Abstract
A clonal strain of Klebsiella pneumoniae producing the plasmid-encoded cephalosporinase DHA-1 was isolated from four patients admitted to the teaching hospital of Clermont-Ferrand, France, in 2006. It was responsible for severe infections in three of the patients; the fourth was colonized only in the gastrointestinal tract. The strain had at least two plasmids encoding resistance to antibiotics (quinolones, aminoglycosides, chloramphenicol, sulfonamides, and trimethoprim), as shown by disk diffusion assay, and harbored only a few genes for virulence factors (wabG and mrkD), as shown by PCRs. DHA-1 synthesis is regulated by an upstream, divergently transcribed gene, ampR, which is also involved in the expression of virulence factors in Pseudomonas aeruginosa. To investigate the role of AmpR in K. pneumoniae, we cloned the wild-type ampR gene from the DHA-1 clonal isolate into a previously characterized K. pneumoniae background plasmid-cured strain, CH608. ampR was also introduced into a CH608 isogenic mutant deleted of ampD, in which AmpR is present only in its activator form, resulting in constitutive hyperproduction of the β-lactamase. We showed that ampR was involved in the upregulation of capsule synthesis and therefore in resistance to killing by serum. AmpR also modulated biofilm formation and type 3 fimbrial gene expression, as well as colonization of the murine gastrointestinal tract and adhesion to HT-29 intestinal epithelial cells. These results show the pleiotropic role of ampR in the pathogenesis process of K. pneumoniae.
INTRODUCTION
Klebsiella pneumoniae is responsible for a wide range of nosocomial infections, including pneumonia, bacteremia, and urinary tract infections (41). The reservoir for K. pneumoniae is the gastrointestinal tract of patients (12). This bacterium produces a number of virulence factors that contribute to pathogenesis and/or gastrointestinal colonization. Thus, the capsule is considered to be the dominant virulence property mediating resistance to phagocytosis and killing by serum (41), but it also plays a critical role in biofilm maturation (2) and colonization of the large intestine of mice (20). The composition of capsular polysaccharides is highly strain dependent, since at least 77 distinct polysaccharides (K antigens) have been reported for K. pneumoniae (37). Epidemiological findings have shown that over 70% of all cases of Klebsiella bacteremia are caused by only 25 of 77 different serotypes (11), with capsular serotypes K1 and K2 being predominant among the virulent strains (47). K. pneumoniae pyogenic liver abscesses, sometimes complicated by endophthalmitis or meningitis, have recently emerged in Taiwan and other Asian countries, and on other continents (18), mostly due to K1 serotype strains (22). The hypermucoviscosity phenotype observed with most of these strains is frequently associated with the expression of the virulence gene magA, which encodes serotype K1 cps polymerase (17). In addition, the mucoid phenotype frequently coexists with production of a siderophore (48), either aerobactin, a hydroxamate-type siderophore, or the kfu-encoded system, a Klebsiella ferric iron uptake system that leads to higher virulence in a mouse model (36). Gastrointestinal tract colonization involves several additional factors, including surface bacterial adhesins such as CF29K, a nonfimbrial adhesive factor encoded by the TEM-5 extended-spectrum-β-lactamase (ESBL)-producing plasmid (13) and some K1 strains (6). Other structures and functions, including fimbriae, membrane transport, the tripartite efflux pump, and metabolic pathways such as urea or allantoin metabolism (8, 10, 14, 34), are also involved in the physiopathologic process.
K. pneumoniae strains are frequently multiresistant to antibiotics, notably by the production of ESBLs. Since the 1980s, plasmid-mediated AmpC-type β-lactamases belonging to class C have been reported in clinical strains of K. pneumoniae and other members of the family Enterobacteriaceae lacking inducible chromosomal AmpC enzymes (40). The first DHA-type enzyme was identified in 1992, in Dhahran, Saudi Arabia, in a clinical isolate of Salmonella enterica serovar Enteritidis (21, 23). Unlike that of most plasmidic ampC genes, blaDHA expression is regulated by ampR (4). When bound to the UDP-Mur-NAc pentapeptide, AmpR represses ampC expression and its own transcription, and this inactivation can be relieved by the presence of β-lactams such as cefoxitin, clavulanic acid, and imipenem or by mutations in the ampD gene (27). Interestingly, the role of AmpR is not restricted to the regulation of β-lactamase synthesis, since in Pseudomonas aeruginosa AmpR was also shown to be involved in the expression of several virulence factors, such as pyocyanin, LasA protease, and LasB elastase (30). AmpR protein contains a DNA-binding motif in its N terminus that binds to consensus sequences located in the ampR-ampC intergenic regions (33). However, in P. aeruginosa, no AmpR-binding site was found upstream of all the AmpR-regulated genes, and its mechanism of action remains unclear (30).
In this study, we describe an epidemic clinical DHA-1-producing K. pneumoniae strain and address the role of ampR in the expression of virulence factors. We show that ampR is involved, in addition to the regulation of DHA-1 synthesis, in capsule synthesis, resistance to killing by serum, biofilm formation, fimbrial synthesis, and intestinal colonization.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and determination of hypermucoviscosity phenotype.
The microorganisms used in this study are listed in Table 1. Strains were grown in lysogeny broth (LB) or agar at 37°C for 18 to 24 h. Biofilm experiments were performed in triplicate in Dulbecco's modified Eagle's medium (DMEM; Cambrex Bioscience, Paris, France) at 37°C. The plasmid pDrive (PCR cloning kit; Qiagen, Courtaboeuf, France) and the plasmid vector pBK-CMV (Stratagene, Amsterdam, The Netherlands) were used for cloning experiments. Where needed, media were supplemented with the following antibiotics, as relevant: kanamycin (30 μg/ml), gentamicin (50 μg/ml), spectinomycin (50 μg/ml), and ceftazidime (2 μg/ml). Each isolate was classified phenotypically as mucoid or nonmucoid by the colony loop lift test. The mucoid phenotype was defined as present when a string-like growth was observed to attach to the loop as it was lifted from the plate.
Table 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Strains | ||
| Klebsiella pneumoniae strains | ||
| CH719 | Wild-type DHA-1-producing isolate 1 (harboring the DHA-1-expressing 61.5-kb plasmid [pCH7191]) | This study |
| CH719.2 | Wild-type DHA-1-producing isolate 2 | This study |
| CH719.3 | Wild-type DHA-1-producing isolate 3 | This study |
| CH719.4 | Wild-type DHA-1-producing isolate 4 | This study |
| CH608 | Wild-type strain; ampD positive and ampR negative; Apr (SHV positive) Nalr | 24 |
| CH864 | CH608 containing the DHA-1-expressing 61.5-kb native plasmid (pCH7191) (ampD positive and ampR positive) | This study |
| CH671 | CH608 containing the pBK-CMV plasmid cloning vector; Apr Kmr (ampD positive and ampR negative) | This study |
| CH791 | CH608 containing the pBK-CMV-ampR recombinant plasmid; Apr Kmr (ampD positive and ampR positive) | This study |
| CH816 | CH608 ΔampD::aadA7; Apr Spr (ampD negative and ampR negative) | This study |
| CH865 | CH816 containing the DHA-1 native plasmid, pCH7191 (ampD negative and ampR positive) | This study |
| CH818 | CH816 containing the pBK-CMV cloning vector plasmid; Apr Spr Kmr (ampD negative and ampR negative) | This study |
| CH819 | CH816 containing the pBK-CMV-ampR recombinant plasmid; Apr Spr Kmr (ampD negative and ampR positive) | This study |
| NTUH-K2044 | Capsular serotype K1; magA+rmpA+ mucoviscosity+ | 18 |
| KP52145 | Capsular serotype K2; contains the 180-kb virulence plasmid | 36 |
| Escherichia coli strains | ||
| JM109 | e14− (McrA−) Δ(lac-proAB) thi-1 gyrA96 (Nalr) endA1 hsdR17(rK− mK−) relA1 supE44 recA1 [F′ traD36 proAB laclqZΔM15] | |
| MG1655 | l−rph-l | 42 |
| LG1522 | F−ara fepA lac leu mtl proC rpsL supE thi tonA trpE xyl; carries pColV-K30 iuc | 7 |
| Plasmids | ||
| pKOBEGapra | Plasmid with λ red genes under the control of the arabinose-inducible promoter; Gmr | 3 |
| pDrive-ampR | pDrive with ampR; Kmr | This study |
| pBK-CMV-ampR | pBK-CMV with ampR; Kmr | This study |
| pCH7191 | DHA-1-expressing 61.5-kb native plasmid | This study |
DNA manipulation and construction of an ampD-defective mutant.
Primers specific for ampR and ampC were designed from the sequences of ampR and blaDHA-2 of K. pneumoniae (GenBank accession number AF259520). Primers ampD F and ampD R were designed from the complete sequence of the ampD gene of the K. pneumoniae MGH 78578 strain (GenBank accession number CP000647) (Table 2) and were used to amplify a 564-bp fragment from K. pneumoniae CH608 genomic DNA.
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| ampR F | GTGAGTTTTACGCCGCCG |
| ampR R | ATGGTCAGACGTTATCTCCCC |
| ampC R | TTATTCCAGCGCACTCAAAAT |
| ampR-ampC F | GGTAAAACTGAGATGACGGGC |
| ampR-ampC R | AGGTGTGAGATAATCCCAGCG |
| ampD F | AGTTGAACAAGGGGTGGTTG |
| ampD R | TTTTATCCAGTCAAAGGCCG |
| ampD-aad-5′ | ATGCAGTTGAACAAGGGGTGGTTGGTCGGCGCGCGTCGGGTTCCCTCGCCGCACCATGACGCTTGCATAATGTGCCTGTC |
| ampD-aad-3′ | AGGTCTCCTTTTCTGACGGGGCGCTGAGCAGCGCACGGTATTTTATCCAGTCAAAGGCCGCGGAAGATCACTTCGCAGAA |
| MrkD 2F | FCCACCAACTATTCCCTCGAA |
| MrkD 2R | ATGGAACCCACATCGACATT |
| TM1 | ATGACCAGCCACACTGGAAC |
| TM2 | CTTCCTCCCCGCTGAAAGTA |
| MFQ1 | GATCGTGAAAGCCAGAAAGG |
| MFQ2 | ACGATGCCTGGTAGTTGTCC |
| AAC6′-A | TTGCGATGCTCTATGAGTGGCTA |
| AAC6′-B | CTCGAATGCCTGGCGTGTTT |
| magA-F | GGTGCTCTTTACATCATTGC |
| magA-R | GCAATGGCCATTTGCGTTAG |
| Wzx_K1-F | GTAGGTATTGCAAGCCATGC |
| Wzx_K1-R | GCCCAGGTTAATGAATCCGT |
| Wzx_K2-F | GGAGCCATTTGAATTCGGTG |
| Wzx_K2-R | TCCCTAGCACTGGCTTAAGT |
| Wzy_K2-F | GGATTATGACAGCCTCTCCT |
| Wzy_K2-R | CGACTTGGTCCCAACAGTTT |
| 1416R | CCGTTAGGCAATCCAGAC |
| 336F2 | TCTGATTTA(A/T)CCCACATT |
| rmpA-F | ACTGGGCTACCTCTGCTTCA |
| rmpA-R | CTTGCATGAGCCATCTTTCA |
| cf29a-F | GACTCTGATTGCACTGGCTGTG |
| cf29a-R | GTTATAAGTTACTGCCACGTTC |
| uge-F | GATCATCCGGTCTCCCTGTA |
| uge-R | TCTTCACGCCTTCCTTCACT |
| wabG-F | CGGACTGGCAGATCCATATC |
| wabG-R | ACCATCGGCCATTTGATAGA |
| kfuB-F1179 | GAAGTGACGCTGTTTCTGGC |
| kfuC-R649 | TTTCGTGTGGCCAGTGACTC |
An ampD-defective mutant of strain CH608 (24), a K. pneumoniae strain previously isolated and cured of its native plasmid(s), was created by allelic exchange after replacement of the entire gene by the selectable spectinomycin resistance gene aadA7. The 1,031-bp aadA7 cassette was generated using primers ampD-aad-5′ and ampD-aad-3′ (Table 2), with the MG1655 strain as the template. The resulting aadA7 cassette was electroporated into a K. pneumoniae CH608 strain harboring the lambda red protein-encoding plasmid pKOBEGapra, resulting in individual colonies that were subcultured until they had lost the pKOBEGapra plasmid. The deletion in the ΔampD mutant strain (CH816) was further checked by PCRs performed with primers ampD F and ampD R.
Plasmid DNA extraction of K. pneumoniae strains was performed either by the method of Kado and Liu (29) or by use of a Qiagen Midi filter plasmid kit (Qiagen). The DHA-1-encoding plasmid from strain CH719 was electroporated into strains CH608 and CH816, giving rise to strains CH864 and CH865, respectively.
To determine the influence of ampR expression on the virulence ability of K. pneumoniae, the ampR gene was amplified from CH719, ligated into the pDrive vector, and transformed into Escherichia coli JM109. The HindIII-BamHI-restricted ampR fragment was then subcloned into the pBK-CMV expression vector, and the resulting recombinant plasmid, pBK-CMV-ampR, was electroporated into both the cured strain (CH608) and its ampD-defective mutant, CH816, giving rise to strains CH791 and CH819, respectively. Control strains were constructed in parallel by electroporating the native pBK-CMV vector into both CH608 and CH816, resulting in strains CH671 and CH818, respectively (Table 1).
Nucleotidic sequences for the ampR, ampC, and aac(6′) genes were obtained commercially from Genome Express (Meylan, France).
β-Lactamase-related studies. (i) Susceptibility to β-lactams.
Antibiotic-containing disks were used for antibiotic susceptibility testing by the disk diffusion assay (Mast Diagnostic, Amiens, France). Results of susceptibility testing were interpreted according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM) (9). MICs were determined in duplicate by a microdilution method and were interpreted according to the guidelines of the CASFM (9). The antibiotics were provided as powders by Glaxo Smith Kline (amoxicillin, ceftazidime, and clavulanic acid), Eli Lilly (cephalothin), Sanofi Aventis (cefotaxime), and Bristol-Myers-Squibb (aztreonam).
(ii) Isoelectric focusing.
Isoelectric focusing of β-lactamases was performed with polyacrylamide gels containing ampholines with a pH range of 3.5 to 10.0, as previously described (5), with TEM-39 (pI 5.2), TEM-1 (pI 5.4), TEM-2 (pI 5.6), SHV-1 (pI 7.7), CTX-M-15 (pI 8.6), and CMY-4 (pI 9) as standards. β-Lactamase activity was revealed with agar containing iodine and penicillin G.
(iii) β-Lactamase specific activity.
Stationary-phase cultures were diluted 1:100 into 10 ml of LB broth containing 30 μg/ml of kanamycin and incubated with shaking at 37°C until an optical density at 600 nm (OD600) of 0.6 to 0.8 was attained. The cultures were then incubated for 3 h, with or without induction (15 μg/ml cefoxitin), before harvesting. The cells were washed with 100 mM NaCl buffer and resuspended in the same buffer (20 ml for 5 g of cells). The specific activity against cephalothin was then determined by the microacidimetric method as previously described (46).
Extraction and quantification of capsular polysaccharide and cps genotyping.
Capsular polysaccharides were extracted with Zwittergent 3-14 detergent. The amount of uronic acid was then measured according to the method described by Domenico et al. (15). In parallel, serial dilutions of the bacterial culture were plated to determine the number of CFU, and the concentration of capsular polysaccharide was expressed according to the amount of glucuronic acid (μg) for 109 CFU per ml of sample. Each experiment was performed in triplicate.
cps genotyping was performed by PCR detection of K-serotype-specific wzy and wzx alleles as previously described to identify K1 and K2 alleles, which are associated with virulence (18). Strains NTUH-K2044 and KP52145 were used as positive controls.
Type 1 fimbria activity assays.
The presence of type 1 fimbriae at the bacterial cell surface was assessed using commercial baker's yeast (Saccharomyces cerevisiae) suspended in phosphate-buffered saline (5 mg [dry weight] per ml) as previously described (25). Equal volumes of yeast cell suspension and bacterial suspension from an overnight culture were mixed on a glass slide. Aggregation was monitored visually.
Serum bactericidal assay.
Twenty-five microliters of a suspension containing 2 × 107 bacteria/ml was mixed with 75 μl of human serum (either nontreated or decomplemented). Viable counts of bacteria were determined after an incubation period of 3 h at 37°C (43). The results are expressed as the ratio of CFU per ml obtained in assays performed with nontreated serum to the CFU per ml obtained in assays performed with decomplemented serum. Each experiment was performed in triplicate.
Biofilm assays.
We used a slightly modified version of the microtiter plate assay developed by O'Toole and Kolter (38). Briefly, 4 μl of overnight culture was inoculated into 100 μl of DMEM in a 96-well culture-treated polystyrene microtiter plate (Nunc). After 6 h of incubation at 37°C, surface-adherent biofilm formation was measured by staining bound cells for 15 min with a 0.5% (wt/vol) aqueous solution of crystal violet. After rinsing, the bound dye was released from stained cells by use of 95% ethanol, and the optical density at 590 nm was determined.
PCRs. (i) RNA extraction and real-time RT-PCR.
RNA was extracted from 1 ml of culture by using a NucleoBond NucleoSpin NucleoTrap kit from Machery-Nagel. The real-time reverse transcription-PCR (RT-PCR) step was carried out with a LightCycler RNA Master SYBR green I kit (Roche Diagnostics, Mannheim, Germany) on LightCycler systems (Roche Diagnostics) with primers MrkD 2 F and 2 R (24) (Table 2). As an internal control, the 16S rRNA gene was amplified with universal primers TM1 and TM2. A 20-μl mixture contained 1 μl of RNA (the RNA preparations were checked for the absence of DNA contamination). To compare relative gene expression in the wild-type strain and its transformant, the term 2[(CPtransformant − CPwild type)TM − (CPtransformant − CPwild type)target gene] was calculated, where CP is the crossing point (32). The experiment was done at least three times.
(ii) ERIC-PCR.
The genomes of the four K. pneumoniae clinical isolates were compared by enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR) as previously described (16).
(iii) PCRs.
PCRs were performed to check for the presence of 10 genes associated with virulence in K. pneumoniae, as previously described (6). Genomic DNAs from strains NTUH-K2044, KP52145, and MGH 78578 were used as controls (Table 1).
(iv) PCR amplification of plasmidic quinolone resistance genes.
Primers MFQ1 and MFQ2 (39) were used to amplify the qnrB gene, and primers AAC6′-A and AAC6′-B were used to amplify and sequence the aac(6′) gene (Table 2).
Murine intestinal colonization assays.
Female specific-pathogen-free mice (OF1; 8 to 18 weeks old; 20 g) (Iffa Credo, L'Arbresle, France) were used. They were caged individually and given sterile water containing 5 g of kanamycin per liter throughout the experiment and had ad libitum access to feed. After 24 h, 100-μl bacterial suspensions containing about 108 CFU were given intragastrically to groups of five mice. On the first day and subsequently every day after inoculation, feces were collected and serial dilutions were plated onto kanamycin agar.
Cell cultures and adhesion assays.
HT-29-MTX, a mucus-secreting cell line, was cultivated as described elsewhere, and adhesion to the cell line was assayed as described previously (19). These experiments were performed in triplicate.
RESULTS AND DISCUSSION
DHA-1-producing K. pneumoniae strain and clinical features of an outbreak.
Strain CH719 was responsible for an outbreak in the teaching hospital of Clermont-Ferrand, France, between August and October 2006. The strain was intermediate or resistant to all penicillins, alone or in combination with β-lactamase inhibitors, and to cephalothin, cefuroxime, cefoxitin, ceftazidime, aztreonam, and cefotaxime and was susceptible to cefepime and imipenem. The double-disk synergy test was negative, and an antagonism was observed between ceftazidime and imipenem, suggesting the production of an inducible β-lactamase. The strain was also intermediate or resistant to all quinolones (nalidixic acid, pefloxacin, ofloxacin, norfloxacin, ciprofloxacin, and moxifloxacin), to all tested aminoglycosides except for gentamicin (i.e., tobramycin, netilmicin, amikacin, and kanamycin), to chloramphenicol, and to sulfonamides and trimethoprim.
Altogether, four K. pneumoniae isolates harboring the same resistance pattern were isolated during the same period. Three of them caused respiratory tract infections in patients with underlying conditions (Waldenström's macroglobulinemia, lymphoma, and traumatic shock), while one was a colonizer, and the three infected patients died after septic shock, despite imipenem treatment.
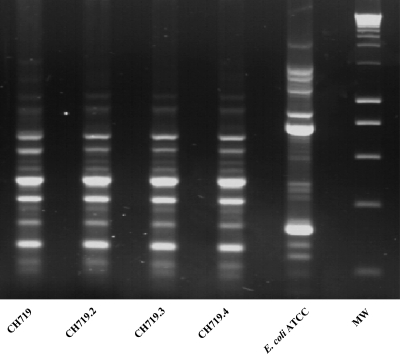
The four isolates were compared by ERIC-PCR: they harbored the same profile (Fig. 1), suggesting the dissemination of a single clone. Isoelectric focusing experiments showed that the four clinical isolates produced similar β-lactamases with a pI of 7.8, in addition to their natural SHV-1 β-lactamase (pI 7.7). PCRs performed with DHA-1-specific primers and DNA sequencing of the resulting amplicons confirmed the presence in these four strains of blaDHA-1 associated with the ampR gene (GenBank accession number HM193083). Plasmid content analysis revealed the presence of at least two plasmids, with approximate sizes of 61.5 kb (pCH7191) and 180 kb (pCH7192) (data not shown). Transformation experiments using plasmidic extracts resulted in clones that harbored the lower-molecular-weight plasmid (pCH7191) and were resistant to most β-lactams specific to DHA-1, to aminoglycosides, to quinolones, and to sulfonamides. The genes encoding DHA-1, an AAC(6′)-Ib cr enzyme, and the QnrB protection system were detected by PCRs using genomic DNAs from the transformants. Transformants harboring the higher-molecular-weight plasmid, pCH7192, were also obtained and showed resistance only to chloramphenicol and trimethoprim.
Fig 1.
ERIC-PCR of the four clinical isolates. Lane 1, CH719; lane 2, CH719.2; lane 3, CH719.3; lane 4, CH719.4; lane 5, E. coli ATCC 25922; lane 6, molecular weight marker (MW).
These strains therefore seemed to combine hyperresistance to antibiotics and enhanced spreading capacities. The four K. pneumoniae isolates were then studied phenotypically and genotypically for their virulence characteristics. They harbored the wabG gene, which encodes a protein that plays a role in colonization and virulence through its involvement in lipopolysaccharide (LPS) synthesis (26), and the mrkD gene, which encodes the type 3 fimbrial adhesin (28), known to play a role in biofilm formation. No other virulence gene markers were detected (6). Specific cps PCRs indicated that none of the isolates belonged to serotype K1 or K2, considered the most virulent serotypes, and their colonies had a nonmucosal appearance, suggesting that the strains produced little capsule, with no hypermucoviscosity phenotype observed. All of the strains expressed type 1 fimbriae, as shown by yeast agglutination. Under iron-deprived growth conditions, no cross-feeding by the isolate was observed with the aerobactin-deficient E. coli strain LG1522, demonstrating the absence of expression of the aerobactin iron uptake system (data not shown). In conclusion, the four isolates showed few known virulence genes, despite the severity of the infections observed in the patients.
The relationship between antibiotic resistance and virulence is still a question of debate. Indeed, bacterial resistance to antimicrobial drugs is frequently reported as difficult to reconcile with bacterial virulence (1), highlighting a biological cost for bacteria. For K. pneumoniae, we showed previously that the presence of ESBL-encoding plasmids alters the basal adhesion capacity of the strain (24). In contrast, other studies showed that acquisition of resistance enhanced virulence. Sahly et al. demonstrated that acquisition of the ESBL SHV-12-encoding plasmid upregulated type 3 fimbria expression in K. pneumoniae (43, 44). However, it has also been demonstrated that ESBL-producing E. coli isolates have fewer urovirulence factors than susceptible isolates (31).
Role of AmpR in virulence of K. pneumoniae.
The ampC gene blaDHA-1 is under the control of the LysR transcription factor family AmpR, also known to regulate the expression of virulence factors in P. aeruginosa (30). In K. pneumoniae, we previously identified OxyR, another member of the LysR regulator family, and showed that it was involved in the expression of several genes, including those associated with biofilm formation, fimbrial synthesis, intestinal colonization, resistance to several gastrointestinal stresses, and resistance to H2O2 exposure (25). In this work, we therefore decided to investigate the potential role of AmpR in the virulence of K. pneumoniae. Since the epidemic strain of this study, CH719, did not express many virulence factors, we chose to analyze the influence of its ampR gene in another K. pneumoniae setting, i.e., a previously characterized clinical strain (24) that did not harbor ampR and was highly virulent, with a high level of capsule production. The ampR gene was cloned from CH719, and the resulting recombinant plasmid, pBK-CMV-ampR, was electroporated into the K. pneumoniae strain CH608, giving rise to CH791. To study the possible role of each form of AmpR (inactive and derepressed), both the pBK-CMV-ampR plasmid and the native 61.5-kb DHA-1-expressing plasmid pCH7191 were also introduced into strain CH816, a ΔampD isogenic mutant of K. pneumoniae CH608, resulting in strains CH819 and CH865, respectively. The DHA-1 cephalosporinase derepression due to the absence of AmpD synthesis in these constructs was confirmed by comparison of their MICs, and the results were confirmed by comparison of the specific activities of crude extracts of these two strains against cephalothin, with or without induction of DHA-1 by cefoxitin (Table 3).
Table 3.
MICs of β-lactam antibiotics for the clinical K. pneumoniae strains CH719 and CH608 and the derivative strains CH864 and CH865a
| β-Lactamb | MIC for Klebsiella pneumoniae strain |
|||
|---|---|---|---|---|
| CH719 (ampR+ampD+) | CH608 (ampR deficient ampD+) | CH864 (ampR+ampD+) | CH865 (ampR+ampD deficient) | |
| Amoxicillin | >512 | 32 | >512 | >512 |
| Amoxicillin-CLA | >512 | 2 | >512 | >512 |
| Cephalothin | >512 | 8 | >512 | >512 |
| Cefotaxime | 4 | <0.06 | 8 | 64 |
| Cefotaxime-CLA | 16 | <0.06 | 16 | 64 |
| Ceftazidime | 32 | 0.25 | 64 | >512 |
| Ceftazidime-CLA | 64 | 0.25 | 256 | >512 |
| Aztreonam | 4 | 0.25 | 4 | 32 |
Specific activities against cephalothin for strains CH719, CH608, CH864, and CH865, without cefoxitin induction, were 1.1, <0.1, 2.2, and 80.0 μmol/min/mg, respectively. Those with induction were 74.7, <0.1, 57.1, and 123.9 μmol/min/mg, respectively. Note that the values for CH608 are identical to those for CH816 (ampD deficient and ampR deficient).
CLA, clavulanic acid at 2 μg/ml.
(i) ampR influences capsule synthesis, biofilm formation, and fimbria-associated phenotype.
Since the capsule is the main virulence factor of K. pneumoniae, we investigated the amounts of capsular polysaccharide produced by CH791, CH819, and their respective controls, CH671 and CH818. The AmpR-producing strains CH791 and CH819 produced significantly more capsule than CH671 and CH818, respectively (Table 4). In addition, they differed in morphology by having large smooth colonies (data not shown). Since capsule production affects biofilm formation (2), we next investigated the capacity of the strains to form biofilms by using a microtiter plate experimental model. The biomass of the biofilm formed by the ampR-positive strain (CH791) was significantly lower than that formed by CH671, whereas there was no significant difference between the ΔampD strains (CH818 and CH819) (Table 4). Since type 3 pili are an adhesive factor facilitating adherence and biofilm formation on abiotic surfaces in K. pneumoniae (14), we also looked for type 3 fimbrial expression by real-time RT-PCR. It was downregulated in the ampR-positive strain CH791, whereas no significant difference was observed between the ΔampD strains CH818 and CH819 (Table 4).
Table 4.
Bacterial characteristics and virulence properties of K. pneumoniae strainsa
| Property | Value for strain |
|||
|---|---|---|---|---|
| CH791 (ampD+ampR+) | CH671 (ampD+ampR deficient) | CH819 (ampD deficient ampR+) | CH818 (ampD deficient and ampR deficient) | |
| Amt of capsuleb | 25.5 ± 5.5 | 14.5 ± 3.0 | 99.0 ± 9.0 | 29.0 ± 5.5 |
| Biofilm capacityc | 0.371 ± 0.046 | 0.709 ± 0.062 | 0.98 ± 0.063 | 0.669 ± 0.021 |
| Type 3 pilus expressiond | 13.55 ± 0.51 | 11.72 ± 0.45 | 14.37 ± 1.55 | 13.199 ± 1.50 |
| Resistance to serum killinge | 95.0 ± 10.0 | 54.5 ± 7.0 | 78.0 ± 9.0 | 38.0 ± 9.0 |
Data are means ± standard deviations.
Bacterial glucuronic acid concentration (μg/109 CFU).
Crystal violet biomass staining (OD590).
CP values from specific RT-PCRs.
Percent survival of cells after 3 h of serum contact (CFU/ml).
(ii) Resistance to killing by serum in relation to presence of ampR.
Since the clinical clonal strain CH719 was responsible for blood infection and the capsule is known to play a role in serum resistance, we studied the serum resistance ability of the strains. The percent survival of cells after 3 h of serum contact was significantly higher in the presence of ampR for both CH671 and its ΔampD mutant, CH818 (Table 4), indicating a role of ampR in serum resistance.
(iii) ampR is involved in intestinal murine colonization and cell adhesion.
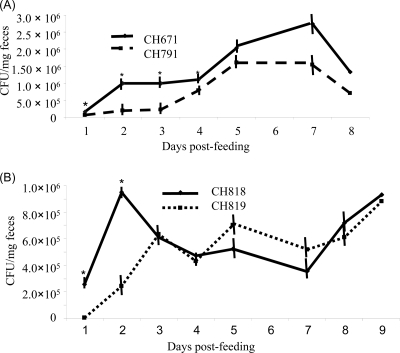
Strains CH671, CH791, CH818, and CH819 were fed to mice individually, and the bacterial counts of K. pneumoniae in feces were recorded over 8 days. The ampR-positive strain CH791 colonized the murine intestinal tract significantly less efficiently than the wild-type strain, mostly during the first 4 days (Fig. 2A). Although the colonization rate of strain CH819 was lower during the first 2 days, there was no significant difference between CH818 and CH819 during the rest of the experiment (Fig. 2B).
Fig 2.
Gastrointestinal colonization of K. pneumoniae strains (A) and their ampD mutants (B) in a murine model. Groups of five mice were fed individually with 108 CFU of an ampR-negative (CH671 or CH818) or -positive (CH791 or CH819) strain ml−1, and feces were collected for 8 or 9 days, starting from 1 day after feeding. *, statistically significant difference (Student's t test; P < 0.05).
Strains CH671, CH791, CH818, and CH819 were also compared for the ability to adhere to mucus-secreting intestinal HT-29 cells. The ampR-positive strains CH791 and CH819 were significantly less adherent than their parental strains, CH671 and CH818, with 2.17-fold ± 0.52-fold and 1.49-fold ± 0.34-fold less adhesion to HT-29 cells, respectively (for CH671 versus CH791, P < 0.01; for CH818 versus CH819, P < 0.05).
In conclusion, whatever the ampD background, the expression of ampR led to increased synthesis of capsule, together with a decreased adherence to mucus-producing cells in vitro, as described elsewhere (19, 44, 45). We also showed that the ampR-expressing strains had increased serum resistance, consistent with the fact that the clinical strain caused severe bloodstream infections in the patients. An impairment in biofilm formation capacity associated with reduced expression of the type 3 pilus-encoding mrkD gene was observed only in the strain harboring functional ampD (CH819), which suggests a possible specific role of AmpR in surface colonization capacity.
The expression of plasmidic ampC in Salmonella Typhimurium caused a decrease in strain virulence that was not detected when an ESBL gene was coexpressed or when ampC was regulated by AmpR (35). The clinical DHA-1-producing strain in our study did not show virulence factors usually associated with the K. pneumoniae species, but its ampR gene was able to act as a regulator of virulence. We speculate that in the absence of β-lactam antibiotics such as cefoxitin, clavulanic acid, and imipenem, cephalosporinase expression is repressed and associated with increased virulence expression. In contrast, in the presence of β-lactams, the active form of AmpR is likely to induce the expression of ampC and to modulate strain virulence, which points to a potential role of this LysR regulator factor in the adaptation of the bacterium to its environment. The multiple antibiotic treatments used in these patients may have influenced, through AmpR, the expression of still unknown genes and allowed the strain to resist the hostile environment, to express virulence factors, and to spread. It is noteworthy that imipenem, an ampR-inducing antibiotic, was used to treat the patients' infections and might thus have increased the virulence of the in vivo strains.
In conclusion, the study presented here provides new insights into the role of AmpR as a global regulator involved in the expression of many virulence factors and highlights the potential role of antibiotic administration in the expression of bacterial virulence factors.
Footnotes
Published ahead of print 10 October 2011
REFERENCES
- 1. Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489–493 [DOI] [PubMed] [Google Scholar]
- 2. Balestrino D, Ghigo JM, Charbonnel N, Haagensen JA, Forestier C. 2008. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10:685–701 [DOI] [PubMed] [Google Scholar]
- 3. Balestrino D, Haagensen JA, Rich C, Forestier C. 2005. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187:2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnaud G, et al. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonnet R, et al. 2000. A novel class A extended-spectrum β-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob. Agents Chemother. 44:3061–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brisse S, et al. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carbonetti NH, Williams PH. 1984. A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect. Immun. 46:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chou HC, et al. 2004. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 72:3783–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comité de l'Antibiogramme de la Société Française de Microbiologie 2010. Communiqué 2010. Société Française de Microbiologie, Paris, France: http://www.sfm.asso.fr/publi/general.php?pa=1 [Google Scholar]
- 10. Coudeyras S, Nakusi L, Charbonnel N, Forestier C. 2008. A tripartite efflux pump involved in gastrointestinal colonization by Klebsiella pneumoniae confers a tolerance response to inorganic acid. Infect. Immun. 76:4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cryz SJ, Jr, Mortimer PM, Mansfield V, Germanier R. 1986. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J. Clin. Microbiol. 23:687–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Champs C, et al. 1989. Prospective survey of colonization and infection caused by expanded-spectrum-β-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J. Clin. Microbiol. 27:2887–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Martino P, et al. 1995. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 63:4336–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A. 2003. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154:9–16 [DOI] [PubMed] [Google Scholar]
- 15. Domenico P, Schwartz S, Cunha BA. 1989. Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect. Immun. 57:3778–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dumarche P, et al. 2002. TEM derivative-producing Enterobacter aerogenes strains: dissemination of a prevalent clone. Antimicrob. Agents Chemother. 46:1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199:697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang CT, et al. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45:284–293 [DOI] [PubMed] [Google Scholar]
- 19. Favre-Bonte S, Joly B, Forestier C. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Favre-Bonte S, Licht TR, Forestier C, Krogfelt KA. 1999. Klebsiella pneumoniae capsule expression is necessary for colonization of large intestines of streptomycin-treated mice. Infect. Immun. 67:6152–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fortineau N, Poirel L, Nordmann P. 2001. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:207–210 [DOI] [PubMed] [Google Scholar]
- 22. Fung CP, et al. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50:420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaillot O, Clement C, Simonet M, Philippon A. 1997. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J. Antimicrob. Chemother. 39:85–87 [DOI] [PubMed] [Google Scholar]
- 24. Hennequin C, Forestier C. 2007. Influence of capsule and extended-spectrum β-lactamases encoding plasmids upon Klebsiella pneumoniae adhesion. Res. Microbiol. 158:339–347 [DOI] [PubMed] [Google Scholar]
- 25. Hennequin C, Forestier C. 2009. OxyR, a LysR-type regulator involved in Klebsiella pneumoniae mucosal and abiotic colonization. Infect. Immun. 77:5449–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Izquierdo L, et al. 2003. The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 185:7213–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacobs C, Frere JM, Normark S. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell 88:823–832 [DOI] [PubMed] [Google Scholar]
- 28. Jagnow J, Clegg S. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397–2405 [DOI] [PubMed] [Google Scholar]
- 29. Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong KF, et al. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 49:4567–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lavigne JP, et al. 2006. Virulence genotype and nematode-killing properties of extra-intestinal Escherichia coli producing CTX-M β-lactamases. Clin. Microbiol. Infect. 12:1199–1206 [DOI] [PubMed] [Google Scholar]
- 32. Lee HW, et al. 2008. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14:49–54 [DOI] [PubMed] [Google Scholar]
- 33. Lindquist S, Lindberg F, Normark S. 1989. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J. Bacteriol. 171:3746–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maroncle N, Rich C, Forestier C. 2006. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res. Microbiol. 157:184–193 [DOI] [PubMed] [Google Scholar]
- 35. Morosini MI, Ayala JA, Baquero F, Martinez JL, Blazquez J. 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44:3137–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nassif X, Fournier JM, Arondel J, Sansonetti PJ. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orskov I. 1955. Serological investigations in the Klebsiella group. I. New capsule types. Acta Pathol. Microbiol. Scand. 36:449–453 [DOI] [PubMed] [Google Scholar]
- 38. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 39. Pai H, Seo MR, Choi TY. 2007. Association of QnrB determinants and production of extended-spectrum β-lactamases or plasmid-mediated AmpC beta-lactamases in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 51:366–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roux A, Beloin C, Ghigo JM. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 187:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sahly H, et al. 2004. Increased serum resistance in Klebsiella pneumoniae strains producing extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 48:3477–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sahly H, et al. 2000. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 68:6744–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schembri MA, Dalsgaard D, Klemm P. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sirot D, et al. 1997. A complex mutant of TEM-1 β-lactamase with mutations encountered in both IRT-4 and extended-spectrum TEM-15, produced by an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 41:1322–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wacharotayankun R, et al. 1993. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 61:3164–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu VL, et al. 2007. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg. Infect. Dis. 13:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]