Abstract
Emergence of artemisinin resistance in Cambodia highlights the importance of characterizing resistance to this class of drugs. Previously, intermediate levels of resistance in Plasmodium falciparum were generated in vitro for artelinic acid (AL) and artemisinin (QHS). Here we expanded on earlier selection efforts to produce levels of clinically relevant concentrations, and the resulting lines were characterized genotypically and phenotypically. Recrudescence assays determined the ability of resistant and parent lines to recover following exposure to clinically relevant levels of drugs. Interestingly, the parent clone (D6) tolerated up to 1,500 ng/ml QHS, but the resistant parasite, D6.QHS340×3, recovered following exposure to 2,400 ng/ml QHS. Resistant D6, W2, and TM91c235 parasites all exhibited elevated 50% inhibitory concentrations (IC50s) to multiple artemisinin drugs, with >3-fold resistance to QHS and AL; however, the degree of resistance obtained with standard methods was remarkably less than expected for parasite lines that recovered from 2,400-ng/ml drug pressure. A novel assay format with radiolabeled hypoxanthine demonstrated a greater degree of resistance in vitro than the standard SYBR green method. Analysis of merozoite number in resistant parasites found D6 and TM91c235 resistant progeny had significantly fewer merozoites than parent strains, whereas W2 resistant progeny had significantly more. Amplification of pfmdr1 increased proportionately to the increased drug levels tolerated by W2 and TM91c235, but not in resistant D6. In summary, we define the artemisinin resistance phenotype as a decrease in susceptibility to artemisinins along with the ability to recover from drug-induced dormancy following supraclinical concentrations of the drug.
INTRODUCTION
Malaria is the most important parasitic disease of humans, causing considerable mortality and morbidity on four continents. The most recent World Malaria Report (58) reported data for 2009, when there were an estimated 225 million cases of malaria and 781,000 deaths, of which 91% occurred in Africa. During the past 5 decades, Plasmodium falciparum, the most lethal malaria parasite infecting humans, has developed resistance to almost every available antimalarial drug. The emergence and spread of resistance has underscored the urgent need for new effective drugs and drug combinations.
Artemisinin (QHS) drugs have been used for the treatment of malaria for centuries, and they are effective against all stages of Plasmodium spp. Artemisinin, the active component of Artemesia annua, is a sesquiterpene trioxane lactone that contains an endoperoxide ring that is critical for antimalarial activity (21). Artemisinin drugs produce faster parasite and fever clearance times in vivo than any other antimalarial drug, and they reduce gametocyte carriage (52), thereby effectively reducing transmission of malaria. The World Health Organization (WHO) has recommended the use of artemisinin combination therapy (ACT) to treat uncomplicated P. falciparum malaria on a worldwide basis (57). Artemisinin drugs also have relatively short half-lives in the body, a property that may be related to the frequent recrudescence observed in patients after treatment with a single artemisinin drug (53). Recrudescent parasites remain susceptible to artemisinin in vitro (32), suggesting that the high frequency of treatment failure is not due to conventional resistance mechanisms. Recent studies have reported emerging artemisinin resistance in Southeast Asia (3, 19, 38). Resistance has been characterized by reduced susceptibility to both artemisinins and ACTs and by prolonged parasite clearance times (PCT). Although these studies acknowledged the potential of artemisinin resistance, they also noted that it is not a widespread phenomenon.
Current understanding of the mechanism of action of artemisinin drugs is unclear, although many mechanisms have been postulated. It has been proposed that artemisinins exert their effect via the generation of reactive oxygen species, becoming activated by free ferrous or heme iron (reviewed in reference 29), disrupting mitochondrial function (51), or by inhibiting a calcium-dependent ATPase (PfATP6) that is similar to mammalian sarco/endoplasmic reticulum Ca2+ ATPases (20).
Similarly, a consensus for the mechanism of artemisinin resistance has not emerged. Currently, there are genes that have tentative associations with the site of action or reduced susceptibility to artemisinins (or cross-resistance with other antimalarials). These include pfatp6 (27, 48), P. falciparum multidrug resistance transporter 1 (pfmdr1) (2, 11, 12, 42, 54), P. falciparum transitionally controlled tumor protein (pftctp) (10), Plasmodium chabaudi multidrug transporter 1 (pcmdr1) (45), and P. chabaudi ubiquitin carboxyl-terminal hydrolase (pcubp1) (24). However, the association between artemisinin resistance and pfatp6 is questionable, as others did not find mutations or an association with susceptibility change (12, 19, 25, 38, 50). Also, the link between pfmdr1 (or its homolog in murine malaria) and artemisinin resistance is unclear, as resistance cannot be explained by point mutations or gene amplification (1, 12, 19, 25, 38).
Previous in vitro studies have provided compelling evidence that some antimalarial drugs are not completely parasiticidal but may cause parasites to enter a dormant state before they recrudesce in culture (36). It has been suggested that recrudescence after artemisinin treatment is attributable to parasites surviving in a hidden, protected state (23). Our group has found that ring-stage parasites enter a dormant state after treatment with artemisinin drugs (16, 46), and these parasites are capable of resuming growth after drug pressure is removed. Based on these data, we believe that dormant ring-stage parasites may be the forms that survive treatment with artemisinin drugs and recover to initiate a recrudescent infection. Witkowski et al. (55) reported that parasites enter a quiescent state after exposure to high levels of artemisinin, and those authors suggested that the ability to enter the quiescent state was a hallmark of resistance. It is unlikely that this mechanism of dormancy and reemergence is responsible for extended PCT in patients.
In order to study mechanisms of resistance, investigators have attempted to establish artemisinin resistance models, including models for P. falciparum and Toxoplasma gondii in vitro and rodent malaria in vivo (1, 9, 13, 24, 26, 45). Although these significant studies proved that artemisinin resistance can be induced in different models, some studies noted problems with resistance stability and/or loss of resistance.
Previous studies from our group found that multiple rounds of parasite exposure to artemisinin drugs, followed by dormancy and recrudescence, select for parasites with reduced susceptibility to this class of antimalarial drugs. This included selection of P. falciparum resistance to AL and QHS in vitro, detection of reduction in susceptibility to artemisinin and other drugs, and molecular changes occurring during the selection (12; L. Gerena and D. E. Kyle, unpublished data). In the present study, higher levels of QHS and AL resistance in P. falciparum strains were induced in order to further dissect artemisinin-induced dormancy and artemisinin resistance. Resistant parasites were characterized by a variety of methods, including quantitative recrudescence assays, two types of antimalarial drug susceptibility testing, and analysis of growth/merozoite development. Furthermore, we expanded on previous molecular methods by focusing on genes with putative links to artemisinin resistance in a variety of resistant strains. Through these methods of characterization, we defined an artemisinin-resistant phenotype that will be useful for future resistance studies.
MATERIALS AND METHODS
Plasmodium falciparum parasites and in vitro culture.
Parental lines and respective progeny resistant to QHS (W2 [Indochina], D6 [Sierra Leone]) or AL (TM91c235 [Thailand]) were maintained in culture by using previously described methods (47). Cultures were maintained at 1 to 15% parasitemia in complete medium supplemented with 10% heat-inactivated A+ human plasma (Interstate Blood Bank, Memphis, TN) and 4% A+ human red blood cells (RBCs; Interstate Blood Bank), except where noted, below. For the recrudescence, drug susceptibility, and growth assays described below, cultures were synchronized to ring stage by using 5% (wt/vol) d-sorbitol (Sigma-Aldrich, St. Louis, MO) (30).
Development of resistant parasite lines in vitro.
Induction of resistance to QHS and AL in D6, W2, and TM91c235 lines was previously reported (12). Briefly, discontinuous exposure to AL produced W2, D6, and TM91c235 parasites resistant to 80 ng/ml AL (W2.AL80, D6.AL80, and TM91c235.AL80). W2.AL80 was subsequently cloned by limiting dilution, and one clone was subjected to increasing pressure with QHS up to a concentration of 200 ng/ml (W2.QHS200). D6.AL80 was subjected to QHS treatment up to 80 ng/ml (D6.QHS80) (Gerena and Kyle, unpublished).
In this work, the above method was modified to produce parasites resistant to higher levels of QHS and AL. Asynchronous cultures grown at 1.4 to 3.9% parasitemia were exposed to drug as follows. D6.QHS80 was first exposed to 80 ng/ml QHS. TM91c235.AL80 did not tolerate 80 ng/ml of AL after thawing from cryopreservation, so 40 ng/ml AL was applied to start. At 48 h post-drug exposure, cultures were washed with RPMI medium to remove drug. After parasites recrudesced to normal growth and morphology (≥1% parasitemia), they were treated 1 to 3 times at the current drug level. Parasites were then treated with an increasing amount of drug (20- or 40-ng/ml increments), and the process was repeated. During induction of resistance in D6, D6.QHS300×2 was treated with 200 ng/ml QHS prior to a cloning attempt. Induction of resistance was later continued at the 300-ng/ml level. W2.QHS200 was treated with 200 ng/ml QHS once as described above (W2.QHS200×1) to verify resistance. After this culture reached >3% parasitemia, it was cloned by limiting dilution. These clones were then exposed to 200 ng/ml QHS again. One of these clones was picked for further analyses and is referred to as W2.QHS200×2. A clone of TM91c235 was used as the parental strain in all experiments that involved comparisons with TM91c235 parasites resistant to AL.
Quantitative recrudescence assays. (i) W2 assay.
Dihydroartemisinin (DHA; final concentration of 200 ng/ml) or dimethyl sulfoxide (DMSO) was added to flasks containing 2% parasitized RBCs. At 6 h post-drug exposure, cultures were washed with RPMI to remove DHA or DMSO. Blood smears were made before drug treatment and approximately every 24 h after drug exposure. Cultures were monitored until parasitemia with morphologically normal parasites exceeded 2.5% (165.5 h post-drug exposure). At each time point, parasitemia was determined as the total number of parasites/total RBCs counted (over 650) from a thin smear. Parasites were classified as dead, dormant, ring, trophozoite, or schizont. Recovery was expressed as the ratio of morphologically normal parasites/total parasites, and the ratio of dormant/total parasites was also calculated. Photomicrographs of W2 and W2.QHS200×2 were taken pre-drug exposure and every 24 h post-drug exposure.
(ii) D6 assay.
QHS (final concentrations of 80 to 340 ng/ml) was added to microtiter plates containing 2% parasitized RBCs. At 48 h post-drug exposure, QHS was washed out. Thick and thin smears were made before drug treatment and every 24 h post-drug exposure. Parasites were followed to a point beyond when normal growth and morphology had returned (over 300 h post-drug exposure). Recovery was monitored by inspecting thick smears. Because dormant and dead parasites could not be separated, they were scored together on each smear. Approximately 700 to 900 total parasites were counted and classified as dead/dormant, ring, trophozoite, or schizont. Recovery was calculated as described above.
(iii) Adapting D6 parasites beyond clinically relevant concentrations.
QHS (final concentration of 80 to 2,400 ng/ml) was added to microtiter plates containing 2% parasitized RBCs. This experiment was conducted as described above. The most resistant parasite that survived (D6.QHS2400×1) was treated three more times with 2,400 ng/ml QHS after this experiment to verify resistance, and then D6.QHS2400×4 was cloned by limiting dilution. One of these clones was treated with 2,400 ng/ml QHS once more, and it is referred to as D6.QHS2400×5 in this work.
Parasite growth assay and merozoite enumeration for parent and resistant lines.
Cultures containing 0.1% parasitized RBCs were monitored for 97 h (W2), 105.5 h (D6), and 144 h (TM91c235). Thin smears were made twice a day, and both parasitemia and the percentages of parasites at the different stages were calculated. Merozoites were counted in each of 60 segmenting schizonts for each strain over the complete time course. Photomicrographs of segmenting schizonts were taken at various time points. The mean, median, mode, standard deviation, and 95% confidence interval (CI) of merozoites were calculated, and a Mann-Whitney rank sum test (SigmaPlot; Systat Software, Chicago, IL) was used for testing statistical significance (α = 0.05).
In vitro drug susceptibility testing. (i) SYBR green drug susceptibility assay.
Stock solutions of DHA, QHS, artesunate (AS), artemether (AM), AL, chloroquine (CQ), mefloquine (MQ), and atovaquone (ATOV) were made at 1 mg/ml or 10 mg/ml (CQ). Drug stocks were diluted to 625 ng/ml (all artemisinins and ATOV), 2,500 ng/ml (MQ), or 6,250 ng/ml (CQ), and a Biomek 3000 laboratory automation work station (Beckman Coulter, Brea, CA) was used to serially dilute drugs 1:2 in a 96-well plate. The Biomek then transferred 15 μl of drug to a 96-well plate, and 135 μl/well of 0.5% parasitized RBCs at 1.5% hematocrit was added. Positive controls included 25 μg/ml of QHS and DHA; cultures without drug were used as negative controls. Drug susceptibility plates were incubated in a humidified modular incubator chamber (Billups-Rothenberg, Del Mar, CA) for 72 h at 37°C. Parasite lysates were added to SYBR green lysis buffer (4). Fluorescence intensity was measured with a Spectra Max-M2 plate reader (Molecular Devices, Sunnyvale, CA). The mean concentration and standard deviation for inhibition of parasite growth by 50% and 90% (IC50 and IC90) were determined for each parasite line (≥2 assays were performed per strain and drug tested). IC50 and IC90 values were estimated based on linear and nonlinear regression curves calculated with Plate Manager software (DataAspects Corporation, Glencoe, CA). Data were exported into and analyzed with Microsoft Excel (Microsoft, Redmond, WA).
(ii) Drug susceptibility assay using a radioisotope microdilution technique.
The Biomek apparatus serially diluted drugs 1:2 from 1,000 ng/ml to 0.98 ng/ml (DHA, QHS, and AL) or from 6,250 ng/ml to 6.10 ng/ml (CQ) in a 96-well plate. The Biomek apparatus transferred 10 μl of drug to a new plate, and 90 μl/well of 1% parasitized RBCs at 2% hematocrit was added. Each concentration of drug was run in triplicate per parasite tested. Each plate included parasitized erythrocytes without drug and nonparasitized erythrocyte controls (each in duplicate per strain tested). Immediately after drugs were added to cultures, [3H]hypoxanthine monohydrochloride (equivalent to 1 μCi/well; Perkin-Elmer, Waltham, MA) was added to each well of the test plate. At 6, 24, and 48 h post-drug exposure, thick and thin smears were made. Plates from 24 and 48 h post-drug exposure were washed and harvested in a FilterMate cell harvester (Perkin-Elmer). Next, 50 μl/well of OptiPhase scintillation cocktail mix (Perkin-Elmer) was added, and plates were read in a TopCount NXT scintillation counter (Perkin-Elmer). Data (in counts per minute [cpm]) from the scintillation counter were imported into Microsoft Excel. For each parasite and time point, the mean cpm for each drug concentration was calculated. Data were imported into TableCurve 2D and SigmaPlot for curve fitting analysis and IC50 calculations.
Real-time quantitative PCR (QPCR). (i) Genomic DNA extraction.
Parasitized red blood cells were treated with 0.05% saponin to free parasites. Genomic DNA was extracted from parasite pellets by using a QiAmp DNA blood minikit (Qiagen, Valencia, CA). Purified genomic DNA was quantitated using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
(ii) Determination of gene copy number by real-time QPCR.
The relative copy numbers of pfmdr1, pfmdr2, PFE1050w, and PF11_0466 were determined by using lactate dehydrogenase (ldh) as the normalizer. A Stratagene (La Jolla, CA) MX3000P real-time quantitative PCR system measured the threshold cycle (CT) in triplicate for each template and primer set (see Table S1 in the supplemental material). Each 25-μl QPCR reaction mixture consisted of 2 μl DNA, Brilliant QPCR SYBR green 2× master mix (Stratagene), 30 nM ROX, and a final primer concentration of 200 nM (pfmdr2, PFE1050w, and PF11_0466) or 400 nM (pfmdr1 and ldh). Fluorescence data for 40 cycles were collected three times at the end of each annealing step and averaged. Dissociation curves after the amplification segment in the thermal profile verified that the correct products were produced. A 96-well plate was used for each experiment that included a five-point standard curve using serially diluted D6 genomic DNA for the gene of interest (GOI) and for ldh. Reports containing CT values were produced using Stratagene MXPro software and exported for further analyses into Microsoft Excel. The mean CT value per triplicate versus log2 concentration of D6 DNA for each standard (ldh and GOI) was plotted, and the slope and y intercept for the GOI and ldh were determined from a trend line. The slope and y intercept were used to convert CT values for unknown templates to a relative copy number with respect to the D6 standard {[(average CT for ldh − ldh y intercept)/−(slope ldh)] × [(average CT for the GOI − GOI y intercept)/−(slope GOI)]}. The mean copy numbers and standard deviations were calculated from ≥2 assays for each parasite line and GOI, and 95% CIs for mean copy numbers were determined. For testing of statistical significance, an unpaired two-tailed Student's t test was used (α = 0.05).
RESULTS
Discontinuous drug pressure generates P. falciparum lines resistant to increased levels of artelinic acid and artemisinin in vitro.
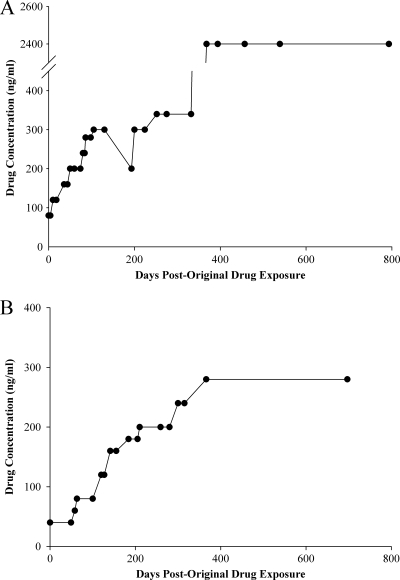
Resistance was initially induced by exposing strains of D6 and TM91c235 that tolerated 80 ng/ml of QHS (D6.QHS80) and AL (TM91c235.AL80), respectively, to increasing concentrations of drug in a stepwise manner (Fig. 1A and B; see also Tables S2 and S3 in the supplemental material). Resistance appeared to be stable, because after a strain was treated with a specific concentration of drug, it tolerated successive treatments at the same concentration. This was also observed when strains were grown off drug pressure for weeks or after thawing from cryopreservation. During the procedure, D6.QHS340×2 and TM91c235.AL80 were treated with lower drug levels and grew back normally before being treated with 340 ng/ml QHS and 80 ng/ml AL, respectively, to continue the induction of resistance. In less than a year using this method, D6.QHS80 tolerated 340 ng/ml QHS; TM91c235.AL80 tolerated 280 ng/ml AL in approximately a year (see Tables S2 and S3). W2.QHS200 was treated with 200 ng/ml QHS, and it recrudesced to 4% parasitemia by 7 days (data not shown). W2.QHS200×1 was cloned, and a single clone was treated with 200 ng/ml of QHS once more. W2.QHS200×2 reached ≥2% parasitemia 10 days after drug exposure.
Fig 1.
Adaptation of D6 and TM91c235 to increasing amounts of artemisinin drugs. Lines D6 and TM91c235 were previously adapted to 80 ng/ml AL and QHS (12) and used to initiate higher levels of resistance. (A) D6 resistant to 80 ng/ml of QHS (D6.QHS80) was discontinuously treated with increments of 20 to 40 ng/ml QHS to increase drug tolerance. Individual points in the graph correspond to a single treatment at a particular drug concentration level. During the procedure, D6.QHS300×2 was treated with 200 ng/ml QHS prior to a cloning attempt. Induction of resistance was later continued with 300 ng/ml QHS. (B) TM91c235 adapted to 80 ng/ml of AL (TM91c235.AL80) was treated discontinuously with increments of 20 to 40 ng/ml AL in order to increase drug tolerance.
Each time QHS or AL was applied to a culture, the majority of parasites had a morphology that was distinct from dead parasites beginning at 24 h (Fig. 2). These dormant parasites were equivalent to what has been reported elsewhere (46; M. S. Tucker et al., submitted for publication). They appeared rounded on a Giemsa-stained blood smear and differed from the collapsed nuclei of pyknotic bodies due to the retention of a small amount of blue cytoplasm and condensed red chromatin. Although it difficult to discern, we believe these characteristics separate dormant parasites from dead parasites, which lack distinct organization of chromatin and cytoplasm and stain purplish-red/pink with Giemsa stain (Fig. 2). After a few days, morphologically normal parasites were observed in cultures, which corresponded with the reduction of dormant forms.
Fig 2.
Comparison of dormant and dead parasites after a typical exposure to artemisinin drugs. (A) After ring-stage parasites were treated with artemisinin drugs, they became dormant, developed a rounded morphology, and retained blue cytoplasm and red chromatin on a Giemsa smear. (B) Dead parasites lacked distinct chromatin and cytoplasm, appearing globular, and they appeared red/purple on a Giemsa smear.
Resistant progeny of D6 and W2 recover faster than parent strains after exposure to clinically relevant concentrations of artemisinin.
After parasites were adapted to certain levels of artemisinin drugs, we quantitatively dissected differences in recrudescence between parent and resistant strains. D6 and W2 strains were treated with concentrations of QHS that are considered clinically relevant (39). Recovery rates of W2 and W2.QHS200×2 were determined by exposing parasites to 200 ng/ml DHA for 6 h and monitoring parasitemia over approximately a week (Fig. 3A). In both strains, the initial parasitemia decreased below 2% at 24 h post-drug exposure, and all parasites observed were classified as dormant or dead. Dormant parasites persisted through the end of the assay for each strain, but there was a greater dormant/total parasite ratio in W2.QHS200 versus W2 up to 72 h (Fig. 3B). The ratio decreased more sharply for W2.QHS200 than for W2, and it correlated with the appearance of morphologically normal parasites at 72 h post-drug exposure for W2.QHS200×2 (6.7% of parasites counted) (Fig. 3C). Morphologically normal parasites were not observed until 96 h for W2 (12.5% of parasites counted) (Fig. 3C). The ratio of normal parasites between the two strains equilibrated at 144 h post-drug exposure. Matched DMSO controls grew normally. The assay was terminated at 165.5 h post-drug exposure after parasitemia of normal parasites was ≥2.5%.
Fig 3.
Comparison of recrudescence in W2 parent and resistant lines after dihydroartemisinin treatment. (A) Parent W2 and resistant line W2.QHS200×2 were exposed to 200 ng/ml DHA for 6 h, and the parasitemia level was determined approximately every 24 h after drug exposure (up to 165.5 h post-drug exposure). (B) The dormant/total parasite ratio was calculated at each time point post-drug exposure. (C) The normal/total parasite ratio was calculated at each time point. Corresponding photomicrographs of parasites during the time course (through 120 h post-drug exposure) are shown below the graph.
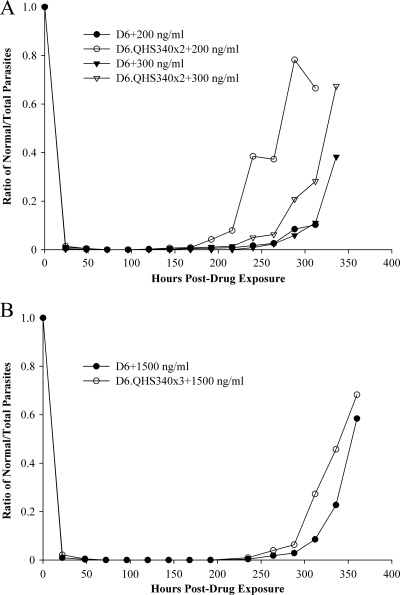
The process of treating D6 lines discontinuously with QHS produced D6.QHS340×2. D6 and D6.QHS340×2 were then exposed to a range of QHS (80 to 340 ng/ml) for 48 h. At each drug concentration, dormant parasites were observed in D6 and D6.QHS340×2 after 24 h, but morphologically normal parasites also persisted. At 48 h post-drug exposure, the majority of parasites (at least 99%) observed for each strain were dormant or dead. Figure 4A shows a representative comparison of recovery rates for D6 versus D6.QHS340×2 after exposure to 200 and 300 ng/ml of QHS. After 72 h of treatment with 200 ng/ml QHS, only dead or dormant parasites were observed for both strains. This remained until 120 h, when morphologically normal parasites were observed for both strains. However, a higher ratio of normal parasites was observed for D6.QHS340×2 versus D6 (0.25% versus 0.13%). This trend continued up to 312 h post-drug exposure (66.5% for D6.QHS340×2 versus 10.2% for D6). Treatment of cultures with 300 ng/ml QHS induced dormancy for a longer period of time than treatment with 200 ng/ml QHS. Morphologically normal parasites were observed at 144 h post-drug exposure for D6.QHS340×2 (0.61% of total parasites), whereas they were not observed until 240 h for D6 (0.86% of total parasites). At 336 h, the ratio of normal parasites was ∼2-fold greater for D6.QHS340×2 (67.4%) versus D6 (38.3%).
Fig 4.
Comparison of recrudescence in D6 parent and resistant lines after artemisinin treatment. (A) Normal/total parasite ratio for D6 and D6.QHS340×2 after 48 h of exposure to 28.2 to 340 ng/ml QHS (data shown are for 200 ng/ml and 300 ng/ml). Cultures were monitored up to 336 h post-drug exposure. (B) The normal/total parasite ratio for D6 and D6.QHS340×3 after 48 h of exposure to 80 to 2,400 QHS (shown is a representative example, at 1,500 ng/ml). Cultures were monitored up to 360 h post-drug exposure.
The most resistant strain produced from the assay described above was D6.QHS340×3. In a separate assay, D6 and D6.QHS340×3 were treated with QHS at a level that surpassed what would typically be found in plasma of malaria patients treated with artemisinin drugs. This produced D6 parasites that tolerated 80 to 1,500 ng/ml QHS and D6.QHS340×3 parasites that tolerated 80 to 2,400 ng/ml QHS. Figure 4B shows results for D6 and D6.QHS340×3 exposed to 1,500 ng/ml QHS. After 48 h post-drug exposure, >99% of observed parasites were dead or dormant. Both D6 and D6.QHS340×3 remained dead or dormant until 235 h post-drug exposure, when normal parasites were observed (0.94% D6.QHS340×3 versus 0.37% D6). After this point, there was a greater ratio of normal parasites for D6.QHS340×3 than D6 (e.g., at 360 h, 68.2% D6.QHS340×3 versus 58.4% D6). At the end of this assay, the most resistant D6 line was D6.QHS2400×1. This parasite was treated three more times with 2,400 ng/ml (Fig. 1A), and each time, parasites recovered to at least 2% parasitemia within 11 days (data not shown). D6.QHS2400×4 was cloned, and one clone was treated a final time with 2,400 ng/ml QHS, yielding D6.QHS2400×5.
Analyses of growth rates and merozoite numbers revealed differences between parental and resistant parasites.
Growth and merozoite numbers in parent and resistant strains were investigated, because previous data suggested that early drug-selected D6 parasites grow slower than D6 when off drug pressure (data not shown). A growth assay was conducted over a period of 105.5 h with D6 and D6.QHS2400×5. At each time point beyond 25 h post-sorbitol exposure, the parasitemia for D6.QHS2400×5 was lower than for D6 (see Fig. S1A in the supplemental material). Furthermore, D6.QHS2400×5 lagged behind D6 in development during the erythrocytic cycle. At 51 h post-sorbitol treatment (equivalent to the early to middle part of the second cycle [61 to 67 h into the life cycle]), there was a noticeable difference in the percentages of the parasite stages. For D6 there were 42.9% rings/57.1% trophozoites, but there were 90.9% rings/9.1% trophozoites in D6.QHS2400×5. This indicated a slower transition from rings to trophozoites or slower invasion, initiating the next cycle of growth. The same experiment with W2 and W2.QHS200×2 did not reveal as much of a difference in parasitemia as with the D6 pair, but W2.QHS200×2 did reach higher parasitemia at 92 h (5.4% versus 4.1%) (see Fig. S1B). We did not notice a difference in parasite development during the life cycles for the W2 pair. For the TM91c235 series, both the parent and resistant parasites had similar growth up to 96 h post-sorbitol exposure, when the parasitemia of TM91c235.AL280×2 spiked compared to TM91c235 (see Fig. S1C). Based on parasite stages, it was not clear why the resistant parasite line grew faster at this point in the assay. The parasitemias by both strains eventually equilibrated around 119 h.
In order to determine if the variation in growth rates between the parent/resistant pairs was related to merozoite development, the mean numbers of merozoites in segmenting schizonts for parent versus resistant strains were determined (Fig. 5A and B; see also Table S4 in the supplemental material). D6 had more merozoites per schizont (19.3; 95% CI, 18.7 to 20.0) than D6.QHS2400×5 (16.3; 95% CI, 15.7 to 16.9; P < 0.001). TM91c235 also had more merozoites (20.7; 95% CI, 20.1 to 21.3) than TM91c235.AL280×2 (19.7; 95% CI, 19.1 to 20.2; P = 0.007). However, W2 had fewer merozoites per schizont (15.3; 95% CI, 14.7 to 15.9) than W2.QHS200×2 (19.7; 95% CI, 19.1 to 20.4; P < 0.001). It was unclear why a lesser number of merozoites was associated with reduced growth for D6.QHS2400×5 versus D6, whereas a higher number of merozoites in W2.QHS200×2 was not associated with an increased growth rate compared to W2. Also, TM91c235 had more merozoites than TM91c235.AL280×2 (like the D6 pair), but the two parasite lines had similar growth until a point where the resistant parasite grew faster than the parent.
Fig 5.
Growth and merozoite numbers in parent and resistant progeny of D6, W2, and TM91c235 lines. (A) The mean merozoite numbers in segmenting schizonts of D6 and resistant D6.QHS2400×5, W2 and resistant W2.QHS200×2, and TM91c235 and resistant TM91c235.AL280×2 are shown. The number of merozoites was lower in D6.QHS2400×5 (*, P < 0.001) and TM91c235.AL280×2 (***, P = 0.007) than in the respective parent strains. The number of merozoites per schizont was greater in resistant W2 parasites than in the parental W2 parasites (**, P < 0.001). (B) Photomicrographs of typical segmenting schizonts of D6, W2, and TM91c235 parental and resistant parasites. Examples of a segmenting schizont containing the mean, minimum, and maximum (left to right) numbers of merozoites for each strain are shown.
Resistant parasites exhibit reduced susceptibility to artemisinins and mefloquine but increased susceptibility to chloroquine.
Parent and resistant pairs of D6, W2, and TM91c235 lines were tested against five artemisinin drugs and three standard antimalarial drugs (Tables 1 and 2). Compared to parental strains, all resistant strains showed a general decrease in susceptibility to all artemisinins. Also, resistant strains were generally the least susceptible to QHS and AL (>3-fold-higher resistance based on the IC50). For all artemisinins tested, D6.QHS2400×5 displayed the greatest degree of resistance (across all strains) compared to its parent. Resistance in D6.QHS2400×5 and W2.QHS200×2 was induced by QHS and AL, yet high levels of changes in the IC50 compared to parent strains also existed with other artemisinins. D6.QHS2400×5 was >9-fold more resistant to QHS than D6 but also >9-fold more resistant to DHA as well. W2.QHS200×2 was >3-fold more resistant to QHS than W2 and >6-fold more resistant to AL. As expected, TM91c235.AL280×2 displayed a high degree of resistance to AL, but this was not as great as for the D6 and W2 lines. It was also ∼4-fold more resistant to QHS and AM as well. TM91c235.AL280×2 displayed an ∼3-fold reduction in susceptibility to CQ compared to its parent. However, resistant D6 and W2 lines showed an increase in susceptibility to CQ compared to the respective parent strains. Resistant lines of D6 and W2 had >2-fold reductions in susceptibility to MQ compared to the respective parent strains. MQ susceptibility did not seem to differ in TM91c235 strains. W2 and TM91c235 parent/resistant pairs were equally susceptible to ATOV, but D6.QHS2400×5 was ∼2-fold more resistant than D6.
Table 1.
In vitro susceptibility testing of parental and resistant lines: artemisinin drugs
| Parasite line | Inhibitory concna (± SD) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DHA |
QHS |
AS |
AM |
AL |
||||||
| IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | |
| W2 | 0.69 ± 0.34 | 1.6 ± 0.95 | 1.3 ± 0.71 | 3.2 ± 0.80 | 0.40 ± 0.16 | 0.94 ± 0.50 | 0.73 ± 0.38 | 1.5 ± 0.56 | 2.0 ± 0.75 | 3.3 ± 1.2 |
| W2.QHS200×2 | 0.89 ± 0.33 | 2.3 ± 0.70 | 4.2 ± 2.0 | 9.6 ± 4.1 | 1.2 ± 0.36 | 2.2 ± 0.58 | 1.6 ± 0.42 | 3.4 ± 1.4 | 12.3 ± 3.35 | 18.4 ± 5.93 |
| D6 | 0.15 ± 0.028 | 0.34 ± 0.19 | 0.92 ± 0.10 | 4.1 ± 0.75 | 0.28 ± 0.080 | 0.68 ± 0.17 | 0.50 ± 0.070 | 1.3 ± 0.44 | 2.8 ± 0.66 | 11.5 ± 4.80 |
| D6.QHS2400×5 | 1.4 ± 0.30 | 4.0 ± 0.44 | 8.8 ± 1.0 | 19.3 ± 2.95 | 1.2 ± 0.040 | 2.0 ± 0.13 | 2.7 ± 0.30 | 5.8 ± 0.27 | 18.0 ± 1.56 | 22.2 ± 4.32 |
| TM91c235 | 1.2 ± 0.78 | 2.7 ± 0.81 | 2.2 ± 1.8 | 5.7 ± 2.1 | 0.93 ± 0.77 | 1.4 ± 0.57 | 0.91 ± 0.73 | 2.3 ± 0.67 | 3.2 ± 1.5 | 22.3 ± 15.9 |
| TM91c235.AL280×2 | 1.7 ± 1.2 | 3.7 ± 1.2 | 8.7 ± 5.4 | 16.5 ± 6.2 | 1.7 ± 0.82 | 4.0 ± 2.1 | 3.5 ± 2.1 | 6.0 ± 2.1 | 16.0 ± 12.0 | 37.6 ± 19.7 |
IC values are means and standard deviations from ≥2 assays per parasite line tested (in ng/ml).
Table 2.
In vitro susceptibility testing of parental and resistant lines: other antimalarial drugs
| Parasite line | Inhibitory concna (± SD) |
|||||
|---|---|---|---|---|---|---|
| CQ |
MQ |
ATOV |
||||
| IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | |
| W2 | 140.1 ± 43.68 | 193.3 ± 46.99 | 2.7 ± 0.94 | 5.7 ± 1.7 | 0.21 ± 0.0904 | 1.3 ± 0.96 |
| W2.QHS200×2 | 101 ± 7.44 | 142.6 ± 10.28 | 5.9 ± 1.3 | 22.3 ± 4.36 | 0.21 ± 0.067 | 0.75 ± 0.31 |
| D6 | 2.6 ± 0.26 | 4.3 ± 0.72 | 0.93 ± 0.17 | 3.1 ± 1.2 | 0.022 ± 0.0040 | 0.11 ± 0.045 |
| D6.QHS2400×5 | 1.8 ± 0.19 | 2.8 ± 0.31 | 3.8 ± 0.37 | 7.8 ± 0.77 | 0.041 ± 0.0050 | 0.25 ± 0.044 |
| TM91c235 | 23.2 ± 5.54 | 51.3 ± 10.6 | 8.0 ± 3.0 | 91.3 ± 37.9 | 0.26 ± 0.15 | 2.0 ± 1.2 |
| TM91c235.AL280×2 | 66.5 ± 40.4 | 126.1 ± 74.10 | 8.7 ± 4.5 | 51.7 ± 22.0 | 0.29 ± 0.19 | 1.9 ± 0.88 |
IC values are the means and standard deviations from ≥2 assays per parasite line tested (in ng/ml).
Resistant ring-stage parasites exhibit greater tolerance shortly after drug exposure.
Although drug susceptibility differences could be detected in parent versus resistant parasites, the degree of resistance was not as great as expected. We attributed this to the formation of dormant parasites and assessment at 72 h, before total recovery could occur. To quantitatively assess these differences more effectively, hypoxanthine incorporation was measured in the D6 and W2 lines during exposure to DHA, QHS, AL, or CQ. After exposure to an artemisinin drug, the resistant parasite strain in each pair tolerated more drug than the parent strain at 24 and 48 h (Fig. 6A to C and E to G). In addition, elevated IC50s were observed for both parental and resistant parasites in the 48-h assay compared to at 24 h (data for 24 h not shown). IC50 values were validated by blood smears that showed resistant parasites tolerated higher drug doses than parent strains. There was a large separation between D6 and D6.QHS2400×5 after treatment with AL and QHS (Fig. 6B and C), but the difference between the strains was greater with AL. After CQ treatment, D6 tolerated more drug than D6.QHS2400×5 (Fig. 6D). For the W2 pair, the greatest separation between parent and resistant parasites occurred with AL, followed by DHA and QHS (Fig. 6E to G). However, there was not as much of a shift between parent and resistant parasites compared to the D6 pair. We did not observe a difference in the CQ response for the W2 parasites in this assay (Fig. 6H).
Fig 6.
Drug susceptibility assay utilizing hypoxanthine incorporation and parent and resistant progeny of D6 and W2 lines. D6 and W2 lines were treated with QHS, DHA, AL, and CQ, and tritiated hypoxanthine was immediately added. The percent parasite growth at 48 h was plotted as a function of drug concentration (in ng/ml). The graphs show results for D6 plus DHA (A), D6 plus AL (B), D6 plus QHS (C), D6 plus CQ (D), W2 plus DHA (E), W2 plus AL (F), W2 plus QHS (G), and W2 plus CQ (H).
Real-time QPCR of genes implicated in prior molecular studies.
Preliminary attempts at the molecular characterization of artemisinin resistance from our group focused on transcriptional analysis of artemisinin-resistant W2 strains exposed to DHA (Q. Cheng et al., unpublished data). These results revealed sets of genes that were significantly upregulated (e.g., pfmdr1 and PFE1050w) or downregulated (e.g., pfmdr2 and PF11_0466) in resistant lines compared to W2. Since other data from our group have implicated pfmdr1 copy number in AL and QHS resistance (12, 14), we examined copy numbers of pfmdr1, PFE1050w, PF11_0466, and pfmdr2 in D6, W2, and TM91c235 lines, including different drug pressure levels from the stepwise selection of D6 and TM91c235 resistant parasites. The copy number of pfmdr1 (Fig. 7; see also Table S5 in the supplemental material) was approximately 1 in all D6 lines, even after adaptation to 2,400 ng/ml QHS (e.g., D6, 1.01 copies with a 95% CI of 1.00 to 1.01; D6.QHS2400×5, 1.10 copies with a 95% CI of 1.06 to 1.15). W2 had approximately 1 copy of pfmdr1 (1.21; 95% CI, 1.14 to 1.27). However, copy number increased from 1 to >2 at 200 ng/ml QHS (W2.QHS200, 2.24 copies; 95% CI, 2.12 to 2.35; P < 0.001). Copy number also increased after multiple treatments with 200 ng/ml QHS (W2.QHS200×2, 2.55 copies; 95% CI, 2.49 to 2.62; P = 0.001). We chose a single TM91c235 clone for comparisons to AL-resistant TM91c235 strains. This clone is a descendant of TM91c235 that was initially isolated from a patient with MQ treatment failure and it contains multiple copies of pfmdr1 (12). Although clones of this line can lose pfmdr1 copies during long-term culture, we found the copy number to be approximately 2 (2.20; 95% CI, 2.07 to 2.32). The copy number was greater at the 80-ng/ml level (TM91c235.AL80, 2.82 copies; 95% CI, 2.63 to 3.01; P = 0.002). As AL pressure increased from 80 to 240 ng/ml, the copy number increased above 3 (TM91c235.AL240×2, 3.60 copies; 95% CI, 3.31 to 3.88; P = 0.003). The effect of increasing drug concentration and a concomitant increase in pfmdr1 copy number diminished after parasites were adapted to 280 ng/ml AL (TM91c235.AL280×2, 3.56 copies; 95% CI, 3.41 to 3.71). QPCR assays on pfmdr2, PFE1050w, and PF11_0466 found all strains had approximately one copy of each gene (Fig. 7; see also Table S5).
Fig 7.
pfmdr1 copy numbers in parent and resistant progeny of D6, W2, and TM91c235 lines. Mean copy numbers of pfmdr1 and pfmdr2 in parental and resistant strains (D6, W2, and TM91c235) relative to D6, as determined by SYBR green real-time QPCR, are shown. Copy number assays were run ≥2 times for each parasite line tested.
DISCUSSION
In the study presented here, we expanded on previous work regarding the discontinuous induction of drug resistance to artemisinins in several lines of P. falciparum. The hallmark of in vitro resistance is the ability for parasites to recover after treatment with increasing concentrations of an artemisinin drug (levels considered clinically relevant) over many generations. Parasites entered dormancy, recrudesced, and tolerated successive, increasing doses of artemisinin drugs. In less than a year, derivatives of D6 and TM91c235 were generated that tolerated 340 ng/ml QHS and 280 ng/ml AL, respectively. We were able to induce higher levels of resistance during recrudescence assays (D6.QHS2400), and we believe it may be possible to select resistance to even greater levels than we report here. The resistance that was induced was stable, since parasites tolerated multiple exposures to drug levels to which they were made resistant. Also, after periods of no drug pressure and after thawing from cryopreservation, parasites could tolerate the dose to which they were resistant. SYBR green susceptibility testing found that resistant parasites showed the greatest tolerance for AL, but they also exhibited a high degree of resistance to QHS. We expected to find reduced susceptibility to these drugs, because they were used to induce resistance in these strains. However, when in vitro resistance to a certain artemisinin drug developed, resistance was also observed to other artemisinin derivatives not utilized in the generation of resistance in a particular line. Our results also showed resistant parasites generally had decreased susceptibility to MQ but were more susceptible to CQ, which is in agreement with other studies (12, 41, 42).
An important note is that it is possible to select resistance in a drug-sensitive background (D6), compared to conclusion from other studies in which it was thought that multiple resistance phenotypes must preexist (1, 43). To our knowledge, the current study and that of Witkowski et al. (55) are unique in that P. falciparum artemisinin-susceptible and -resistant lines generated in vitro survived levels of drug well above those typically found in patient sera. The latter study showed that after resistance was induced in F32-Tanzania to 9 μM QHS, it could recover from a 48-h treatment with 70 μM QHS. The parent survived 9 and 18 μM QHS after 48 h. Although these levels are higher than what we report here (D6 tolerated 5.3 μM and D6.QHS340×3 tolerated 8.5 μM), drug levels greatly exceeded the peak drug concentrations in treated patients. Also intriguing is the ability of wild-type parasites, D6 and F32, to tolerate 5.3 to 18 μM QHS without any prior selection pressure. Both parasites are from Africa, which is interesting since most previous successful in vitro resistance studies were accomplished with Asian parasites (see below). A key difference between this work and that of Witkowski et al. is that we are not implicating dormancy as an absolute artemisinin resistance mechanism. This is because both artemisinin-sensitive and -resistant parasites entered dormancy and recrudesce after artemisinin treatment. Indeed, recent work from our group showed that non-drug-adapted W2, D6, S55, H3, and PH1 recovered from DHA-induced dormancy or were unaffected by the drug (46), reflecting an intrinsic ability of P. falciparum strains to tolerate artemisinin drugs.
Clinical resistance to artemisinin is expressed as a delay in PCT or reduction in parasite clearance rate, yet thus far a clear association between the in vivo and in vitro phenotypes has not emerged (19). Similarly, we and others have not observed a dramatic shift in the in vitro drug susceptibility profile for parasites from patients ex vivo (11, 19) or in vitro with parasites selected for resistance (12, 55). These studies emphasize a problem of in vitro growth studies for detection of artemisinin resistance. Although it is possible to detect differences in IC50 levels (parent versus resistant strains) after artemisinin treatment, it is essential to recognize that dormant parasites persist beyond the time that typical in vitro drug susceptibility assays assess viability (48 to 72 h). Furthermore, SYBR green testing with artemisinin drugs found resistant progeny had reduced susceptibilities compared to parental parasites, as evidenced by IC50s and IC90s, yet the values for AL and QHS in resistant lines were lower than drug concentrations that parasites could endure in vitro. Also, longer recrudescence assays demonstrated that although resistant parasites recovered either before parent parasites or in higher numbers at the same time point, there was not always a great difference in the time of appearance (or number) of normal parasites. Thus, it appears that the treatment of parasites with different amounts of drug may not yield an appreciable difference in drug recovery in a typical drug susceptibility assay. This is most likely due to the ability of both wild-type and artemisinin-resistant parasites to enter dormancy in the ring stage.
Because of these issues, we investigated the use of a tritiated hypoxanthine (Hx) method as an alternative to assess antimalarial drug susceptibility. Although very useful, Hx assays are subject to variables, such as starting parasitemia/hematocrit, isotope pulse time, time of Hx addition, and duration in culture (7, 15, 18). Parasite viability after exposure to low levels of artemisinin was assessed during the first 48 h after treatment. Hx was added immediately after drug exposure, because we theorized that IC50s might be underestimated (due to dormancy) if Hx were added later. Also, assays utilized 1% parasitemia/2% hematocrit, which is in agreement with the above studies. Resistant parasites tolerated higher levels of AL, DHA, and QHS than parental parasites at 24 to 48 h after Hx was added, possibly due to a greater number of resistant dormant parasites incorporating Hx or more resistant parasites proceeding through the life cycle as normal (confirmed by smear). If this is true, then resistant strains have the ability to produce a greater number of dormant parasites and/or exit dormancy faster than parent parasites. Therefore, this may be the reason that in longer-duration susceptibility assays and recrudescence assays the artemisinin-resistant parasites recover at a higher rate than parent strains.
The Hx assay we employed has utility for exploring artemisinin resistance (and resistance to other antimalarials) for several reasons. It can utilize even lower hematocrit (1%) and parasitemia (0.5%) levels when hypoxanthine is added at time zero (K. Sparks et al., unpublished data). This assay would be suitable for ring stages, which primarily exist in the circulation of infected patients. It would also allow low-level parasitemias to be tested, making it amenable to testing parasites grown for short periods in vitro after they are obtained from patients. Since the majority of Hx incorporation occurs in the trophozoite stage and reflects the rapid increase in DNA replication and transcription, the assay would also detect a suitable ring/trophozoite maturation period. This would make it useful for detecting exit of dormancy or parasites that have reduced susceptibility to (or are unaffected by) artemisinins and other drugs.
Recent studies found that artemisinin-resistant parasites have prolonged PCT (3, 11, 19, 38). It may be that the extended PCT is due to a delay in the life cycle of resistant parasites that are circulating in malarious areas (34). We determined that resistant D6 parasites have reduced growth rates compared to the parental parasite, marked by a longer life cycle, delay in the progression from rings to trophozoites, and a decrease in the number of merozoites in segmenting schizonts. Interestingly, W2.QHS200x2 had more merozoites than W2, without an associated difference in growth. Reilly et al. (44) reported that Dd2 (a drug-resistant descendant of W2) had a shorter cycle time (faster growth and more rapid ring-to-trophozoite transition time), more merozoites per segmenting schizont, and a higher invasion rate than HB3. Perhaps long-term QHS pressure in D6 selected for genetic changes that manifested in growth inhibition. Recent studies by Balu et al. (5, 6) found reduced growth rates in transposon-generated P. falciparum mutants. The most attenuated mutants contained insertions in genes encoding RNA binding/recognition proteins and protein serine/threonine phosphatase 2C (PP2C), implicating the importance of these proteins in parasite development. PP2Cs participate in many cellular functions in eukaryotes, including growth factor-dependent signal transduction, transcription, pre-mRNA splicing, DNA replication, and the DNA damage response. These phosphatases also contribute to cell cycle and developmental check points (22). Other work we have performed suggests that genes and related proteins are downregulated in D6.QHS2400×5 (Tucker et al., submitted). It is possible that the D6 QHS-resistant parasites we generated displayed impaired growth due to mutations/reduced expression of RNA binding proteins or genetic alterations in cell cycle checkpoints. However, if present, these genetic changes did not appear to affect the abilities of resistant parasites to exit dormancy. These findings may be important for the relationship between extended PCT and resistance.
Our group previously found W2 parent and resistant parasites exposed to artemisinins displayed differential regulation of certain genes, implicating the genes in increased AL and QHS resistance. Some of these (pfmdr2 and PF11_0466) encode proteins of the ATP binding cassette (ABC) transporter superfamily (28), which are localized to the parasite food vacuole membrane or plasma membrane and can decrease intracellular drug accumulation by pumping out drugs. We did not detect changes in copy numbers of pfmdr2 or PF11_0466 in any resistant strains, but the relation of these genes to pfmdr1 and other transporters warrants studies to investigate any role(s) they may have in artemisinin susceptibility.
Two interesting genes were overexpressed in resistant progeny (PFE1050w and pfmdr1). PFE1050w encodes S-adenosyl homocysteine hydrolase (SAHH), which catalyzes reversible hydrolysis of S-adenosyl homocysteine (SAH) to adenosine and homocysteine, and it is necessary for active methylation of different biomolecules (17). A recent study found PFE1050w and enzymes involved in protein synthesis and parasite respiration were downregulated after P. falciparum was treated with AM (33). More recent research found PF10_0121 (which encodes hypoxanthine phosphoribosyltransferase) was downregulated in QHS-selected or AS-treated P. falciparum parasites (37, 55). Although PFE1050w does not appear to be involved in artemisinin resistance we report herein, SAHH and nucleic acid metabolism in general may be important drug targets for ongoing antimalarial drug development efforts. Molecular studies by our group have implicated pfmdr1 in artemisinin resistance generated in vitro (12). Interestingly, W2 and TM91c235 resistant lines have increased pfmdr1 copy number/expression levels, but resistant D6 parasites do not display these characteristics. Expanding on these data, we observed that the pfmdr1 copy number increased with multiple drug treatments at a single level (W2.QHS200×2) or with higher drug pressure (e.g., TM91c235.AL280). It appears that pfmdr1 amplification can be associated with stable resistance to AL and QHS in vitro, but clearly pfmdr1 amplification is not the only mechanism involved in artemisinin resistance. Another study from our group found that a rapid reduction in pfmdr1 copy number in the W2.AL80 line was associated with parasite growth in the absence of drug pressure (14). The reduction in pfmdr1 copy number involved the deamplification of an entire amplicon (19 genes) and associated reversal of resistance to AL and increased susceptibility to MQ. Based on these analyses, artemisinin resistance appears multifactorial, but it can be enhanced by amplification of pfmdr1, which also confers resistance to MQ (40–42, 54). It is well known that pfmdr1 is associated with drug resistance in Asia (2, 11, 31, 35, 41, 42). However, relatively fewer studies have reported on amplification of pfmdr1 in Africa (8, 49, 56). Perhaps there is significant copy number variation in Africa, as we did not detect a pfmdr1 copy number change in D6, which is from West Africa. Also, there may be a unique mechanism for resistance in the D6 resistant lines we generated, since parasites tolerating 2,400 ng/ml QHS still retained one copy of pfmdr1. We are in the process of identifying genetic determinants of artemisinin resistance in the D6 selected lines exhibiting the highest resistance.
Based on our investigations with parental and resistant pairs of parasites, we have defined an artemisinin resistance phenotype. The phenotype is partially characterized by the ability of resistant parasites to recover before parental strains after artemisinin drug pressure or the ability to recrudesce at higher numbers than parent strains. Resistant parasites may exhibit delayed progression through the normal erythrocytic cycle, with a noticeable extended transition from rings to trophozoites. SYBR green susceptibility testing showed resistant parasites had reduced susceptibility to multiple artemisinins compared to parent strains, yet artemisinin resistance phenotypes may not be accurately determined by this assay. Hypoxanthine incorporation assays allowed the determination of differences in parasite susceptibilities from the time of drug exposure through 24 to 48 h post-drug exposure, a time period when dormant parasites are typically observed. By using this measure of susceptibility, an earlier window of drug activity could be examined (entry into dormancy), making it possible to gauge if resistant parasites tolerated higher levels of drug as a result of a greater capacity to produce dormant parasites. Interestingly, artemisinin-resistant parasites tolerated higher drug concentrations than parental strains (at levels surpassing clinical significance); yet, in vitro susceptibility data did not correlate with the levels of drug to which the parasites were adapted. Overall, the results of artemisinin drug treatment indicate that resistant parasites tolerate more drug than parent strains, possibly due to the increased ability to exit dormancy and transition to normal growth at a greater rate than parent strains. This may be because dormant resistant parasites have a higher survival rate from dormancy; if a greater number of dormant parasites existed in resistant cultures, recrudescence would occur earlier. The in vitro resistance to artemisinin drugs that our group has described is different from that displayed with conventional antimalarial drugs, for which molecular mechanisms have been well described (e.g., CQ and MQ). Resistance is not completely explained by candidate gene amplifications or mutations, as genes with tentative associations with artemisinin resistance (pftctp, pfatp6, and pfmdr1) may not contain mutations or increases in copy number in all resistant progeny (reference 12 and the current study).
Artemisinin drugs are essential tools for controlling the world's most important parasitic disease. The recent WHO decision to introduce ACT globally makes the discovery of artemisinin resistance markers more important than ever before. Our group has demonstrated that resistance to artemisinin drugs can be induced in P. falciparum in vitro. Although long-term stability studies at high levels of drug exposure have not been completed, our data suggest that stable artemisinin resistance was selected for by multiple pulse exposures to drug over time. An alarming fact is that parasites in the field that are continually exposed to suboptimal levels of artemisinin drugs may begin to tolerate high levels of these drugs, leading to a greater distribution of drug resistance. The artemisinin-resistant parasites we generated represent unique reagents for molecular and cellular studies of the artemisinin resistance mechanism(s) and markers, mechanism-of-action studies, determination of the role of drug-induced dormancy as a mechanism of recrudescence, and optimization of ACT drug partners. It is tempting to speculate about the nature of artemisinin-induced dormancy as it relates to an artemisinin resistance mechanism, as some lines of evidence point in this direction. The ability of parent and resistant parasites to enter dormancy only to resume growth at a later time suggests artemisinin-induced dormancy is a novel resistance mechanism that can result in parasite recrudescence. Furthermore, it is possible that induction and selection of artemisinin-resistant parasites enhance recrudescence rates (i.e., extended PCT) and could explain a resistance mechanism where an increased proportion of parasites recover from dormancy following the removal of drug pressure, by decreasing the duration of dormancy, or both. Future research will focus on dissecting whole-genome sequence, transcription, and proteomic data of parental and resistant parasites to further elucidate mechanisms of resistance to artemisinin drugs. These studies will define the relationship between resistance and changes in the recovery rates or duration of dormancy. It is important to determine if the inherent susceptibility to artemisinin is reduced after multiple dormancy episodes or if the recovery rate increases or duration of dormancy decreases. Either of these outcomes would confirm a unique parasite survival mechanism to the most important class of antimalarial drugs. Clearly, more studies are required to identify the mechanism(s) by which resistance to artemisinin emerges and to find molecular markers that can be used for epidemiological studies to track the possible emergence of artemisinin resistance.
Supplementary Material
ACKNOWLEDGMENTS
The project was supported by the National Institutes of Health (NIAID R01 AI058973).
We thank Jennifer Peters, Marina Chavchich, and Carly Romero for their assistance in this research.
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Afonso A, et al. 2006. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob. Agents Chemother. 50:480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alker AP, et al. 2007. pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am. J. Trop. Med. Hyg. 76:641–647 [PubMed] [Google Scholar]
- 3. Anderson TJ, et al. 2010. High heritability of malaria parasite clearance rate indicates a genetic basis for artemisinin resistance in western Cambodia. J. Infect. Dis. 201:1326–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bacon DJ, et al. 2007. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob. Agents Chemother. 51:1172–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balu B, et al. 2009. piggyBac is an effective tool for functional analysis of the Plasmodium falciparum genome. BMC Microbiol. 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balu B, Singh N, Maher SP, Adams JH. 2010. A genetic screen for attenuated growth identifies genes crucial for intraerythrocytic development of Plasmodium falciparum. PLoS One 5:e13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basco LK. 2004. Molecular epidemiology of malaria in Cameroon. XX. Experimental studies on various factors of in vitro drug sensitivity assays using fresh isolates of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 70:474–480 [PubMed] [Google Scholar]
- 8. Basco LK, Le BJ, Rhoades Z, Wilson CM. 1995. Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from sub-Saharan Africa. Mol. Biochem. Parasitol. 74:157–166 [DOI] [PubMed] [Google Scholar]
- 9. Berens RL, Krug EC, Nash PB, Curiel TJ. 1998. Selection and characterization of Toxoplasma gondii mutants resistant to artemisinin. J. Infect. Dis. 177:1128–1131 [DOI] [PubMed] [Google Scholar]
- 10. Bhisutthibhan J, et al. 1998. The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. J. Biol. Chem. 273:16192–16198 [DOI] [PubMed] [Google Scholar]
- 11. Carrara VI, et al. 2009. Changes in the treatment responses to artesunate-mefloquine on the northwestern border of Thailand during 13 years of continuous deployment. PLoS One 4:e4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chavchich M, et al. 2010. Role of pfmdr1 amplification and expression in induction of resistance to artemisinin derivatives in Plasmodium falciparum. Antimicrob. Agents Chemother. 54:2455–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chawira AN, Warhurst DC, Peters W. 1986. Qinghaosu resistance in rodent malaria. Trans. R. Soc. Trop. Med. Hyg. 80:477–480 [DOI] [PubMed] [Google Scholar]
- 14. Chen N, et al. 2010. Deamplification of pfmdr1-containing amplicon on chromosome 5 in Plasmodium falciparum is associated with reduced resistance to artelinic acid in vitro. Antimicrob. Agents Chemother. 54:3395–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chulay JD, Haynes JD, Diggs CL. 1983. Plasmodium falciparum: assessment of in vitro growth by [3H]hypoxanthine incorporation. Exp. Parasitol. 55:138–146 [DOI] [PubMed] [Google Scholar]
- 16. Codd A, Teuscher F, Kyle DE, Cheng Q, Gatton ML. 2011. Artemisinin-induced parasite dormancy: a plausible mechanism for treatment failure. Malar. J. 10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Creedon KA, Rathod PK, Wellems TE. 1994. Plasmodium falciparum S-adenosylhomocysteine hydrolase. cDNA identification, predicted protein sequence, and expression in Escherichia coli. J. Biol. Chem. 269:16364–16370 [PubMed] [Google Scholar]
- 18. Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dondorp AM, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eckstein-Ludwig U, et al. 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424:957–961 [DOI] [PubMed] [Google Scholar]
- 21. Golenser J, Waknine JH, Krugliak M, Hunt NH, Grau GE. 2006. Current perspectives on the mechanism of action of artemisinins. Int. J. Parasitol. 36:1427–1441 [DOI] [PubMed] [Google Scholar]
- 22. Heideker J, Lis ET, Romesberg FE. 2007. Phosphatases, DNA damage checkpoints and checkpoint deactivation. Cell Cycle 6:3058–3064 [DOI] [PubMed] [Google Scholar]
- 23. Hoshen MB, Na-Bangchang K, Stein WD, Ginsburg H. 2000. Mathematical modelling of the chemotherapy of Plasmodium falciparum malaria with artesunate: postulation of ‘dormancy’, a partial cytostatic effect of the drug, and its implication for treatment regimens. Parasitology 121:237–246 [DOI] [PubMed] [Google Scholar]
- 24. Hunt P, et al. 2010. Experimental evolution, genetic analysis and genome re-sequencing reveal the mutation conferring artemisinin resistance in an isogenic lineage of malaria parasites. BMC Genomics 11:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imwong M, et al. 2010. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob. Agents Chemother. 54:2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inselburg J. 1985. Induction and isolation of artemisinine-resistant mutants of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 34:417–418 [DOI] [PubMed] [Google Scholar]
- 27. Jambou R, et al. 2005. Resistance of Plasmodium falciparum field isolates to in vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366:1960–1963 [DOI] [PubMed] [Google Scholar]
- 28. Koenderink JB, Kavishe RA, Rijpma SR, Russel FG. 2010. The ABCs of multidrug resistance in malaria. Trends Parasitol. 26:440–446 [DOI] [PubMed] [Google Scholar]
- 29. Krishna S, Uhlemann AC, Haynes RK. 2004. Artemisinins: mechanisms of action and potential for resistance. Drug Resist. Update 7:233–244 [DOI] [PubMed] [Google Scholar]
- 30. Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418–420 [PubMed] [Google Scholar]
- 31. Lim P, et al. 2009. pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar. J. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Looareesuwan S, et al. 1997. Open randomized trial of oral artemether alone and a sequential combination with mefloquine for acute uncomplicated falciparum malaria. Am. J. Trop. Med. Hyg. 56:613–617 [DOI] [PubMed] [Google Scholar]
- 33. Makanga M, Bray PG, Horrocks P, Ward SA. 2005. Towards a proteomic definition of CoArtem action in Plasmodium falciparum malaria. Proteomics 5:1849–1858 [DOI] [PubMed] [Google Scholar]
- 34. Mok S, et al. 2011. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics 12:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nair S, et al. 2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 24:562–573 [DOI] [PubMed] [Google Scholar]
- 36. Nakazawa S, Maoka T, Uemura H, Ito Y, Kanbara H. 2002. Malaria parasites giving rise to recrudescence in vitro. Antimicrob. Agents Chemother. 46:958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Natalang O, et al. 2008. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics 9:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noedl H, et al. 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359:2619–2620 [DOI] [PubMed] [Google Scholar]
- 39. Nosten F, White NJ. 2007. Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. Hyg. 77:181–192 [PubMed] [Google Scholar]
- 40. Peel SA, Bright P, Yount B, Handy J, Baric RS. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648–658 [DOI] [PubMed] [Google Scholar]
- 41. Pickard AL, et al. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price RN, et al. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rathod PK, McErlean T, Lee PC. 1997. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 94:9389–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. 2007. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int. J. Parasitol. 37:1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodrigues LA, et al. 2010. Experimental evolution of resistance to artemisinin combination therapy results in amplification of the mdr1 gene in a rodent malaria parasite. PLoS One 5:e11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teuscher F, et al. 2010. Artemisinin induced dormancy in Plasmodium falciparum: duration, recovery rates, and implications in treatment failure. J. Infect. Dis. 202:1362–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 48. Uhlemann AC, et al. 2005. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 12:628–629 [DOI] [PubMed] [Google Scholar]
- 49. Uhlemann AC, Ramharter M, Lell B, Kremsner PG, Krishna S. 2005. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J. Infect. Dis. 192:1830–1835 [DOI] [PubMed] [Google Scholar]
- 50. Valderramos SG, Scanfeld D, Uhlemann AC, Fidock DA, Krishna S. 2010. Investigations into the role of the Plasmodium falciparum SERCA (PfATP6) L263E mutation in artemisinin action and resistance. Antimicrob. Agents Chemother. 54:3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J, et al. 2010. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS One 5:e9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White NJ. 2008. Qinghaosu (artemisinin): the price of success. Science 320:330–334 [DOI] [PubMed] [Google Scholar]
- 53. White NJ, Olliaro P. 1998. Artemisinin and derivatives in the treatment of uncomplicated malaria. Med. Trop. (Madr.). 58:54–56 (In Spanish.) [PubMed] [Google Scholar]
- 54. Wilson CM, et al. 1993. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol. Biochem. Parasitol. 57:151–160 [DOI] [PubMed] [Google Scholar]
- 55. Witkowski B, et al. 2010. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 54:1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Witkowski B, et al. 2010. Plasmodium falciparum isolates with increased pfmdr1 copy number circulate in West Africa. Antimicrob. Agents Chemother. 54:3049–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. World Health Organization 2010. Guidelines for the treatment of malaria, 2nd ed World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/publications/atoz/9789241547925/en/index.html [Google Scholar]
- 58. World Health Organization 2010. World malaria report 2010. World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/world_malaria_report_2010/en/index.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.