Abstract
An Ambler class A β-lactamase gene, blaCIA-1, was cloned from the reference strain Chryseobacterium indologenes ATCC 29897 and expressed in Escherichia coli BL21. The blaCIA-1 gene encodes a novel extended-spectrum β-lactamase (ESBL) that shared 68% and 60% identities with the CGA-1 and CME-1 β-lactamases, respectively. blaCIA-1-like genes were detected from clinical isolates. In addition to the metallo-β-lactamase IND of Ambler class B, C. indologenes has a class A ESBL gene, blaCIA-1, located on the chromosome.
TEXT
The organism Chryseobacterium indologenes is the most common flavobacterium in clinical specimens and is associated with various types of infections, such as intra-abdominal infections, biliary tract infections, wound sepsis, catheter-related bacteremia, sepsis, and pneumonia (1, 11, 15, 16). However, no clonal outbreaks have been reported. C. indologenes is resistant to nearly all penicillins, restricted-spectrum cephalosporins, and carbapenems (3). It has been speculated that this resistance is due to metallo-β-lactamase IND (6).
Previously, two Ambler class A β-lactamases, CGA-1 and CME, were characterized in Chryseobacterium gleum and Elizabethkingia meningoseptica (formally Chryseobacterium meningosepticum), respectively (5, 17). CGA-1 and CME exhibit broad-spectrum profiles and are chromosomally encoded. Zeba et al. reported a single β-lactamase band at pI 9.0 in addition to IND from C. indologenes, using isoelectric focusing, and the band at pI 9.0 probably included an active site serine enzyme (19). However, no class A β-lactamases have been identified from C. indologenes. The aims of this study were to perform molecular characterization of the Ambler class A β-lactamase produced by C. indologenes and to investigate its distribution among other strains.
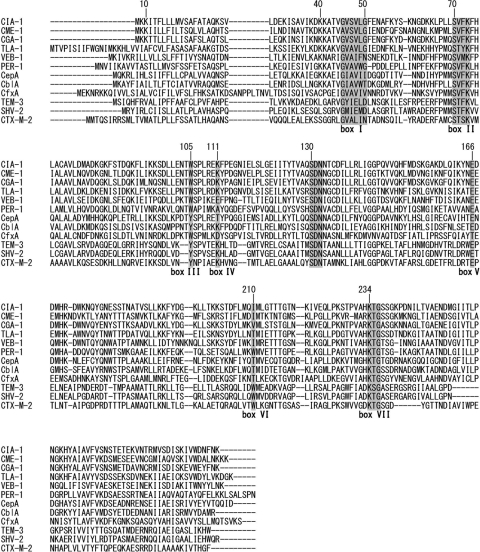
The C. indologenes reference strain (ATCC 29897) used in this study was purchased from the American Type Culture Collection. The MICs of the antimicrobial agents were determined using an agar dilution technique on Mueller-Hinton plates (BBL Microbiology Systems, Cockeysville, MD), with an inoculum of 104 CFU/spot in accordance with the performance standards for antimicrobial susceptibility testing in the guidelines published by the Clinical and Laboratory Standards Institute (8). We confirmed the blaIND from C. indologenes ATCC 29897 by PCR (3). Conjugation experiments failed to transfer any β-lactam resistance marker from C. indologenes ATCC 29897 to rifampin-resistant Escherichia coli CSH2 (10), and extraction of plasmid DNA from C. indologenes ATCC 29897 was attempted; however, plasmids were not detected. Genomic DNA from C. indologenes ATCC 29897 was extracted, and fragments from genomic DNA, which were partially digested with PstI, were ligated into the PstI-restricted phagemid pBK-CMV (Stratagene, La Jolla, CA) by using a previously reported method (7). The recombinant clone E. coli BL21(pCIA-1) was obtained after selection on amoxicillin (30 μg/ml)- and kanamycin (30 μg/ml)-containing Mueller-Hinton agar plates. The recombinant plasmid was purified and sequenced on both strands. An 879-bp open reading frame, which encoded a 292-amino-acid protein, was identified. This open reading frame showed the highest similarity to other class A β-lactamases in a BLAST search. This novel CIA-1 β-lactamase had identities of 68% to CGA-1 (5), 60% to CME-1 (17), 51% to TLA-1 (18), 48% to CSP-1 (9), and 44% to VEB-1 (14) (Fig. 1).
Fig 1.
Amino acid sequence of β-lactamase CIA-1 with 11 class A ESBLs. Boxes I through VII correspond to amino acid sequences described by Joris et al. (12). Dashes show gaps used to optimize the alignments.
A serine active site, characteristic of β-lactamases, was found within the mature protein sequence of CIA-1 (Fig. 1) (12). CIA-1 had 4 conserved elements of class A β-lactamases (2, 12): a Ser-X-X-Lys consensus active site serine residue at position 70, an SDN loop at position 130, a conserved Glu166, and a KTG sequence at position 234.
The G+C content of blaCIA-1 was 36.41%, which is typical of Chryseobacterium species. This result, along with the negative conjugation and plasmid isolation attempts, suggests a chromosomal location for blaCIA-1.
Genomic DNAs of C. indologenes clinical isolates (SH187, SH520, and SH3157) were investigated to clarify the distribution of the blaCIA-1 gene, using PCR screening with a set of designed primers, CIA-F (GCGAGAATAAACTCAGAGTACAT) and CIA-R (AGCATGAACTTCCATAAGAGATC). Three specific amplicons were amplified by standard PCR (denaturation for 10 min at 95°C with 35 cycles of 1 min at 95°C, 1 min at 58°C, and 2 min at 72°C and a final extension for 10 min at 72°C), and analyses of predicted amino acid sequences were performed with the ClustalW program. The blaCIA-1 gene had 98% amino acid identity to blaCIA-1-like genes from SH187, SH520, and SH3157.
The blaCIA-1 open reading frame encodes a putative protein comprising 292 amino acids with a molecular mass of ∼32.5 kDa (Fig. 1). The mature peptide has a theoretical pI of 9.0, which corresponded to the pI of CIA-1 by isoelectric focusing (data not shown). It is probably the same β-lactamase band observed by Zeba et al. (19). The susceptibility of C. indologenes ATCC 29897 to β-lactams was similar to that reported for C. indologenes (13). The MICs of several β-lactams reported previously (5) are shown in Table 1. C. indologenes ATCC 29897 was resistant to amino- and carboxypenicillins, narrow-spectrum cephalosporins, cefotaxime, cefoperazone, carbapenems, and aztreonam; however, it was susceptible to ceftazidime and cefepime (Table 1). The antibiotic effects of piperacillin, ceftazidime, and cefepime were decreased 2- to 32-fold in the presence of clavulanic acid (Table 1). E. coli BL21(pCIA-1) was resistant to amoxicillin, ticarcillin, carbenicillin, narrow-spectrum cephalosporins, and ceftazidime and had reduced susceptibility to cefotaxime, cefoperazone, cefepime, and aztreonam. Clavulanic acid reduced the MICs of β-lactams for E. coli BL21(pCIA-1) (Table 1). The MIC of C. indologenes ATCC 29897 decreased to below 1/8 by the addition of piperacillin or cefepime with clavulanic acid. The antibiotic susceptibility pattern of the C. indologenes clinical isolate SH187 was similar to that of C. indologenes ATCC 29897. The expression of class B β-lactamase IND can explain the resistance of C. indologenes ATCC 29897 to cefoxitin and carbapenems (3).
Table 1.
Antimicrobial susceptibilities of C. indologenes ATCC 29897, C. indologenes SH187, E. coli BL21(pCIA-1), and reference strain E. coli BL21
| β-Lactam | MIC (μg/ml) |
|||
|---|---|---|---|---|
| C. indologenes ATCC 29897 | E. coli BL21 (pCIA-1) | C. indologenes SH187 | E. coli BL21 | |
| Penicillin G | 64 | 128 | 16 | 4 |
| Amoxicillin | 256 | 256 | 64 | 1 |
| Carbenicillin | >256 | >256 | 256 | 2 |
| Ticarcillin | >256 | >256 | >256 | 1 |
| Piperacillin | 256 | 16 | 32 | 0.5 |
| Cefazolin | 256 | 16 | 256 | 1 |
| Cephalothin | 256 | 32 | 128 | 1 |
| Cephalexin | 256 | 64 | >256 | 4 |
| Cefuroxime | >256 | 32 | 128 | 0.5 |
| Cefotaxime | 64 | 1 | 16 | ≤0.125 |
| Ceftazidime | 8 | 32 | 8 | ≤0.125 |
| Ceftriaxone | 128 | 1 | 64 | ≤0.125 |
| Cefoperazone | 32 | 1 | 16 | ≤0.125 |
| Cefepime | 4 | 0.5 | 4 | ≤0.125 |
| Cefoxitin | 16 | 1 | 8 | 1 |
| Moxalactam | 128 | ≤0.125 | 128 | ≤0.125 |
| Imipenem | 128 | ≤0.125 | 2 | ≤0.125 |
| Aztreonam | 256 | 8 | 256 | ≤0.125 |
| Amoxicillin + CLAa | 256 | 2 | 4 | 1 |
| Piperacillin + CLA | 8 | 0.5 | 0.25 | 0.5 |
| Ceftazidime + CLA | 4 | ≤0.125 | 4 | ≤0.125 |
| Cefepime + CLA | 0.5 | ≤0.125 | 0.5 | ≤0.125 |
CLA, clavulanic acid at 2 μg/ml.
In this study, we demonstrated the presence of the blaCIA gene in the C. indologenes chromosome by the absence of conjugation transfer, determined the nucleotide sequence of blaCIA-1 (G+C content, 36%) and detected blaCIA-1-like genes in clinical isolates. C. indologenes was shown to possess chromosomally encoded class A extended-spectrum β-lactamase (ESBL) CIA, in addition to class B β-lactamase IND, and the class A β-lactamase of CIA-1 shared functional and structural similarities with CME-1, CME-2, and CGA-1.
Nucleotide sequence accession number.
The nucleotide sequences of the complete blaCIA genes from C. indologenes ATCC 29897, SH187, SH520, and SH3157 that have been reported in this study have been submitted to the GenBank and EMBL databases under accession no. AB639753, AB674566, AB674567, and AB674568, respectively.
ACKNOWLEDGMENT
The work is partially supported by funding from Nagano Society for the Promotion of Science.
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Akay M, Gunduz E, Gulbas Z. 2006. Catheter-related bacteremia due to Chryseobacterium indologenes in a bone marrow transplant recipient. Bone Marrow Transplant. 37:435–436 [DOI] [PubMed] [Google Scholar]
- 2. Ambler RP, et al. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellais S, Leotard S, Poirel L, Naas T, Nordmann P. 1999. Molecular characterization of a carbapenem-hydrolyzing beta-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 171:127–132 [DOI] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Bellais S, Naas T, Nordmann P. 2002. Molecular and biochemical characterization of Ambler class A extended-spectrum beta-lactamase CGA-1 from Chryseobacterium gleum. Antimicrob. Agents Chemother. 46:966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellais S, Poirel L, Leotard S, Naas T, Nordmann P. 2000. Genetic diversity of carbapenem-hydrolyzing metallo-beta-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob. Agents Chemother. 44:3028–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. 2000. Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 44:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S20, vol 30, no. 1 National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 9. Guillon H, Eb F, Mammeri H. 2010. Characterization of CSP-1, a novel extended-spectrum beta-lactamase produced by a clinical isolate of Capnocytophaga sputigena. Antimicrob. Agents Chemother. 54:2231–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horii T, et al. 1993. Plasmid-mediated AmpC-type beta-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum beta-lactams, including moxalactam. Antimicrob. Agents Chemother. 37:984–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsueh PR, et al. 1997. Increasing incidence of nosocomial Chryseobacterium indologenes infections in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 16:568–574 [DOI] [PubMed] [Google Scholar]
- 12. Joris B, et al. 1991. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob. Agents Chemother. 35:2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin XH, Xu YH, Cheng J, Li T, Wang ZX. 2008. Heterogeneity of bla(IND) metallo-beta-lactamase-producing Chryseobacterium indologenes isolates detected in Hefei, China. Int. J. Antimicrob. Agents 32:398–400 [DOI] [PubMed] [Google Scholar]
- 14. Poirel L, et al. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ray P, Sharma K, Gautam V. 2005. Chryseobacterium indologenes bacteremia: a case report. J. Commun. Dis. 37:259–260 [PubMed] [Google Scholar]
- 16. Reynaud I, et al. 2007. A severe form of Chryseobacterium indologenes pneumonia in an immunocompetent patient. Med. Mal. Infect. 37:762–764 [DOI] [PubMed] [Google Scholar]
- 17. Rossolini GM, et al. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaA(CME)) encoding an extended-spectrum class A beta-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER beta-lactamases. Antimicrob. Agents Chemother. 43:2193–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silva J, et al. 2000. TLA-1: a new plasmid-mediated extended-spectrum beta-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeba B, et al. 2009. IND-6, a highly divergent IND-type metallo-beta-lactamase from Chryseobacterium indologenes strain 597 isolated in Burkina Faso. Antimicrob. Agents Chemother. 53:4320–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]