavage of the targeted mRNA (5, 8). HIV-1 can be inhibited effectively and specifically by RNAi in vitro. Many HIV-1 genes have been targeted by transfected siRNAs or intracellular expressed short hairpin RNAs (shRNAs), and combinatorial RNAi strategies can durably inhibit HIV-1 replication (2, 11, 31). Antiviral drugs and siRNAs can also be combined (9). One successful RNAi-based approach concerns the use of second-generation shRNAs designed to target the favorite escape variants that are selected under pressure of first-generation shRNAs, thus skewing virus evolution (23). In this study, we designed second-generation shRNAs to counter the evolution of clinically relevant drug-resistant HIV-1 variants.
We investigated the potential of combining two anti-HIV strategies. Protease inhibitors (PIs) that successfully suppress HIV-1 replication were combined with second-generation shRNAs to block the favorite viral escape routes. To do so, we first designed second-generation shRNAs and tested them in reporter, HIV-1 production, and virus replication assays. We selected the most active and specific shRNAs. Subsequently, we performed virus evolution studies to monitor the selection of PI-resistant HIV-1 variants in cells that express second-generation or control shRNAs. In this way, we attempted to block virus evolution or to drive evolution in a direction that yields virus mutants with reduced replication fitness.
MATERIALS AND METHODS
Plasmid construction.
shRNA-D30N targets the PI-resistant D30N variant and is expressed from a pSUPER plasmid (OligoEngine, Seattle, WA) with the human H1 polymerase III promoter. The shRNA-L90M variant targets the L90M escape virus and is based on pSilencer 2.0-U6 (Ambion, Austin, TX) with the human U6 polymerase III promoter. The shRNA expression plasmids were constructed as described previously (23). The shRNA-D30N and shRNA-L90M cassettes were combined to generate the shRNA-combi construct. The lentiviral vector JS1 (pRRLcpptpgkgfppreSsin) and the construction of shRNA derivatives were described previously (23). The shRNA-D30N, shRNA-L90M and shRNA-combi cassettes were cloned into the lentiviral vector. The full-length HIV-1 molecular clone pLAI (17) was used to produce wild-type (wt) virus and to study its inhibition by antiviral drugs and shRNAs. The D30N and L90M mutated HIV-1 LAI molecular clones were generated as follows. pLAI was digested with ApaI and SalI, and the 2058–5869 protease fragment was cloned in pBSK to generate pBSK-pr. Mutations were introduced into pBSK-pr by site-directed mutagenesis and verified by sequence analysis, and the mutated ApaI-SalI fragment was subsequently cloned back into pLAI. Firefly luciferase (Luc) reporter plasmids with an HIV-1 target sequence (wt, D30N, and L90M) were constructed by insertion of a 50- to 70-nucleotide HIV-1 fragment, with the 19-nucleotide target sequence in the center, in the EcoRI and PstI sites of pGL3.
Cell culture.
Human embryonic kidney 293T adherent cells were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified chamber at 37°C and 5% CO2. SupT1 suspension T cells were grown in Advanced Roswell Park Memorial Institute medium (Invitrogen, Carlsbad, CA) supplemented with l-glutamine, 1% fetal calf serum, penicillin (30 U/ml), and streptomycin (30 μg/ml) in a humidified chamber at 37°C and 5% CO2.
Transfection experiments.
Cotransfections of pLAI or pGL-3 (firefly luciferase reporter) with the shRNA vector were performed in the 96-well format. Per well, 2 × 104 293T cells were seeded in 100 μl DMEM with 10% FCS without antibiotics. The next day, 100 ng pLAI (or 25 ng of pGL-3) and 0, 1, 5, or 10 ng shRNA vector and 0.5 ng pRL (Renilla luciferase) were transfected with 0.5 μl Lipofectamine 2000 in a reaction volume of 50 μl according to the manufacturer's instructions (Invitrogen). Two days after pLAI transfection, the supernatant was harvested, virus was inactivated, and a CA-p24 enzyme-linked immunosorbent assay (ELISA) was performed. The cells were lysed for Renilla luciferase activity measurements with the Renilla luciferase assay system (Promega). To correct for transfection variation, the CA-p24 values were divided by the Renilla values. We set the condition that for an experiment to be valid the ratio between the highest and the lowest Renilla values should differ by less than a factor of 2. Two days after pGL-3 transfection, cells were lysed to measure firefly and Renilla luciferase activities with the Dual-Luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's instructions.
Lentiviral vector production and T-cell transduction.
The lentiviral vector was produced as previously described (31). Briefly, the vector was made by cotransfection of lentiviral vector plasmid and packaging plasmids pSYNGP, pRSV-rev, and pVSV-g with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After transfection, the medium was replaced with Opti-MEM (Invitrogen, Carlsbad, CA). The lentiviral vector-containing supernatant was collected after 2 days, and aliquots were stored at −80°C. Next, SupT1 cells were transduced at a multiplicity of infection (MOI) of 0.15. Two to 3 days after transduction, live cells were sorted with fluorescence-activated cell sorting (FACS), and green fluorescent protein (GFP)-positive cells were selected.
HIV-1 infection.
HIV-1 LAI and the escape virus variants D30N and L90M were produced by transfection of the molecular clones in 293T cells. Virus production was measured by CA-p24 enzyme-linked immunosorbent assay. SupT1 cells (5-ml cultures with 2.5 × 106 cells or 24-well plate with 2 × 105 cells per well in 1 ml) were infected with the wt or D30N/L90M escape variants (viral input ranged from 0.1 to 1 ng CA-p24). Virus spread was monitored by syncytium formation and CA-p24 production.
The median inhibitory concentration (IC50) was determined as follows. SupT1 cells were seeded in a 96-well plate at 50,000 cells per well. Virus (1 ng CA-p24) was mixed with either nelfinavir (NFV) or saquinvir (SQV) in a concentration range of 0, 0.8, 1.3, 1.9, 2.5, 4.0, 5.5, 7.4, 22.2, 66.7, 200, and 600 nM. This mixture was added to the cells, and we longitudinally scored syncytium formation and CA-p24 levels in the culture supernatant to monitor viral replication. The median inhibitory concentration (IC50) was calculated by the nonlinear regression method, with the variable slope and IC50 curves plotted with GraphPad Prism 5. The IC30 and IC90 values were also determined.
HIV-1 drug resistance evolution.
The HIV-1 wt, D30N, and L90M variants were mixed in a 10:1:1 ratio (based op CA-p24), which was determined in pilot experiments as being optimal for these evolution studies. SupT1-shRNA cells and SupT1 control cells were infected with the virus mixture (1 ng/ml CA-p24) under drug pressure (NFV at IC30 and IC90 and SQV at IC50). Virus replication was monitored by syncytium formation and CA-p24 production. When virus replication was observed, cell-free virus was passaged to uninfected SupT1-shRNA or control SupT1 cells and virus replication was monitored. Cell and supernatant samples were stored at −80°C at peak infection or used directly for sequence analysis of the viral target regions. DNA of the infected cells with the integrated provirus was isolated as described previously (23). Integrated proviral DNA sequences were PCR amplified with the primer pair PROT sense (GCAAATACTGGAGTATTGTATGGATTTTCAGG) and antisense (AGGCTAATTTTTTAGGGAAGATCTGGCCTTCC). The PCR products were sequenced by Sanger sequencing to score the outgrowth of the wt/D30N/L90M genotype. This “population” sequence is able to detect minority sequences when present at approximately 20%.
RESULTS
Design of second-generation shRNAs specific for drug-resistant HIV-1 variants.
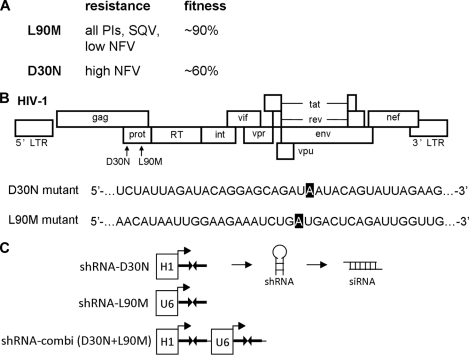
To test whether we could control the evolution of drug-resistant HIV-1 variants, we deliberately chose the PI antiviral drugs NFV and SQV. The L90M mutation in the protease gene (PR) of HIV-1 confers resistance against both drugs, but high NFV levels require the alternative substitution D30N (20, 25, 26). Selection of these two escape mutants is thus determined by the specific drug applied and its concentration (Fig. 1A). The D30N virus provides high-level NFV resistance but exhibits a significantly reduced replication efficiency or fitness (13, 18, 28). Both evolutionary scenarios involve guanine (G)-to-adenine (A) mutations in the respective PR codon (Fig. 1B) that are frequently selected in treated patients (4, 15, 16). In a previous study, we demonstrated that specific viral escape routes that occur under RNAi pressure can be efficiently blocked with second-generation shRNAs (23). Here, we designed second-generation shRNAs to block the D30N and L90M escape routes and cloned shRNA-L90M and shRNA-D30N in expression constructs under the control of the polymerase III promoters H1 and U6, respectively (Fig. 1C). These cassettes were individually cloned in the lentiviral vector JS1. In addition, both cassettes were combined in the shRNA-combi vector. The use of different promoter elements is required to avoid recombination on repeat sequences during transduction of the lentiviral vector. We previously demonstrated equal shRNA expression levels for these vector constructs using reporter assays and Northern blotting (30). We now generated effective second-generation reagents and subsequently tested their specificity in reporter and virus replication assays before we could perform the final test of trying to determine the outcome of virus evolution under PI pressure by second-generation shRNAs.
Fig 1.
Design of second-generation shRNAs for HIV-1 D30N and L90M variants. (A) Resistance of the D30N and L90M viruses against nelfinavir (NFV) and saquinvir (SQV). Fitness was compared to that of the wt virus. These data were derived from previous publications (13, 18, 28). (B) Position of the D30N and L90M mutations in the HIV-1 genome. The exact A residue that is mutated to G in the D30N or L90M variant is marked in black. (C) The expression cassettes of shRNA-D30N and shRNA-L90M under the control of the polymerase III promoters H1 and U6, respectively. Both expression cassettes were combined in shRNA-combi.
Efficient target knockdown by second-generation shRNAs.
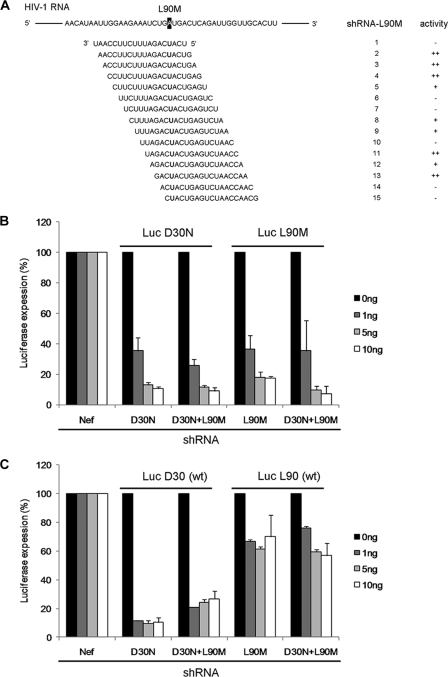
It remains important to screen several candidate shRNAs because the available siRNA design algorithms do not apply for shRNA design (19, 22, 29). We therefore designed 15 overlapping shRNAs for the L90M target (Fig. 2A). Their inhibitory capacity was measured on a luciferase reporter construct with the HIV-1 L90M target sequence inserted in the 3′ untranslated region (UTR). Although all shRNAs are perfectly complementary to this HIV-1 target, shRNAs 2, 3, 4, 11 and 13 were the most effective shRNAs (results not shown; summarized in Fig. 2A). Based on titration experiments, we selected shRNA 3 as the most potent shRNA-L90M for the subsequent experiments. A very active shRNA-D30N reagent was already available (32).
Fig 2.
Efficient target inhibition by second-generation shRNAs. (A) A nested set of 15 shRNA-L90M candidates were designed and screened for their knockdown activity. The silencing activity was measured on a luciferase reporter: ++, very good; +, moderate; −, weak. (B) The inhibitory effect of the second-generation shRNAs was measured on a luciferase reporter with the HIV-1 D30N or L90M target sequence. 293T cells were cotransfected with 25 ng firefly luciferase reporter plasmid, 0.5 ng of Renilla luciferase plasmid, and 0, 1, 5, and 10 ng shRNA constructs. Relative luciferase activity was determined as the ratio of firefly and Renilla luciferase expression. Values are shown as percentages of the transfection with control shRNA-Nef. Averages and standard deviations represent at least three independent transfections that were performed in quadruplicate. (C) The selectivity of the second-generation shRNAs was determined with a luciferase reporter with the wt HIV-1 sequence (D30 or L90).
The activity and sequence specificity of the selected shRNA-D30N and shRNA-L90M were further investigated in cotransfection experiments in 293T cells with luciferase reporter constructs. The latter property is critical, as second-generation shRNAs should ideally target the mutated but not the wild-type (wt) HIV-1 sequence. An additional construct that expresses both inhibitors (shRNA-combi) was tested. Two different luciferase reporters were constructed with the D30N or L90M target sequences inserted in the 3′ UTR, and the inhibitory profile of shRNAs-D30N and L90M was determined. A Renilla luciferase reporter plasmid was cotransfected to control for the transfection efficiency. The relative luciferase expression was determined as the ratio of the firefly and Renilla luciferase activity. We transfected three amounts of the shRNA construct (1, 5, and 10 ng), and the luciferase values obtained with an unrelated control shRNA inhibitor (shNef) were set at 100% for each reporter construct (Fig. 2B). Both shRNA constructs show effective knockdown of the fully complementary target by approximately 90%, and shRNA-combi inhibited both the D30N and L90M targets.
We next determined the inhibitory profile of the shRNAs on two corresponding wt luciferase reporters that have a single mismatch with the second-generation shRNAs (Fig. 2C). Reduced luciferase expression was measured with shRNA-D30N, indicating that this shRNA is also active on the wt target and thus not strictly sequence specific. A similar knockdown pattern was observed for shRNA-combi on the wt sequence. The shRNA-L90M behaved ideally, as luciferase expression did not decrease significantly. Thus, both second-generation shRNAs are active inhibitors and shRNA-L90M acts in a sequence-specific manner, whereas shRNA-D30N exhibits some cross-activity on the mismatched wt target.
Inhibition of drug-resistant HIV-1 variants by the second-generation shRNA.
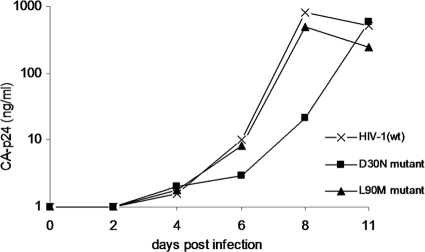
We generated variants of the HIV-1 LAI molecular clone, a CXCR4-using primary virus isolate, with the D30N or L90M mutation in the PR gene. Virus stocks were prepared by transfection of the molecular clones in 293T cells. The SupT1 T cell line was infected with a fixed amount (1 ng CA-p24) of the wt, D30N, or L90M virus. Virus replication was monitored over time by measuring the CA-p24 concentration in the culture supernatant using ELISA (Fig. 3). The wt virus was the first to reach peak infection, followed by the L90M mutant, and the D30N mutant showed a significant delay. This result confirms previous reports that the L90M virus has good replication capacity (90% of the fitness of wt) and the D30N variant a significantly reduced fitness (60% of wt) (13, 18).
Fig 3.
Replication of the drug-resistant HIV-1 D30N and L90M variants. SupT1 cells were infected with HIV-1 wt, D30N, or L90M. Virus replication was monitored over time by measuring CA-p24 production.
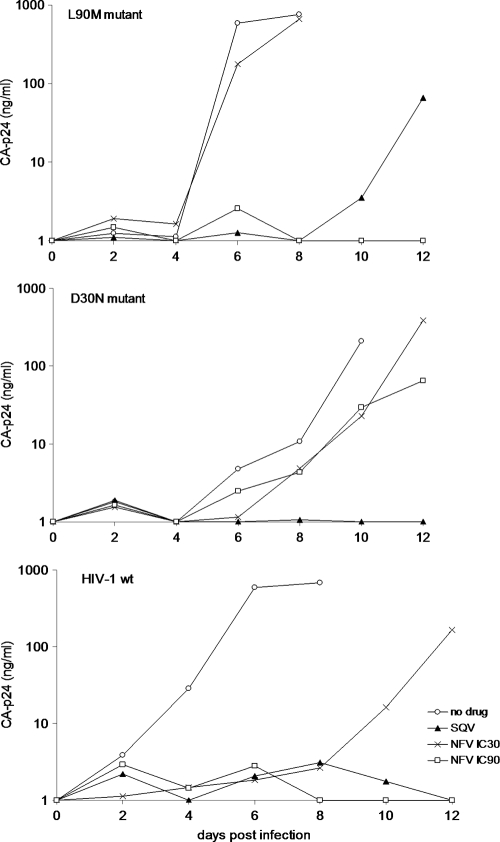
We also infected SupT1 cells with these three virus variants under selective PI drug pressure (Fig. 4). SQV was used at the IC50 (17 nM), and for NFV we used a low (IC30; 7 nM) and a high (IC90; 81 nM) drug concentration. HIV-1 (wt) was unable to replicate with SQV and NFV at the IC90 and showed delayed replication with NFV at the IC30. The D30N virus replicated under both NFV conditions but not under SQV pressure. In contrast, the L90M mutant did replicate under SQV and NFV IC30 pressure, but not at the NFV IC90. These results are fully consistent with literature findings (13, 18).
Fig 4.
Virus replication at differential drug pressure. SupT1 cells were cultured in the presence of PIs; NFV (IC30), NFV (IC90), and SQV (IC50). Cells were infected with HIV-1 wt, D30N, or L90M, and virus replication was monitored over time by measuring CA-p24 production.
Next, we monitored viral replication in the presence of the second-generation shRNAs. We infected SupT1 cells that stably express the relevant shRNAs, and cells transduced with the empty lentiviral vector (JS1) were used as a control (Fig. 5). Replication of the D30N and L90M viruses was inhibited when the SupT1 cells expressed the corresponding shRNA. The cell line with shRNA-combi inhibited both viruses. The wt virus replicated on all cell lines, although replication was slightly delayed in cells that express shRNA-D30N. This is probably due to cross-reactivity of shRNA-D30N on the wt virus as observed in the reporter assay. From these results we conclude that the second-generation shRNAs can effectively inhibit the replication of drug-resistant HIV-1 variants. This sets the stage for the final test: whether PI resistance evolution can be manipulated by second-generation shRNAs.
Fig 5.
Selective inhibition of HIV-1 variant replication by second-generation shRNAs. SupT1 cell lines expressing the shRNA inhibitors (shRNA-D30N, shRNA-L90M, or combined) were infected with wt, D30N, or L90M virus. SupT1 cells with the empty lentiviral vector JS1 were used as the positive control. Virus replication was monitored over time by measuring CA-p24 concentration in the culture supernatant using ELISA.
Directed HIV-1 evolution by combining PIs and second-generation shRNAs.
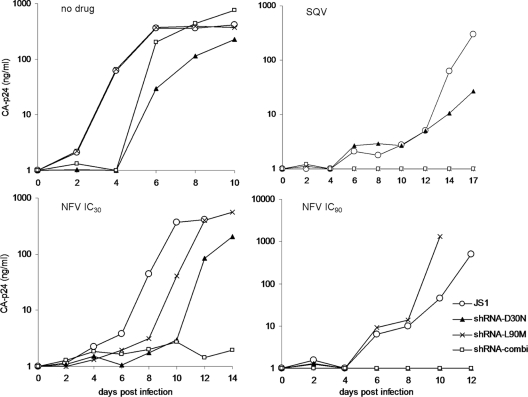
We demonstrated that second-generation shRNAs can be designed that specifically target PI-resistant virus variants. Next, we tested whether these shRNAs can determine the outcome of viral evolution under PI pressure. As input for virus selection, we used a virus mixture of wt-D30N-L90M at the ratio of 10:1:1. This makes the virus evolution experiment independent of the initial mutation step, which otherwise would create much experimental variation, as mutation is a chance process (3, 10). SupT1 cells expressing second-generation shRNA-D30N, -L90M, or -combi (D30N and L90M) were infected with this virus mixture, and cells expressing no shRNA (JS1) served as a control. The cultures were maintained without drug or with SQV, NVF at the IC30, and NVF at the IC90. Virus replication was monitored over time by measuring the CA-p24 concentration in the culture supernatant using ELISA (Fig. 6). Viral breakthrough replication may indicate the selection of a PI drug-resistant HIV-1 variant. To confirm this, the virus collected at peak infection was used to infect fresh SupT1 cells (with the same shRNA and PI regimen). All reinfections resulted in rapid viral outgrowth (5 to 7 days postinfection; results not shown). HIV-1 proviral sequences were PCR amplified from infected cells, and this population was subsequently sequenced to determine which HIV-1 variant was selected. A truly selected variant will become dominant in the viral quasispecies and should thus be present as the majority sequence. Table 1 provides a summary of the outcome of HIV-1 evolution under PI pressure and directed by second-generation shRNAs. In the absence of PI drugs, HIV-1 wt remained the dominant virus species (Fig. 6, Table 1). Delayed replication was apparent in cultures expressing shRNA-D30N, indicating partial inhibition of HIV-1 wt by this shRNA, which is consistent with the results of the luciferase reporter and virus production assays. SQV selects the L90M variant as expected, but no virus variant was selected in the presence of shRNA-L90M (L90M and combi), which blocks this SQV-resistant L90M virus. With NFV at the IC30, the L90M variant was selected in the control and D30N cultures. L90M replication was delayed in the latter culture, probably because of the partial inhibitory effect of shRNA-D30N. The outcome of the evolution experiment was switched to the D30N variant in the shRNA-L90M cultures, because the shRNA blocks replication of the L90M variant and the drug inhibits the wt virus. With shRNA-combi, no viral breakthrough was observed due to the combined drug and shRNA pressure. Virus replication was observed under NFV IC90 pressure in the control and SupT1-L90M cells. Both cultures selected the D30N virus, which is known to replicate at high NFV levels. Most importantly, no viral escape was possible with high levels of NFV and shRNA-D30N pressure. These combined results demonstrate that virus evolution can be blocked or forced into a predefined direction by a combination of antiviral drugs and predesigned second-generation shRNAs.
Fig 6.
Mixed infection of SupT1 cells expressing second-generation shRNAs under PI pressure. SupT1 cells that express the indicated shRNA inhibitor were infected with a mixture of wt-D30N-L90M viruses at the ratio of 10:1:1. These cultures were maintained without drugs or with SQV, NVF at its IC30, and NVF at its IC90. Cultures expressing no shRNA (JS1) served as a positive control. Virus replication was monitored over time by measuring CA-p24 concentration in the culture supernatant using ELISA. After a second round of infection, the provirus was sequenced to determine the most dominant virus variant present in the culture. These results were plotted in Table 1.
Table 1.
HIV-1 evolution under PI and shRNA pressurea
| SupT1 cell line and shRNA inhibitor type | Dominant virus in culture with: |
|||
|---|---|---|---|---|
| No drug | SQV (IC50) | NFV (IC30) | NFV (IC90) | |
| JS1 | wt | L90M | L90M | D30N |
| shRNA-D30N | wt | L90M | L90M | |
| shRNA-L90M | wt | D30N | D30N | |
| shRNA-combi | wt | |||
HIV-1 input mixture: wt-D30N-L90M at a 10:1:1 ratio.
DISCUSSION
Inhibition of HIV-1 by small-molecule drugs or RNAi-based antiviral strategies can fail because of the emergence of drug-resistant virus variants with mutations in the targeted viral protein or RNA, respectively. Clinical success is usually achieved by a combination of antiviral drugs, which raises the genetic threshold for the acquisition of (multidrug) resistance mutations. The combination of a clinically relevant drug with second-generation shRNAs that were specifically designed to target drug resistance mutations was tested in this study for durable inhibition of viral replication. More specifically, we tested whether virus evolution under PI pressure can be skewed into a certain direction or even blocked by such shRNA molecules. We focused on the PI drugs SQV and NFV because they select for overlapping but not for identical resistance mutations. The resistant L90M virus is preferentially selected with SQV and low NFV concentrations, whereas high NFV selects for the D30N virus. These mutations in the protease gene are the most commonly observed resistance mutations in patients on NFV and SQV (13, 21, 28).
SQV combined with shRNA-L90M resulted in a complete block of viral replication because the wt and D30N viruses are inhibited by the drug and L90M is blocked by the shRNA. A similar scenario was observed at high NFV. In combination with shRNA-D30N, the wt and L90M viruses cannot replicate due to drug pressure and the shRNA prevents the emergence of D30N, thus leading to an effective blockade. In contrast, the pressure of shRNA-L90M results in the selection of the D30N variant because both the drug and shRNA are ineffective against this virus. Low NFV (IC30) selects for L90M as expected. When the drug was combined with shRNA-L90M, we observed the outgrowth of the D30N variant and L90M was effectively blocked. We were thus able to skew viral evolution toward variants with reduced replication capacity. These results demonstrate the ability to steer virus evolution with the second-generation shRNAs.
Sequence specificity is an important property of second-generation shRNA because they should ideally target the mutant and not the wt sequence, which differ only by one or a few nucleotides (23, 32, 33). We performed luciferase assays with mismatched wt targets and observed that RNAi activity of the shRNA-L90M was significantly impaired, consistent with the finding that a single mismatch can lead to a complete loss of silencing (1, 7, 24). For shRNA-30N some cross-reactivity on the wt HIV-1 was scored, indicating that for future in vivo applications an improved shRNA-D30N needs to be designed that acts in an exquisitely sequence-specific manner.
The selective pressure of RNAi on the virus genome can be used to direct viral evolution. A recent report used HIV-1 isolate-specific siRNAs to block two parental viruses and thus forced the selection of intersubtype HIV-1 recombinants (6). We demonstrate that virus evolution can be forced toward less fit viruses by a combination of PIs and second-generation shRNAs and that viral escape can even be blocked. This approach can be extended to all classes of antiretroviral drugs and the corresponding virus escape variants but also to direct cytotoxic-T-lymphocyte escape toward virus variants that are less fit (12, 27).
Directing viral evolution using second-generation shRNAs in combination with antiretroviral drugs can be interesting for future therapeutic purposes. Patients failing on certain drug regimens could be treated with drugs combined with second-generation shRNAs. At the start of antiviral treatment, it may be beneficial to combine ART with multiple second-generation siRNAs in order to block the emergence of escape virus mutants. A major drawback of the use of siRNAs to date is the delivery, especially as repeated doses will be required in cases of chronic HIV-1 infection. The emergence of multidrug-resistant HIV-1 has serious clinical implications. Future combinations of antiretroviral drugs with second-generation shRNAs may lead to a new therapeutic approach in the treatment of drug-resistant HIV-1 infection and improve the quality of life of HIV-1-infected individuals.
ACKNOWLEDGMENTS
We thank Stephan Heynen for performing CA-p24 ELISA.
This research was sponsored by ZonMw (Translational Gene Therapy Program) and NWO-CW (Top grant).
Footnotes
Published ahead of print 7 November 2011
REFERENCES
- 1. Amarzguioui M, Holen T, Babaie E, Prydz H. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J, et al. 2007. Safety and efficacy of a lentiviral vector containing three anti-HIV genes—CCR5 ribozyme, tat-rev siRNA, and TAR decoy—in SCID-hu mouse-derived T cells. Mol. Ther. 15:1182–1188 [DOI] [PubMed] [Google Scholar]
- 3. Das AT, Berkhout B. 2010. HIV-1 evolution: frustrating therapies, but disclosing molecular mechanisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doualla-Bell F, et al. 2006. Impact of human immunodeficiency virus type 1 subtype C on drug resistance mutations in patients from Botswana failing a nelfinavir-containing regimen. Antimicrob. Agents Chemother. 50:2210–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elbashir SM, et al. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 6. Gao Y, et al. 2011. Enrichment of intersubtype HIV-1 recombinants in a dual infection system using HIV-1 strain-specific siRNAs. Retrovirology 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gitlin L, Stone JK, Andino R. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296 [DOI] [PubMed] [Google Scholar]
- 9. Huelsmann PM, Rauch P, Allers K, John MJ, Metzner KJ. 2006. Inhibition of drug-resistant HIV-1 by RNA interference. Antiviral Res. 69:1–8 [DOI] [PubMed] [Google Scholar]
- 10. Keulen W, Back NK, van Wijk A, Boucher CA, Berkhout B. 1997. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71:3346–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar P, et al. 2008. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 134:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leslie AJ, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 13. Martinez-Picado J, Savara UV, Sutton L, D'Aquila RT. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montagnier L. 2010. 25 years after HIV discovery: prospects for cure and vaccine. Virology 397:248–254 [DOI] [PubMed] [Google Scholar]
- 15. Ntemgwa M, Brenner BG, Oliveira M, Moisi D, Wainberg MA. 2007. Natural polymorphisms in the human immunodeficiency virus type 2 protease can accelerate time to development of resistance to protease inhibitors. Antimicrob. Agents Chemother. 51:604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parera M, Fernandez G, Clotet B, Martinez MA. 2007. HIV-1 protease catalytic efficiency effects caused by random single amino acid substitutions. Mol. Biol. Evol. 24:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672 [DOI] [PubMed] [Google Scholar]
- 18. Perrin V, Mammano F. 2003. Parameters driving the selection of nelfinavir-resistant human immunodeficiency virus type 1 variants. J. Virol. 77:10172–10175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reynolds A, et al. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326–330 [DOI] [PubMed] [Google Scholar]
- 20. Rhee SY, et al. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhee SY, et al. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 54:4253–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schopman NC, Liu YP, Konstantinova P, Ter Brake O, Berkhout B. 2010. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 86:204–211 [DOI] [PubMed] [Google Scholar]
- 23. Schopman NC, Ter Brake O, Berkhout B. 2010. Anticipating and blocking HIV-1 escape by second generation antiviral shRNAs. Retrovirology 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schubert S, Grunweller A, Erdmann VA, Kurreck J. 2005. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J. Mol. Biol. 348:883–893 [DOI] [PubMed] [Google Scholar]
- 25. Shafer RW. 2006. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 194(Suppl. 1):S51–S58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shafer RW, Schapiro JM. 2008. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 10:67–84 [PMC free article] [PubMed] [Google Scholar]
- 27. Smith SM. 2004. HIV CTL escape: at what cost? Retrovirology 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sugiura W, et al. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taxman DJ, et al. 2006. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ter Brake O, et al. 2008. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 16:557–564 [DOI] [PubMed] [Google Scholar]
- 31. Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. 2006. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 14:883–892 [DOI] [PubMed] [Google Scholar]
- 32. von Eije KJ, Ter Brake O, Berkhout B. 2008. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 82:2895–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33:796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]