Abstract
Nasal carriage of methicillin-resistant coagulase-negative staphylococci (MR-CoNS) is highly prevalent in community subjects, but its dynamic has been little investigated. Nasal swabbing was performed in 2006 and 2008 in 154 Amerindians living isolated in French Guiana. MR-CoNS strains were identified and characterized by non-β-lactam susceptibility testing and staphylococcal cassette chromosome mec element (SCCmec) typing, characterizing the associations of ccr and mec gene complex allotypes, and for MR Staphylococcus epidermidis (MRSE), multilocus variable number of tandem repeats analysis (MLVA) was used. The impact of sociodemographic and medical characteristics on the persistence of MR-CoNS carriage was assessed by bivariate analysis. Prevalence of MR-CoNS carriage was 50.6% in 2006 and 46.8% in 2008. The 274 MR-CoNS isolates, including S. epidermidis (n = 89, 62 MLVA patterns), Staphylococcus haemolyticus (n = 78), and Staphylococcus hominis (n = 72), exhibited 41 distinct ccr and mec gene complex associations. Persistent carriage (in 2006 and 2008), intermittent carriage (either in 2006 or 2008), and noncarriage were documented in 25.3, 47.4, and 27.3% of the participants, respectively. Persistent carriage of a given MRSE isolate was rarely observed (n = 8 isolates). Furthermore, no epidemiological factor, including antibiotic exposure, was associated with persistent carriage. The high diversity of MRSE clones and their ccr and mec gene complex associations contrasted with the high carriage rates in this isolated community, which might reflect the occurrence of SCCmec rearrangement and the generation of new MR-CoNS strains.
INTRODUCTION
The emergence of methicillin resistance (MR) in staphylococci results from the acquisition of the mecA-encoded penicillin-binding protein 2a, a transpeptidase conferring broad-spectrum β-lactam resistance (21). A mobile genetic element designated staphylococcal cassette chromosome mec (SCCmec) carries both the mec gene complex, i.e., mecA and its regulatory genes, and the ccr gene complex that encodes the recombinases involved in its chromosomal integration/excision (17, 21). Eight major SCCmec types (I to VIII) have been described in methicillin-resistant Staphylococcus aureus (MRSA), differing in allotypic combinations of the mec and ccr gene complexes, with SCCmec IVa and V being currently the most prevalent types in community-acquired MRSA (CA-MRSA) strains (17, 21, 34). Furthermore, methicillin-resistant coagulase-negative staphylococci (MR-CoNS) display a higher diversity of SCCmec elements with frequent nontypeable patterns, including ccr-mec complex combinations that do not fit the classification proposed for MRSA and nontypeable or multiple ccr allotypes (1, 14, 22, 41).
MR-CoNS, most notably MR Staphylococcus epidermidis (MRSE), probably act as a reservoir of SCCmec for S. aureus, although the mechanisms of transfer are not yet elucidated. Indeed, similar SCCmec patterns were observed in MRSA and MR-CoNS isolates from the same health care environment (2, 13, 20, 48, 51). Moreover, SCCmec IVa sequences from MRSE display >98% identity with those carried by MRSA (3, 49), including when CA-MRSE and CA-MRSA strains were compared, suggesting that SCCmec transfer can occur in the community (1).
S. epidermidis and other CoNS species, such as Staphylococcus hominis and Staphylococcus haemolyticus, are major components of the human skin and mucosal flora, including the nasal microbiota (26). Recent studies highlighting the community spread of MR-CoNS have raised concerns, because of the probable role of MR-CoNS as a source of SCCmec for CA-MRSA and the increasing prevalence of CoNS in community-acquired diseases, such as native valve endocarditis and late infections of indwelling prosthetic devices (6, 30, 46). Nasal colonization by MR-CoNS has been documented in 11 to 31% of outpatients from contrasting geographic areas (1, 41). Strikingly, high carriage rates were observed in subjects without any previous exposure to the health care system (1). However, little is known about the long-term dynamic of MR-CoNS nasal carriage in the community. A single study assessed this issue and found that 4% of 339 Japanese children were potential persistent MRSE carriers (22). Data from other community environments are lacking, and whether this dynamic is impacted by sociodemographic characteristics or antibiotic exposure remains unknown. The latter points are difficult to assess in open populations, where subjects are exposed to multiple antibiotic sources, and where interindividual contacts and thus cross-transmission are untraceable. In this study, we investigated the dynamic of MR-CoNS nasal carriage in the community, with a special focus on MRSE carriage, by using serial nasal samples previously gathered in a cohort of healthy adults from an isolated population and whose antibiotic exposure and sociodemographic characteristics were precisely documented (37, 50).
MATERIALS AND METHODS
Study population and design.
Two campaigns of nasal swabbing were performed 16 months apart, in October 2006 and June 2008, in 154 adult Wayampi Amerindians, who were part of a traditional, ethnically homogeneous community of 525 individuals living in Trois-Sauts, a very isolated village in the southernmost area of French Guiana, with limited contacts with the outside world (28, 29). Precise characteristics of this study population have been previously described (12, 37, 50). Carriage rates of methicillin-susceptible S. aureus (MSSA) were 42.2% in 2006 and 57.8% in 2008. No MRSA was found in 2006, while two subjects carried MRSA in 2008 (37). The study design was approved by the ethical committee in charge of French Guiana (Comité de Protection des Personnes Sud-Ouest et Outre Mer III, authorization no. 2006/0498-DGS and 2008/C07-44 for the 2006 and 2008 surveys, respectively).
Isolation of MR-CoNS isolates from nasal swab samples.
The methodologies used for sampling, isolation, and characterization of MR-CoNS isolates were strictly the same for the 2006 and 2008 campaigns. Samples from our previous S. aureus carriage survey (37) were defrosted in a batch, and 100 μl of the suspension was plated on chromID MRSA agar (bioMérieux), a chromogenic agar selective for MR staphylococci (cefoxitin, 4 μg/liter) with a >90% sensitivity and a 94 to 100% specificity to discriminate mecA-positive and mecA-negative CoNS isolates, including for the Staphylococcus saprophyticus species (10, 35, 36). After incubation for 48 h at 37°C, four white colonies were randomly selected from each plate and subcultured on Trypticase soy agar. The DNA of each isolate was extracted with a MagNA Pure LC instrument (Roche, Germany) and stored at −80°C. The isolates confirmed as MR-CoNS (i.e., mecA positive) by real-time triplex PCR were retained for further characterization (39).
Antimicrobial susceptibility testing of MR-CoNS isolates.
Susceptibility to the antibiotics listed in Table 1 was determined by disk diffusion as recommended by the French Society for Microbiology (http://www.sfm.asso.fr/). A resistance score was calculated for each MR-CoNS carrier (32).
Table 1.
Prevalence of non-β-lactam antibiotic resistances in 274 carriage isolates of MR-CoNS isolated from 154 adult Wayampi Amerindians during the two sampling campaigns (2006 and 2008)a
| Antibiotic | No. (%) of resistant MR-CoNS isolates |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All species |
S. epidermidis |
S. haemolyticus |
S. hominis |
S. saprophyticus |
Othersb |
|||||||
| 2006 (141 isolates) | 2008 (133 isolates) | 2006 (56 isolates) | 2008 (33 isolates) | 2006 (36 isolates) | 2008 (42 isolates) | 2006 (44 isolates) | 2008 (28 isolates) | 2006 (2 isolates) | 2008 (25 isolates) | 2006 (3 isolates) | 2008 (5 isolates) | |
| Kanamycin | 50 (33) | 39 (29) | 21 (33) | 7 (21) | 12 (32) | 18 (43) | 17 (39) | 12 (43) | 0 | 0 | 0 | 2 (40) |
| Tobramycin | 37 (25) | 35 (26) | 18 (28) | 6 (18) | 9 (24) | 17 (40) | 10 (23) | 10 (36) | 0 | 0 | 0 | 2 (40) |
| Gentamicin | 16 (11) | 21 (16) | 8 (13) | 2 (6) | 7 (18) | 11 (26) | 1 (2) | 6 (21) | 0 | 0 | 0 | 2 (40) |
| Erythromycin | 51 (34) | 45 (34) | 31 (48) | 11 (33) | 3 (8) | 3 (7) | 13 (30) | 5 (18) | 2 (100) | 22 (88) | 2 (67) | 4 (80) |
| Lincomycin | 14 (9) | 15 (11) | 12 (19) | 3 (9) | 0 | 0 | 1 (2) | 0 | 0 | 9 (36) | 1 (33) | 3 (60) |
| Pristinamycin | 0 | 1 (1) | 0 | 1 (3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tetracycline | 37 (25) | 30 (23) | 15 (23) | 5 (15) | 10 (26) | 20 (48) | 12 (27) | 1 (4) | 0 | 4 (16) | 0 | 0 |
| Ofloxacin | 3 (2) | 20 (15) | 2 (3) | 2 (6) | 0 | 1 (2) | 0 | 0 | 0 | 15 (60) | 1 (33) | 2 (40) |
| Fosfomycin | 51 (34) | 71 (53) | 15 (23) | 5 (15) | 3 (8) | 17 (40) | 30 (68) | 21 (75) | 2 (100) | 25 (100) | 3 (100) | 3 (60) |
| Rifampin | 39 (26) | 49 (37) | 23 (36) | 14 (42) | 2 (5) | 16 (38) | 12 (27) | 9 (32) | 0 | 6 (24) | 2 (67) | 4 (80) |
| Cotrimoxazole | 35 (23) | 40 (30) | 17 (27) | 13 (39) | 0 | 3 (7) | 17 (39) | 11 (39) | 1 (50) | 13 (52) | 0 | 0 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 1 (1) | 0 | 0 | 0 | 1 (2) | 0 | 0 | 0 | 0 | 0 | 0 |
P = 0.0002 for the comparison of ofloxacin resistance rates between 2006 and 2008 (P = NS for all other antibiotics).
Including S. kloosii (n = 3 isolates in both 2006 and 2008), S. capitis (n = 1 isolate in 2008), and S. cohnii (n = 1 isolate in 2008).
Species identification of MR-CoNS isolates.
The isolates were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Autoflex MALDI-TOF MS spectrometer, Bruker Daltonics, Wissembourg, France) after ethanol extraction, according to the manufacturer's recommendations. Mass spectra were analyzed using the Flex Control software (Bruker Daltonics). Quality indices (QI) were allocated to assess the accuracy of species identifications (4, 15). Only spectra with high QI (>2) were considered reliable for identification. Isolates with QI of ≤2 were identified by comparison of an internal 1,350-bp fragment of the 16S rRNA gene with sequences available in the National Center for Biotechnology Information databases, using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast) (38).
Isolates from a given patient were considered duplicates and not analyzed further if they belonged to the same species and shared the same antibiotic susceptibility patterns.
SCCmec typing.
SCCmec elements were typed by multiplex PCR (M-PCR) amplification of the ccr (M-PCR1) and mec (M-PCR2) gene complexes (27). Additional primers were incorporated to amplify the ccrA4B4 and C1 mec allotypes (41). SCCmec elements were designated by the association of the ccr gene complex allotype(s) (i.e., 1, 2, 3, 4 for ccrA1B1, ccrA2B2, ccrA3B3, and ccrA4B4, respectively, and 5 for ccrC1) and the mec gene complex allotype (A, B, C1, or C2) (21). MRSA strains COL/SCCmec I (1B), BK2464/SCCmec II (2A), ANS46c/SCCmec III (3A), 300-FPR3757/SCCmec IV (2B), WCH100/SCCmec V (5C2), and HDE288/SCCmec VI (4B) were used as references. Nondetectable ccr gene complexes were defined based on the positivity of the mecA internal control and the lack of a ccr amplicon with M-PCR1. Nontypeable mec gene complexes were defined based on either the lack of an mec amplicon or an amplicon of unreported size with M-PCR2. Nontypeable SCCmec isolates were defined by a nondetectable ccr gene complex and/or the absence of a typeable mec gene allotype or an unreported association between a typeable ccr gene complex and a typeable mec gene complex.
Multilocus variable number of tandem repeats analysis (MLVA) of MRSE isolates.
MRSE isolates were typed by MLVA, a method that assesses the length polymorphism of five chromosomal variable-number tandem repeats designated Se1 to Se5 (23). The phylogenetic relationship between MLVA patterns was evaluated by the unweighted pair group method with arithmetic mean (UPGMA) using the BioNumerics software version 6.0 (Applied Maths, Ghent, Belgium). MRSE isolates with the same number of tandem repeats for each loci were defined as having the same MLVA profile (23).
Classification of MR-CoNS carriers.
Given the absence of a consensual definition for CoNS nasal carriage, subjects in whom no MR-CoNS strain was isolated in 2006 or 2008 were defined as noncarriers, those carrying at least one MR-CoNS isolate in either 2006 or 2008 were considered intermittent carriers, and those carrying at least one isolate in both 2006 and 2008 were defined as persistent carriers.
Epidemiological data.
Demographic, lifestyle, and environmental data as well as medical events and antibiotic consumptions during the year preceding the first swabbing campaign, previously gathered both for the study population and the rest of the villagers (37), were used to investigate factors associated with MR-CoNS carriage (Table 2).
Table 2.
Factors associated with MR-CoNS nasal carriage at study inclusion in 2006 and with persistent nasal carriage of MR-CoNS between 2006 and 2008 in 154 adult Wayampi Amerindians
| Sociodemographic characteristic | No. (%) of isolates |
||||||
|---|---|---|---|---|---|---|---|
| All volunteers (n = 154) | 2006 |
2006–2008 |
|||||
| MR-CoNS carriers (n = 78) | Noncarriers (n = 76) | P/Paa | Persistent MR-CoNS carriers (n = 39) | Other volunteers (n = 115)b | P/Paa | ||
| Female | 79 (51.3) | 41 (52.6) | 38 (50.0) | 0.87/1.00 | 22 (56.4) | 57 (49.6) | 0.58/1.00 |
| Age (yr) | |||||||
| 18-24 | 38 (24.6) | 19 (24.4) | 19 (25.0) | 0.55/1.00 | 9 (23.1) | 29 (25.2) | 0.29/1.00 |
| 25–30 | 37 (24.0) | 22 (28.2) | 15 (19.7) | 9 (23.1) | 28 (24.3) | ||
| 31–40 | 30 (19.5) | 12 (15.4) | 18 (23.7) | 4 (10.3) | 26 (22.6) | ||
| 41–50 | 28 (18.2) | 13 (16.7) | 15 (19.7) | 9 (23.1) | 19 (16.5) | ||
| 51–81 | 21 (13.7) | 12 (15.4) | 9 (11.8) | 8 (20.5) | 13 (11.3) | ||
| Marital status | |||||||
| Single | 23 (15.0) | 15 (19.2) | 8 (10.5) | 0.18/1.00 | 9 (23.1) | 14 (12.2) | 0.12/1.00 |
| Couple | 131 (85.0) | 63 (80.8) | 68 (89.5) | 30 (76.9) | 101 (87.8) | ||
| No. of children | |||||||
| ≤2 | 56 (36.3) | 33 (42.9) | 23 (31.5) | 0.31/1.00 | 16 (42.1) | 40 (35.7) | 0.20/1.00 |
| 3-5 | 57 (37.0) | 28 (36.4) | 29 (39.7) | 10 (26.3) | 47 (42.0) | ||
| 6-10 | 37 (26.7) | 16 (72.0) | 21 (28.8) | 12 (31.6) | 25 (22.3) | ||
| Babysitting children ≤5 yr old | 112 (72.7) | 54 (72.0) | 58 (79.5) | 0.34/1.00 | 25 (67.6) | 87 (78.4) | 0.19/1.00 |
| Hamlet (household distance from the health post) | |||||||
| Hamlet 1 (0 m) | 92 (59.7) | 48 (61.5) | 44 (57.9) | 0.84/1.00 | 29 (74.4) | 63 (54.8) | 0.19/1.00 |
| Hamlet 2 (1,000 m) | 36 (23.4) | 17 (21.8) | 19 (25.0) | 5 (12.8) | 31 (27.0) | ||
| Hamlet 3 (3,500 m) | 12 (7.9) | 7 (9.0) | 5 (6.6) | 2 (5.1) | 10 (8.7) | ||
| Hamlet 4 (5,500 m) | 14 (9.0) | 6 (7.7) | 8 (10.5) | 3 (7.7) | 11 (9.6) | ||
| No. of inhabitants per household | |||||||
| ≤3 | 14 (9.1) | 8 (10.3) | 6 (7.9) | 0.36/1.00 | 5 (12.8) | 9 (7.8) | 0.40/1.00 |
| 4-7 | 80 (51.9) | 44 (56.4) | 36 (47.4) | 17 (43.6) | 63 (54.8) | ||
| 8-12 | 60 (39.0) | 26 (33.3) | 34 (44.7) | 17 (43.6) | 43 (37.4) | ||
| Presence of animals in the household | 125 (81.2) | 63 (80.8) | 62 (81.6) | 1.00/1.00 | 32 (82.1) | 93 (80.9) | 1.00/1.00 |
| Type of drinking water | |||||||
| River | 78 (50.6) | 41 (54.7) | 37 (52.1) | 0.81/1.00 | 19 (50.0) | 59 (54.6) | 0.71/1.00 |
| Cove | 70 (45.4) | 35 (46.7) | 35 (49.3) | 0.87/1.00 | 20 (52.6) | 50 (46.3) | 0.57/1.00 |
| Tap | 27 (17.5) | 14 (18.7) | 13 (18.3) | 1.00/1.00 | 8 (21.1) | 19 (17.6) | 0.63/1.00 |
| Medical characteristics | |||||||
| Intestinal carriage of ESBL-Ef | 27 (17.5) | 14 (18.7) | 13 (18.3) | 1.00/1.00 | 8 (21.1) | 19 (17.6) | 0.63/1.00 |
| Chronic diseasec | 8 (5.2) | 1 (1.3) | 7 (9.2) | 0.03/0.91 | 0 | 8 (7.0) | 0.20/1.00 |
| Pregnancy | 10 (12.6) | 6 (13.0) | 4 (9.1) | 0.74/1.00 | 2 (9.1) | 8 (14.0) | 1.00/1.00 |
| Hospitalizationsd | 23 (14.9) | 10 (12.8) | 13 (17.1) | 0.50/1.00 | 5 (12.8) | 18 (15.7) | 0.80/1.00 |
| Surgeryd | 11 (7.1) | 4 (5.1) | 7 (9.2) | 0.37/1.00 | 2 (5.1) | 9 (7.8) | 0.73/1.00 |
| Antibiotic use by the volunteerd | 70 (45.4) | 38 (48.7) | 32 (42.1) | 0.42/1.00 | 24 (61.5) | 46 (40.0) | 0.03/0.71 |
| No. of antibiotic treatments | 84 (54.5) | 40 (51.3) | 44 (57.9) | 0.61/1.00 | 15 (38.5) | 69 (60.0) | 0.07/1.00 |
| 0 | 26 (16.9) | 15 (19.2) | 11 (14.5) | 10 (25.6) | 16 (13.9) | ||
| 1 | 26 (16.9) | 12 (15.4) | 14 (18.4) | 7 (17.9) | 19 (16.5) | ||
| 2 | 18 (11.7) | 11 (14.1) | 7 (9.2) | 7 (17.9) | 11 (9.6) | ||
| ≥3 | |||||||
| Antibiotic use among volunteer's relativesd,e | 147 (95.4) | 73 (93.6) | 74 (97.4) | 0.44/1.00 | 37 (94.9) | 110 (95.7) | 1.00/1.00 |
Bivariate analysis was performed using the Pearson χ2 test or Fisher test (α = 0.05). Pa, P value after adjustment by Holm's method.
Other volunteers include intermittent carriers (n = 73) and noncarriers (n = 42).
Defined in the Charlson's comorbidities score (5).
Within the year preceding study inclusion, i.e., between October 2005 and October 2006.
Relatives were defined as members of the same family living in the same household (in case of multiple life partners, second/third wives and children were included as relatives even if they lived in a different household), and the number of relatives ranged from 1 to 11.
ESBL-E, extended spectrum β-lactamase-producing Enterobacteriaceae.
Statistical analysis.
Continuous data were presented as the mean (±standard deviation [SD]) or median and range, and categorical data were presented as numbers (proportions). Associations between species, year of sampling, antibiotic susceptibility profile, mec gene complex, ccr gene complex, and SCCmec type were investigated using Fisher's exact test. A P value of <0.05 was considered statistically significant. We compared the epidemiological characteristics of MR-CoNS nasal carriers with others, using the R software (version 2.6.1) (http://cran.r-project.org/). For bivariate analysis, we used Pearson's χ2 test, Fisher's exact test, Student's t test, and McNemar's χ2 test. All the tests were two sided, and the significance level was set at 5%. Because of the large number of explanatory variables, bivariate analyses were adjusted by Holm's test for multiple testing (18, 43).
RESULTS
Study population and overall MR-CoNS carriage rates.
A total of 525 Wayampi Amerindians (238 adults and 287 children) were living in Trois-Sauts at the time of our first campaign in 2006, and 163 adults (68.5%) volunteered to participate. A total of 154 of the 163 volunteers in 2006 (94.5%) were resampled in 2008 (female/male ratio = 1.09, mean age = 35.1 years) and were included in the present study (Table 2). The overall carriage rates of MR-CoNS were 50.6% (78/154 samples) and 46.8% (72/154 samples) for the 2006 and 2008 campaigns, respectively (P = not significant [NS]). Among the MR-CoNS carriers, 30 (38.5%) in 2006 and 34 (47.2%) in 2008 cocarried S. aureus (P = NS).
Isolation and characterization of MR-CoNS isolates.
After exclusion of duplicates, 274 MR-CoNS isolates were analyzed, including 141 and 133 isolates from the 2006 and 2008 campaigns, respectively. The mean numbers of strains per individual were 0.93 ± 1.09 in 2006 and 0.86 ± 1.10 in 2008 (P = NS). Most of the carriers (83% in 2006 and 76% in 2008) carried 1 or 2 MR-CoNS isolates, with the remaining ones carrying 3 to 4 distinct isolates. This distribution did not differ significantly between the two periods.
The MR-CoNS isolates were divided into seven distinct species, including S. epidermidis (n = 89), S. haemolyticus (n = 78), S. hominis (n = 72), Staphylococcus saprophyticus (n = 27), Staphylococcus kloosii (n = 6), Staphylococcus capitis (n = 1), and Staphylococcus cohnii (n = 1) (Table 2). The proportion of S. epidermidis isolates decreased between 2006 and 2008 (39.7% versus 24.8%, respectively; P = 0.012), while that of S. saprophyticus isolates increased (1% versus 19%; P < 0.0001). The proportions of S. haemolyticus and S. hominis isolates did not change significantly.
The overall resistance rates for non-β-lactam antibiotics were high (notably for kanamycin, tobramycin, erythromycin, rifampin, and cotrimoxazole) but remained stable over the study period, except for ofloxacin (2% in 2006 versus 15% in 2008; P = 0.0002) (Table 1). The mean resistance scores for MR-CoNS carriers were 22% ± 16% in 2006 and 27% ± 16% in 2008 (P = NS).
The results of SCCmec typing are shown in Table 3. Half of the MR-CoNS isolates (137/274 [50%]) carried a typeable SCCmec, including type 2A (8/274 [2.9%]), type 2B (24/274 [8.8%]), type 5C2 (65/274 [23.8%]), type 4B (2/274 [0.7%]), and type 4A (38/274 [13.8%]). No SCCmec type 1B was found. The remaining 137 isolates carried a nontypeable SCCmec pattern, including 3 isolates with a single ccr allotype but a ccr-mec combination not described in MRSA, 36 isolates with multiple ccr allotypes, 16 isolates with multiple mec allotypes, and 82 isolates with nondetectable ccr or nontypeable mec allotypes. The complete list of ccr-mec combinations is available in Table S1 in the supplemental material. Associations were noted between SCCmec types and CoNS species, namely, SCCmec type 2B and S. epidermidis (23/89 isolates versus 1/185 isolates for other species; P < 0.0001), SCCmec type 5C2 and S. haemolyticus (40/78 isolates versus 25/196 isolates; P < 0.0001), and SCCmec type 4A and S. hominis (35/72 isolates versus 3/202 isolates; P < 0.0001). All but one MR S. saprophyticus (MRSS) isolate carried a nontypeable SCCmec element (26/27 isolates versus 111/247 isolates from other species; P < 0.0001) (Table 4). The two MRSA isolates collected in 2008 carried SCCmec type 1B.
Table 3.
SCCmec typing of 274 carriage isolates of MR-CoNS isolated from 154 adult Wayampi Amerindians during the 2006 and 2008 sampling campaigns
| SCCmec type/ccr-mec combinations | No. (%) of MR-CoNS isolates |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All species |
S. epidermidis |
S. haemolyticus |
S. hominis |
S. saprophyticus |
Othersa |
|||||||
| 2006 | 2008 | 2006 | 2008 | 2006 | 2008 | 2006 | 2008 | 2006 | 2008 | 2006 | 2008 | |
| 2A | 7 (5) | 1 (1) | 6 (11) | 1 (3) | 0 | 0 | 1 (2) | 0 | 0 | 0 | 0 | 0 |
| 2B | 12 (8) | 12 (9) | 12 (21) | 11 (33) | 0 | 0 | 0 | 1 (4) | 0 | 0 | 0 | 0 |
| 5C2 | 24 (17) | 41 (31) | 11 (19) | 11 (33) | 11 (31) | 29 (69) | 0 | 1 (4) | 0 | 0 | 2 (66) | 0 |
| 4B | 1 (1) | 1 (1) | 0 | 1 (3) | 0 | 0 | 1 (2) | 0 | 0 | 0 | 0 | 0 |
| 4A | 25 (17) | 13 (10) | 0 | 0 | 0 | 1 (2) | 25 (57) | 10 (36) | 0 | 1 (4) | 0 | 1 (20) |
| Nontypeableb | 72 (51) | 65 (49) | 27 (48) | 9 (27) | 25 (69) | 12 (29) | 17 (39) | 16 (57) | 2 (100) | 24 (96) | 1 (33) | 4 (80) |
| Total | 141 | 133 | 56 | 33 | 36 | 42 | 44 | 28 | 2 | 25 | 3 | 5 |
Including S. kloosii (n = 3 isolates in both 2006 and 2008), S. capitis (n = 1 isolate in 2008), and S. cohnii (n = 1 isolate in 2008).
The complete list of nontypeable SCCmec patterns is available in Table S1 in the supplemental material.
Table 4.
Comparison of non-β-lactam resistances and SCCmec types between MR S. saprophyticus and MR-CoNS isolates from other species collected during the 2008 campaign
| Variable | No. (%) of MR-CoNS isolates |
P value | |
|---|---|---|---|
| S. saprophyticus (n = 25 isolates) | Other species (n = 108 isolates) | ||
| Antibiotic resistance | |||
| Kanamycin | 0 (0) | 39 (36) | <0.001 |
| Tobramycin | 0 (0) | 35 (32) | <0.001 |
| Gentamicin | 0 (0) | 21 (19) | 0.013 |
| Vancomycin | 0 (0) | 0 (0) | NS |
| Teicoplanin | 0 (0) | 1 (1) | NS |
| Erythromycin | 22 (88) | 23 (21) | <0.001 |
| Lincomycin | 9 (36) | 6 (6) | NS |
| Pristinamycin | 0 (0) | 1 (1) | NS |
| Tetracycline | 4 (16) | 26 (24) | NS |
| Ofloxacin | 15 (60) | 5 (5) | <0.001 |
| Fosfomycin | 25 (100)a | 46 (43) | <0.001 |
| Rifampin | 6 (24) | 43 (40) | NS |
| Cotrimoxazole | 13 (52) | 27 (25) | 0.014 |
| SCCmec typeb | |||
| Nontypeable | 24 (96) | 41 (38) | <0.001 |
| 2A | 0 | 1 (1) | NS |
| 2B | 0 | 12 (11) | NS |
| 5C2 | 0 | 41 (38) | NS |
| 4B | 0 | 1 (1) | NS |
| 4A | 1 (4) | 12 (11) | 0.0014 |
Natural resistance in S. saprophyticus.
Indicated as type (ccr-mec gene complexes).
MLVA typing of MRSE.
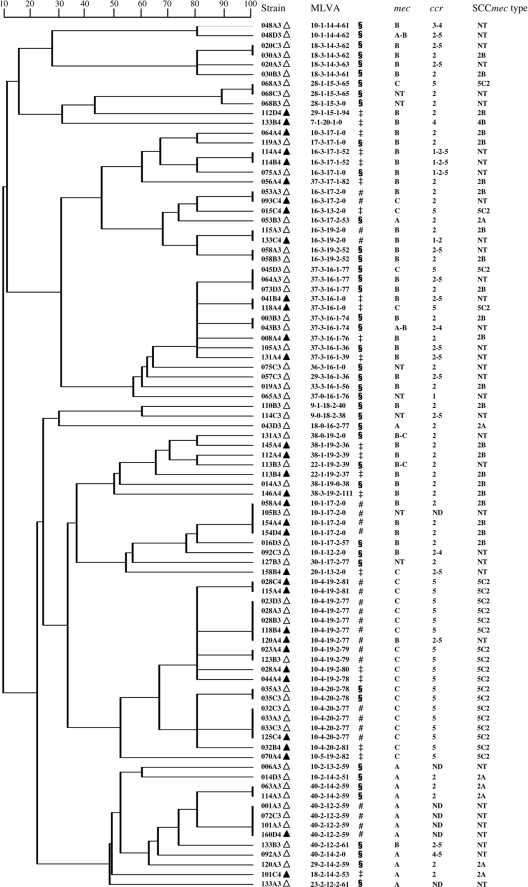
The 89 MRSE isolates showed highly heterogeneous MLVA patterns, regardless of their SCCmec types and year of isolation. Indeed, 62 distinct patterns were observed, including 42 in 2006 and 27 in 2008, with only 7 found in both 2006 and 2008. Forty-five patterns were found in only 1 MRSE isolate (singletons), 12 in 2, and the remaining 5 patterns were found in 3 to 5 isolates. By combining MLVA and SCCmec profiles, we observed that only seven (7.8%) MRSE isolates were carried by more than one volunteer (maximum, n = 4) (Fig. 1). The remaining 82 isolates were found in only one subject.
Fig 1.
Genetic relationship of multiple-locus variable-number tandem repeat analysis (MLVA) patterns of the 89 methicillin-resistant Staphylococcus epidermidis isolates collected during the 2006 (▵) and 2008 (▴) sampling campaigns, as implemented by the BioNumerics version 6.0 software (Applied Maths NV, Sint-Martens-Latem, Belgium) by using the unweighted pair group method with arithmetic mean (UPGMA). The symbols §, ‡, and # indicate MLVA genotypes that were found only in 2006, only in 2008, or both, respectively. Staphylococcal cassette chromosome mec elements (SCCmec) are defined by the ccr-mec allotype combination according to the current classification used in methicillin-resistant S. aureus (21). MLVA patterns are defined as the number of tandem repeats for each of the five Se loci (Se1, Se2, Se3, Se4, Se5) (23). ccr allotypes are classified as types 1 to 4 (i.e., ccrA1B1 to ccrA4B4) and 5 (i.e., ccrC). ND, nondetectable ccr gene complex; NT, nontypeable mec allotypes or SCCmec.
Dynamic of MR-CoNS nasal carriage.
We found that 39 (25.3%), 73 (47.4%), and 42 (27.3%) subjects were persistent carriers, intermittent carriers, or noncarriers, respectively (Table 5). Interestingly, most of the persistent carriers (21/39) carried isolates from different species in 2006 and 2008. Persistent carriage of MRSE, MR S. haemolyticus, and MR S. hominis was documented in only 12 (8%), 3 (2%), and 3 (2%) of the 154 subjects, respectively. However, based on non-β-lactam resistance patterns, SCCmec typing, and MLVA profile (for S. epidermidis), none of these subjects carried the same isolate in 2006 and 2008. No persistent carriage of MRSS or MR S. kloosii was observed.
Table 5.
Type of nasal carriage of MR-CoNS in 154 adult Wayampi Amerindians sampled during the two sampling campaigns (2006 and 2008)
| Type of MR-CoNS nasal carriagea | No. (%) of all subjects | No. (%) who carried MR-CoNS species: |
||||
|---|---|---|---|---|---|---|
| S. epidermidis | S. haemolyticus | S. hominis | S. saprophyticus | Othersb | ||
| Persistent carrier | 39 (25) | 12 (8) | 3 (2) | 3 (2) | 0 | 0 |
| Intermittent carrier | 73 (47) | 43 (28) | 58 (38) | 41 (27) | 15 (10) | 6 (4) |
| Noncarrier | 42 (27) | 99 (64) | 93 (60) | 110 (71) | 139 (90) | 148 (96) |
Noncarrier, subject in whom no MR-CoNS strain (indicated for all species together and then detailed species by species) was isolated in 2006 or 2008; intermittent carrier, subject carrying at least one MR-CoNS isolate in either 2006 or 2008; persistent carrier, subject carrying at least one isolate in both 2006 and 2008.
Including S. kloosii, S. capitis, and S. cohnii.
Factors associated with nasal carriage of MR-CoNS by bivariate analysis.
Strikingly, neither MR-CoNS nasal carriage at inclusion (i.e., in 2006) nor persistent MR-CoNS carriage was associated with sociodemographic or medical characteristics (Table 2). Previous antibiotic use was, at first, significantly associated with persistent carriage (odds ratio [OR], 2.4; 95% confidence interval [CI], 1.1 to 5.5; P = 0.03), but the P value dropped to nonsignificance after Holm's adjustment for multiple testing (P = 0.71).
DISCUSSION
The spread of MR-CoNS out of the hospital setting has been reported in several recent works. However, data on the dynamics of MR-CoNS carriage in community subjects are currently very scarce. Here, we investigated the persistence and biodiversity of MR-CoNS nasal colonization in a population of 154 adult Amerindians forming part of a remote and precisely characterized community.
Prevalence of MR-CoNS carriage was 51% in 2006 and 47% in 2008, which contrasts with previous community-based surveys, where the reported rates ranged from 11 to 30% (1, 22, 41, 44). Rates similar to ours have been reported only in inpatients, notably those hospitalized in long-term-care facilities (20, 25). Promiscuity and suboptimal hygienic conditions in this traditional population may contribute to this finding. Indeed, cross-transmission is a major determinant of MR-CoNS dissemination in the hospital setting (7, 47) and was shown to be an important mechanism for S. aureus dissemination in this community (37). However, the extreme biodiversity (i.e., the lack of patent clonality) of MR-CoNS isolates in our work pleads against this hypothesis. The impact of antibiotic selective pressure is another possible explanation for this high overall prevalence of MR-CoNS carriage. Non-β-lactam coresistances were actually frequent in MR-CoNS isolates, and antibiotic use in this cohort (Table 2) (37) may be higher than that in other populations, including the metropolitan French population (42). Moreover, non-β-lactam resistances might have been underestimated, since we used the CASFM methods, which detect only clinically relevant resistances. Our use of a medium selective for MR staphylococci to ease and enhance MR-CoNS isolation from the subdominant nasal flora may also explain these discrepancies with previous studies (41). Lastly, equatorial climatic conditions in south French Guiana, with precipitations above 80 mm per month and temperatures higher than 20°C all year long, may also play a role. Indeed, an increase in MR-CoNS nasal carriage rates was previously observed during the hot and rainy season (22), even though the determinants of this climatic impact remain to be investigated.
The most striking result of this study is that up to 25.3% of the participating subjects were persistent carriers of MR-CoNS between 2006 and 2008, while 47.4% were intermittent carriers and only 27.3% were noncarriers. Factors associated with persistent carriage of MR-CoNS in the community have not been previously investigated. Here, precise characterization of the study population enabled us to search for such factors. However, we found no association between persistent MR-CoNS carriage and sociodemographic features, such as age and lifestyle. Most notably, persistent carriage was not associated with medical characteristics, namely, chronic diseases, previous hospitalization and surgery, intestinal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae (50), and a history of antibiotic exposure in the sampled subject or his or her relatives. Consequently, we hypothesize that this high frequency of persistent MR-CoNS colonization depends on the overall prevalence of MR-CoNS carriage in the studied population rather than on the individual's characteristics. In this instance, when CoNS species were considered, only 8% of the subjects were persistent MRSE carriers, and according to MLVA data, none of them carried the same isolate twice. Persistent carriage of MR S. haemolyticus and MR S. hominis was even more uncommon (2% for each), and based on non-β-lactam resistance patterns, it involved distinct isolates in all cases. Persistent MR-CoNS carriage thus appears as the result of successive acquisitions of distinct isolates from the community environment and not as the long-term colonization by a given isolate.
S. epidermidis increasingly appears as a major actor of MR dissemination among community strains of staphylococci. This species represents 69 to 84% of the MR-CoNS isolates collected in previous community-based studies (1, 41, 44), and several data strongly support its potential role as a reservoir of SCCmec for S. aureus (1–3, 13, 20, 49), including CA-MRSA (1). Two previous surveys in open communities found an extremely high diversity of MRSE in terms of SCCmec structures and genetic background (1, 22). In our work, the 89 MRSE isolates displayed large polymorphisms of SCCmec elements, including 36 isolates with untypeable patterns, and divided into 62 distinct MLVA profiles. Most of these profiles were found in only one or two MRSE isolates, with only five of them accounting for more than three isolates. Moreover, only six MLVA patterns were found in both 2006 and 2008. These data provide further evidence that, even in a nearly closed population, the community spread of MRSE is not clonal. This situation contrasts with the one observed for CA-MRSA, whose worldwide spread currently involves only a limited number of clones (34) and might result from the easiest SCCmec recombinations and intraspecies transfers in S. epidermidis. Indeed, the few available SCC sequences from S. epidermidis strains suggest that both new SCCmec and SCC non-mec variants may arise by recombination of DNA fragments from distinct SCC elements (1, 8, 51). Moreover, a previous work based on MLST data analysis indicated that SCCmec transfers were probably frequent in S. epidermidis compared to in S. aureus (31). Plasticity of the S. epidermidis genome (11, 52) may constitute an auspicious background for the chromosomal integration of SCCmec elements and allow the frequent rise of new MRSE strains, making this species a wide and dynamic reservoir of SCCmec in the community and explaining this lack of patent clonality.
Regardless of the species, we found that SCCmec elements were highly polymorphous among MR-CoNS. Half of our isolates harbored a nontypeable SCCmec, with a new association between known mec and ccr gene complexes (18%) or a nondetectable ccr and/or nontypeable mec gene complex (31%). Thirty-five (13%) MR-CoNS isolates were found to carry 2 or 3 distinct ccr allotypes, a finding that corroborates those of previous works (14, 20, 41), and one S. hominis strain gave 4 typeable ccr amplicons (ccrA1B1 with ccrA2B2, ccrA3B3, and ccrA4B4). We cannot firmly exclude that a lack of primer specificity might have led to the amplification of several DNA fragments from a single, new ccr variant. However, all MR-CoNS strains that we reported as carrying multiple ccr allotypes gave ccr amplicons with band sizes that matched exactly with those from reference MRSA strains. Further studies including sequencing data are needed to better describe such MR-CoNS strains harboring multiple ccr allotypes. Furthermore, the associations of class A and class B mec gene complexes have been described in health care-associated MR strains of S. epidermidis and S. capitis (19, 20). In this study, we found three strains with new associations of mec gene complexes, namely, class A with C2, with or without C1, and class B with C2. These results, in line with those of other works (1, 11, 14, 22, 41, 44, 52), emphasize the extreme plasticity of SCCmec elements among CoNS, with frequent rearrangements and continuous generation of new composite cassettes.
SCCmec types 2B and 5C2 were carried by 8.7% and 23.7% of the 274 MR-CoNS isolates, respectively. These types account for nearly all of the CA-MRSA lineages described to date (34). Considering the high rate of cocarriage of MSSA and MR-CoNS, the lack of evidence of SCCmec transfer between CoNS and S. aureus in this community is surprising (37). In addition, 92 (61.7%) of the 149 MSSA isolates isolated during the two campaigns belonged to clonal complex 1 (CC1), CC5, CC8, CC30, or CC398, which might constitute suitable phylogenic backgrounds for SCCmec acquisition (9, 24, 34). Indeed, only two MRSA isolates were found (both during the 2008 campaign), and both carried SCCmec type I (1B), this profile being not found in MR-CoNS isolates. However, although the role of MR-CoNS as a reservoir of SCCmec for S. aureus appears undeniable (1–3, 13, 20, 49), this transfer has been directly observed only once (3), suggesting that it may occur at a very low frequency.
The prevalence of MRSS among MR-CoNS isolates increased from 1% to 19% over the study period. Other community-based surveys have recently highlighted the emergence of MR in this species (16, 22), including the nasal carriage isolates (22). The implantation of MRSS in the nasal flora is unexpected, as this uropathogenic species colonizes mainly the female genital tract, with an overall colonization rate of ∼7% (16, 40). This shift toward a novel colonization site might result from a selective advantage in terms of antibiotic pressure, because S. saprophyticus is usually more resistant to non-β-lactam agents than other CoNS species (33), as observed in our work. Besides, all but one of our 27 MRSS strains carried an untypeable SCCmec element, mostly because of a nondetectable ccr gene complex (see Table S1 in the supplemental material). These difficulties to type ccr in MRSS have been previously reported (45) and suggest that S. saprophyticus carries undescribed ccr variants more often than other CoNS species.
In conclusion, this study brings new insights on the biodiversity and dynamic of MR-CoNS carriage in the community and provides further evidence of their role as a large and evolutionary reservoir of SCCmec in this setting. Fortunately, SCCmec transfers between MR-CoNS and the highly pathogenic S. aureus are probably exceptional compared with inter-CoNS exchanges.
Supplementary Material
ACKNOWLEDGMENTS
We thank the partners of the ERAES project (Ecologie de la Résistance aux Antibiotiques d'Escherichia coli et Staphylococcus aureus): R.-M. Bettinger, M.-E. Bougnoux, F. Catzeflis, O. Clermont, C. Dupont, D. Guillemot, S. Jarraud, M. Lescat, and F. Rousset. We also thank the villagers for their help during the study.
All authors declare that they have no conflicts of interest.
The study was supported in part by the Agence Française de Sécurité Sanitaire de l'Environnement et du Travail (AFSSET, grant ES05-001), the Agence Nationale pour la Recherche (ANR, grant 05-9-114), the Institut National de la Santé et de la Recherche Médicale (INSERM, grant C06-18), and the French National Reference Center for Bacterial Resistance in Commensal Flora. C. Angebault was supported by a grant from the Fondation pour la Recherche Médicale.
Footnotes
Published ahead of print 7 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Barbier F, et al. 2010. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202:270–281 [DOI] [PubMed] [Google Scholar]
- 2. Berglund C, Soderquist B. 2008. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden—possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin. Microbiol. Infect. 14:1048–1056 [DOI] [PubMed] [Google Scholar]
- 3. Bloemendaal AL, Brouwer EC, Fluit AC. 2010. Methicillin resistance transfer from Staphylococcus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One 5:e11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carbonnelle E, et al. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 45:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40:373–383 [DOI] [PubMed] [Google Scholar]
- 6. Chu VH, et al. 2008. Emergence of coagulase-negative staphylococci as a cause of native valve endocarditis. Clin. Infect. Dis. 46:232–242 [DOI] [PubMed] [Google Scholar]
- 7. Cimiotti JP, Wu F, Della-Latta P, Nesin M, Larson E. 2004. Emergence of resistant staphylococci on the hands of new graduate nurses. Infect. Control Hosp. Epidemiol. 25:431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diep BA, et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 9. Enright MC, et al. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gallon O, et al. 2011. Performance of a new MicroScan WalkAway PC30 panel and disk diffusion method for detection of oxacillin resistance in Staphylococcus spp. J. Clin. Microbiol. 49:2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill SR, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grenet K, et al. 2004. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg. Infect. Dis. 10:1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanssen AM, Kjeldsen G, Sollid JU. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanssen AM, Sollid JU. 2007. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 51:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris LG, et al. 2010. Rapid identification of staphylococci from prosthetic joint infections using MALDI-TOF mass-spectrometry. Int. J. Artif. Organs 33:568–574 [DOI] [PubMed] [Google Scholar]
- 16. Higashide M, et al. 2008. Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob. Agents Chemother. 52:2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hiramatsu K, Katayama Y, Yuzawa H, Ito T. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67–74 [DOI] [PubMed] [Google Scholar]
- 18. Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6:65–70 [Google Scholar]
- 19. Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin. Microbiol. Infect. 14:1020–1027 [DOI] [PubMed] [Google Scholar]
- 20. Ibrahem S, et al. 2009. Carriage of methicillin-resistant staphylococci and their SCCmec types in a long-term-care facility. J. Clin. Microbiol. 47:32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jamaluddin TZ, et al. 2008. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J. Clin. Microbiol. 46:3778–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johansson A, Koskiniemi S, Gottfridsson P, Wistrom J, Monsen T. 2006. Multiple-locus variable-number tandem repeat analysis for typing of Staphylococcus epidermidis. J. Clin. Microbiol. 44:260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katayama Y, Robinson DA, Enright MC, Chambers HF. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J. Clin. Microbiol. 43:2380–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kernodle DS, Barg NL, Kaiser AB. 1988. Low-level colonization of hospitalized patients with methicillin-resistant coagulase-negative staphylococci and emergence of the organisms during surgical antimicrobial prophylaxis. Antimicrob. Agents Chemother. 32:202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kloos WE, Musselwhite MS. 1975. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 30:381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kondo Y, et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazieres S, et al. 2008. Uniparental (mtDNA, Y-chromosome) polymorphisms in French Guiana and two related populations—implications for the region's colonization. Ann. Hum. Genet. 72:145–156 [DOI] [PubMed] [Google Scholar]
- 29. Mazieres S, et al. 2007. Genetic studies in French Guiana populations: synthesis. Am. J. Phys. Anthropol. 132:292–300 [DOI] [PubMed] [Google Scholar]
- 30. Mermel LA, et al. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murray BE, Mathewson JJ, DuPont HL, Ericsson CD, Reves RR. 1990. Emergence of resistant fecal Escherichia coli in travelers not taking prophylactic antimicrobial agents. Antimicrob. Agents Chemother. 34:515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicolle LE, Harding GK. 1982. Susceptibility of clinical isolates of Staphylococcus saprophyticus to fifteen commonly used antimicrobial agents. Antimicrob. Agents Chemother. 22:895–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Otter JA, French GL. 2010. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Europe. Lancet Infect. Dis. 10:227–239 [DOI] [PubMed] [Google Scholar]
- 35. Perry JD, et al. 2004. Development and evaluation of a chromogenic agar medium for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 42:4519–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pupin H, et al. 2007. Evaluation of moxalactam with the BD Phoenix system for detection of methicillin resistance in coagulase-negative staphylococci. J. Clin. Microbiol. 45:2005–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruimy R, et al. 2010. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J. Infect. Dis. 202:924–934 [DOI] [PubMed] [Google Scholar]
- 38. Ruimy R, et al. 1994. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 44:416–426 [DOI] [PubMed] [Google Scholar]
- 39. Ruimy R, et al. 2008. Accuracy and potential usefulness of triplex real-time PCR for improving antibiotic treatment of patients with blood cultures showing clustered Gram-positive cocci on direct smears. J. Clin. Microbiol. 46:2045–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rupp ME, Soper DE, Archer GL. 1992. Colonization of the female genital tract with Staphylococcus saprophyticus. J. Clin. Microbiol. 30:2975–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruppe E, et al. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sabuncu E, et al. 2009. Significant reduction of antibiotic use in the community after a nationwide campaign in France, 2002–2007. PLoS Med. 6:e1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarkar SK, Chang C-K. 1997. The Simes method for multiple hypothesis testing with positively dependent test statistics. J. Am. Stat. Assoc. 92:1601–1608 [Google Scholar]
- 44. Silva FR, Mattos EM, Coimbra MV, Ferreira-Carvalho BT, Figueiredo AM. 2001. Isolation and molecular characterization of methicillin-resistant coagulase-negative staphylococci from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol. Infect. 127:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soderquist B, Berglund C. 2009. Methicillin-resistant Staphylococcus saprophyticus in Sweden carries various types of staphylococcal cassette chromosome mec (SCCmec). Clin. Microbiol. Infect. 15:1176. [DOI] [PubMed] [Google Scholar]
- 46. Sohail MR, et al. 2007. Management and outcome of permanent pacemaker and implantable cardioverter-defibrillator infections. J. Am. Coll. Cardiol. 49:1851–1859 [DOI] [PubMed] [Google Scholar]
- 47. Terpstra S, Noordhoek GT, Voesten HG, Hendriks B, Degener JE. 1999. Rapid emergence of resistant coagulase-negative staphylococci on the skin after antibiotic prophylaxis. J. Hosp. Infect. 43:195–202 [DOI] [PubMed] [Google Scholar]
- 48. Wielders CL, et al. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus [corrected]. Lancet 357:1674–1675 [DOI] [PubMed] [Google Scholar]
- 49. Wisplinghoff H, et al. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47:3574–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woerther PL, et al. 2010. Emergence and dissemination of extended-spectrum beta-lactamase-producing Escherichia coli in the community: lessons from the study of a remote and controlled population. J. Infect. Dis. 202:515–523 [DOI] [PubMed] [Google Scholar]
- 51. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang YQ, et al. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.