Abstract
The Etest glycopeptide resistance detection identified two potential heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) isolates from a screen of 288 methicillin-resistant Staphylococcus aureus (MRSA) isolates from patients at a Connecticut Veterans Hospital. However, the two isolates did not meet the criteria for hVISA by the population analysis profile-area under the curve analysis, arguing against routine screening for hVISA in this low prevalence population.
TEXT
Vancomycin is a treatment of choice for methicillin-resistant Staphylococcus aureus (MRSA) infections, but MRSA isolates with reduced vancomycin susceptibility are increasingly recognized (11). Heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) strains contain a resistant subpopulation (≤10−5 to 10−6 CFU) yet demonstrate vancomycin sensitivity by standard laboratory testing methods. Modified population analysis profile (PAP) using the area under the curve (AUC), a labor-intensive and time-consuming method, is the gold standard for hVISA detection. hVISA has been associated with vancomycin treatment failures and is also considered the precursor of vancomycin-intermediate S. aureus (2, 3, 7, 9, 12, 13, 17).
The prevalence of hVISA ranges widely from 0 to 74%, depending on the geographic location, study population, and methodology (8, 11). Most studies in the United States have focused on hVISA among isolates causing bacteremia, while only limited data exist regarding the prevalence of hVISA from diverse infection sites (14). This may lead to an underestimation of the true prevalence, as a recent study from Australia found the frequency of hVISA among all clinical isolates of MRSA to be 48% and suggested that hVISA strains are common colonizers in hospitalized patients and are less frequently associated with invasive disease (6). The frequency of hVISA among colonizing strains in American hospitalized patients has not been determined but has important implications for infection control and ensuring appropriate antimicrobial treatment.
We obtained 1,611 consecutive MRSA isolates from clinical and surveillance specimens received by the Department of Veterans Affairs (VA) Connecticut Healthcare System (West Haven, CT) Clinical Microbiology Laboratory, which serves a 230-bed hospital. Isolates were received between February 2008 and January 2010 and stored at −70°C prior to testing. The mean MRSA colonization rate was 16%, and there was an average of 20 episodes of MRSA bacteremia per year during this period. Molecular typing using a DiversiLab Staphylococcus kt (bioMérieux) revealed a predominance of USA100 strains. Isolates from clinical specimens were selected from patients from whom a nasal screening isolate was also obtained. The hVISA phenotype is thought to arise after prolonged periods of infection and treatment with vancomycin (5, 8), and we hypothesized that prior studies which only examined the first isolate during an infection may have underestimated the prevalence of hVISA. Thus, for each patient, we examined the last received isolate to maximize the likelihood of detecting hVISA (n = 146).
MRSA isolates were recovered from the respiratory tract (36%), abscesses and wounds (31%), urine (15%), blood (12%), and other sites (5%). As expected from a population served by the VA health care system, 98% of these isolates were obtained from male patients, with the mean age of 70 years. Thirty-eight percent of the patients had received vancomycin in the 6 weeks prior to collection of the tested specimen. The average number of days of vancomycin treatment was 5.98, with a range of 1 to 33 days, and the standard daily dose of vancomycin was 2 g.
We screened these isolates using Etest glycopeptide resistance detection (GRD) strips (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions and as previously described (19). This is a highly sensitive and specific method for the detection of hVISA which could be easily implemented in the clinical microbiology laboratory (10, 19). hVISA Mu3 (ATCC 700698), the positive-control organism (18), consistently gave a positive screen result.
One of the 146 tested isolates (0.7%) was positive for presumptive hVISA by having Etest GRD MIC values of >32 μg/ml for teicoplanin and 3 μg/ml for vancomycin, with a standard Etest vancomycin MIC of <4 μg/ml. The finding of reduced teicoplanin sensitivity without a vancomycin phenotype is not surprising, as teicoplanin is more sensitive in the detection of the hVISA phenotype than vancomycin (19). This isolate was obtained from a wound culture from a patient without documented vancomycin exposure in the prior 6 weeks. An independent MRSA isolate obtained from nasal carriage surveillance from the same patient was also positive for presumptive hVISA.
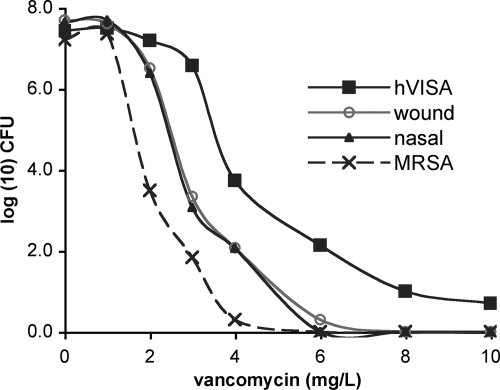
PAP-AUC was performed on these isolates as previously described (18) by serially diluting overnight cultures grown in Trypticase soy broth onto brain heart infusion agar plates containing increasing vancomycin concentrations, incubating for 48 h, and counting subsequent colonies. Although the presumptive hVISA isolates displayed reduced vancomycin sensitivity compared to a vancomycin-sensitive S. aureus (VSSA; ATCC 43300) control (Fig. 1), they did not meet the criteria for hVISA (a PAP-AUC ratio to Mu3 of ≥0.9), with PAP-AUC ratios of 0.54 and 0.58 for the nasal and wound specimens, respectively. The significance of this phenotype in terms of the clinical response to vancomycin treatment is unknown. Therefore, the confirmed hVISA prevalence as detected by Etest GRD from diverse sites of infection in our population is 0%, with the upper 95% confidence limit being 2.2% using the adjusted Wald method (1).
Fig 1.
Population analysis profile curves of two isolates (wound and nasal) from a single patient that were positive for hVISA by Etest GRD. Neither was defined to be hVISA compared to the susceptibility of the Mu3 reference strain.
To specifically investigate the prevalence of hVISA among MRSA strains colonizing the anterior nares, we obtained 121 additional consecutive MRSA isolates from nasal carriage screening between 25 July and 2 October 2009 and tested the last received isolate for each patient using Etest GRD (n = 92). An additional 50 consecutive MRSA isolates obtained from nasal carriage screening between 6 October and 5 December 2009 were tested directly after initial culture, without storage at −70°C, as vancomycin-resistant phenotypes have been suggested to be unstable with potential loss of resistance during prolonged storage (8, 15). None of these isolates was positive for presumptive hVISA. Thus, the prevalence of hVISA nasal colonization detected by Etest GRD in this population was 0%, with the upper 95% confidence limit being 2.3%. Similar to our results, a screen for hVISA among colonizing strains of MRSA in American patients also found no confirmed hVISA isolates (4). However, in this report, screening was performed by simplified population analysis, which has reduced sensitivity compared with Etest GRD (11, 16), making it more likely that potential hVISA may have been missed.
While vancomycin exposure potentially selects for hVISA (5, 8), we found no hVISA isolates, even though a high percentage of patients had prior exposure to vancomycin. We did not screen the same patients over time, so we cannot exclude intermittent colonization with hVISA. This would require prospective studies of a cohort of patients with multiple isolates over time.
In this low prevalence population, screening for hVISA by Etest GRD gave false-positive results that have the potential to cause an unnecessary switch from vancomycin to an alternative agent. Therefore, these data do not support routine screening for hVISA by Etest GRD in low prevalence populations without a confirmatory method. Additional studies are required to determine the clinical utility of routine screening for hVISA in populations with higher point prevalence rates.
ACKNOWLEDGMENTS
We are grateful to Wilson Vientos and the staff of the West Haven VA Connecticut Healthcare System microbiology laboratory for processing the MRSA isolates, to James Drozd, Brian Kotansky, Valerie Smalley, Mary Jane Rubino, Antoinette Towle, and Barbara Welch for tracking infection and antibiotic trends in our hospital, and to Mark Brady for statistical advice.
Footnotes
Published ahead of print 7 November 2011
REFERENCES
- 1. Agresti A, Coull BA. 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52:119–126 [Google Scholar]
- 2. Ariza J, et al. 1999. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587–1588 [DOI] [PubMed] [Google Scholar]
- 3. Charles PGP, Ward PB, Johnson PDR, Howden BP, Grayson ML. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448–451 [DOI] [PubMed] [Google Scholar]
- 4. Eguia JM, et al. 2005. Low colonization prevalence of Staphylococcus aureus with reduced vancomycin susceptibility among patients undergoing hemodialysis in the San Francisco Bay area. Clin. Infect. Dis. 40:1617–1624 [DOI] [PubMed] [Google Scholar]
- 5. Fridkin SK, et al. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429–439 [DOI] [PubMed] [Google Scholar]
- 6. Horne KC, et al. 2009. Prospective comparison of the clinical impacts of heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-susceptible MRSA. Antimicrob. Agents Chemother. 53:3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howden BP, Ward PB, Johnson PDR, Charles PGP, Grayson ML. 2005. Low-level vancomycin resistance in Staphylococcus aureus—an Australian perspective. Eur. J. Clin. Microbiol. Infect. Dis. 24:100–108 [DOI] [PubMed] [Google Scholar]
- 8. Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howden BP, et al. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521–528 [DOI] [PubMed] [Google Scholar]
- 10. Leonard SN, Rossi KL, Newton KL, Rybak MJ. 2009. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63:489–492 [DOI] [PubMed] [Google Scholar]
- 11. Liu C, Chambers HF. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maor Y, et al. 2009. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J. Infect. Dis. 199:619–624 [DOI] [PubMed] [Google Scholar]
- 13. van Hal SJ, Paterson DL. 2011. Systematic review and meta-analysis of the significance of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Hal SJ, et al. 2011. Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J. Clin. Microbiol. 49:1489–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voss A, et al. 2007. A multi-center blinded study on the efficiency of phenotypic screening methods to detect glycopeptide intermediately susceptible Staphylococcus aureus (GISA) and heterogeneous GISA (h-GISA). Ann. Clin. Microbiol. Antimicrob. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walsh TR, et al. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang G, Hindler JF, Ward KW, Bruckner DA. 2006. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J. Clin. Microbiol. 44:3883–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wootton M, et al. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399–403 [DOI] [PubMed] [Google Scholar]
- 19. Yusof A, et al. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J. Clin. Microbiol. 46:3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]