Abstract
One of the most effective and widely used antituberculosis (anti-TB) drugs is isoniazid (INH), a prodrug activated via oxidation that forms an adduct with NAD+ to inhibit NADH-dependent targets of Mycobacterium tuberculosis, such as enoyl-acyl carrier protein reductase (InhA). The metabolic by-products and potentially toxic intermediates resulting from INH therapy have been identified through a large body of work. However, an INH-NAD adduct or structures related to this adduct have not been identified in specimens from human TB patients or animal models of TB. Analyses by mass spectrometry of urine collected from TB patients in a study conducted by the NIAID-funded Tuberculosis Research Unit identified 4-isonicotinoylnicotinamide (C12H9N3O2) as a novel metabolite of INH therapy. This compound was formed by M. tuberculosis strains in a KatG-dependent manner but could also be produced by mice treated with INH independent of an M. tuberculosis infection. Thus, the 4-isonicotinoylnicotinamide observed in human urine samples is likely derived from the degradation of oxidized INH-NAD adducts and provides direct evidence of host INH activation.
INTRODUCTION
Infection of humans with Mycobacterium tuberculosis resulted in an estimated 9.4 million new cases of tuberculosis (TB) worldwide in 2009 (37). The treatment of TB is based on a four-drug regimen that includes isoniazid (INH) as a key component. Further, INH is administered as a prophylactic monotherapy to prevent disease in individuals with an asymptomatic or latent M. tuberculosis infection (32). The wide use of this drug and its potentially severe side effects, particularly hepatotoxicity, have driven efforts to understand its mode of action, metabolism, and pharmacodynamic properties (27, 35). Orally administered INH is absorbed rapidly through the gastrointestinal tract and is metabolized (Fig. 1) in several organs, predominantly the liver, kidney, and brain (11). N-Acetylation of INH by a hepatic N-actyl transferase is the major route of INH metabolism, and the rate of this reaction is genetically determined (27). Some of the N-acetyl isoniazid is subsequently hydrolyzed to isonicotinic acid and acetyl hydrazine (11, 27). Alternatively, INH can be directly hydrolyzed to isonicotinic acid. Additional metabolism results in conjugation of the isonicotinic acid with glycine, and the acetyl hydrazine is further acetylated or deacetylated (11, 27). Besides these major metabolites, various other metabolites of INH, such as oxoacid hydrozones and isonicotinyl glucoronate, are produced (27) (Fig. 1).
Fig 1.
INH metabolism in mammals and mycobacteria. In mammals (left of the dashed line), N-acetyl isoniazid is the most abundant INH metabolite and undergoes hydrolysis, producing isonicotinic acid and acetylhydrazine. Isonicotinic acid and oxoacid hydrazones are also formed directly from INH. Isonicotinic acid conjugates to other molecules, such as glycine. Acetylhydrazine undergoes rapid acetylation to produce diacetylhydrazine or cleavage, yielding ammonia. The boxed metabolites are the major products found in mammals. In M. tuberculosis (right of the dashed line), INH undergoes oxidative activation by KatG, and the resulting isonicotinyl radical forms adducts with NAD+.
Although INH has been used to treat TB since 1952, its mode of action was not well understood until the 1990s (35). INH is a prodrug that undergoes oxidative activation by the catalase peroxidase (KatG) of M. tuberculosis to form an isonicotinoyl radical that reacts nonenzymatically with pyridine nucleotide coenzymes, such as NAD, to form adducts (Fig. 1) (17, 20). The INH-NAD adducts bind to and inhibit enoyl-acyl carrier protein (ACP) reductase (InhA) (2). Two other enzymes, ACP-ketoacyl synthase (KasA) and dihydrofolate reductase, are also reported as possible targets of INH (1, 23). The formation of the INH-NAD adducts is well documented using cell-free assays, and the elucidation of the molecular interactions and kinetics of InhA adduct generation have allowed remarkable insight into the structure-activity relationship of the drug (28, 30). More recently, Wang and colleagues directly demonstrated binding of INH-NAD to recombinant InhA isolated from an Escherichia coli strain coexpressing inhA and katG of M. tuberculosis and exposed to INH (36). In spite of this wealth of in vitro data, no direct evidence exists for in vivo INH-NAD adduct formation in human patients with TB or animal models treated with INH. Likewise, although mammalian peroxidases have been shown to activate INH in cell-free assays (16, 31), demonstration of in vivo INH activation via mammalian enzymes has remained elusive.
The present study was performed based on the rationale that detailed metabolomic analyses of urine, a complex biological specimen, could detect host and pathogen responses to therapeutic intervention for TB, including the formation of drug metabolites. Our analysis of small molecules in the urine of pulmonary TB patients receiving INH therapy has allowed the discovery of a new metabolite, 4-isonicotinoylnicotinamide (4-INN), that results from the formation of an INH-NAD adduct. We further demonstrate not only that this metabolite is generated by M. tuberculosis strains producing a functional KatG protein, but also that it arose in mice treated with INH regardless of whether they were infected with M. tuberculosis. This provides direct evidence that INH is activated by both the host and the pathogen.
MATERIALS AND METHODS
Clinical samples.
Urine samples were obtained from the Tuberculosis Research Unit (TBRU) Specimen Repository. The urine specimens were samples collected in the context of an observational study conducted by the TBRU (18). The TB patients in that study were HIV-infected and uninfected adult Ugandan men and women with newly diagnosed initial episodes of culture-confirmed pulmonary tuberculosis. M. tuberculosis strains isolated from all patients prior to treatment underwent susceptibility testing versus INH, rifampin (RIF), ethambutol (EMB), pyrazinamide (PZA), and streptomycin using standard methods in the Bactec 460 TB system. The critical concentration of INH used in susceptibility testing was 0.1 μg/ml. Isolates from all eight patients evaluated in this study were sensitive to INH. Urine was collected at the initial time of TB diagnosis (before the start of therapy [day 0]) and at 2 weeks (week 2), 8 weeks (week 8), and 6 months (month 6) after the start of standard anti-TB therapy. The specimens were stored unprocessed at −80°C upon collection and were sterilized by gamma irradiation before analysis.
Animal experiments.
Female 6- to 8-week-old specific-pathogen-free immunocompetent C57BL/6 mice (Charles River, Wilmington, MA) were infected via low-dose aerosol exposure to M. tuberculosis Erdman (21). Three mice were euthanized 1 day after low-dose aerosol exposure to verify bacterial uptake of approximately 100 CFU per mouse. Administration of INH (Sigma-Aldrich, St. Louis, MO) and RIF (Sigma-Aldrich) at 100 mg/liter in the drinking water (an approximate dose of 25 mg/kg of body weight) was initiated at 3 weeks postinfection and continued for 12 weeks. Urine samples from all the animals were collected before and at different time points during the treatment.
In a second study, a group of five uninfected mice were treated via intraperitoneal (i.p.) injections with INH at a dose of 25 mg/kg for 5 days per week for 1 month. Another group of five uninfected mice were treated with moxifloxacin (extracted from tablets provided through Tuberculosis Antimicrobial Acquisition and Coordinating Facility NIH contract NO1 AI-95385) (100 mg/kg) for 1 week, followed by INH (25 mg/kg) by gavage for three more weeks. Urine samples from all the animals were collected before the start of drug administration (day 0) and at different time points during the treatment. The urine specimens from M. tuberculosis-infected mice were sterilized by gamma irradiation before analysis.
Bacterial strains and extraction of metabolites.
M. tuberculosis strain H37Rv (ATCC 25618) was obtained through the American Type Culture Collection, Manassas, VA. M. tuberculosis CDC1551 and the M. tuberculosis CDC1551 katG transposon mutant MT1959 (point of insertion 628) (19) were received from the Tuberculosis Vaccine Testing and Research Materials Contract (HHSN266200400091C) at Colorado State University. M. tuberculosis strains were grown in Bacto Middlebrook-7H9 broth (Difco, Detroit, MI) containing 0.2% glycerol and 10% Bacto OADC (oleic acid-albumin-dextrose-catalase) enrichment (Difco). INH was added to exponentially growing M. tuberculosis cells at a final concentration of 0.5 μg/ml. The cultures were allowed to incubate for an additional 24 or 48 h with shaking at 37°C and were harvested by centrifugation at 5,000 × g for 10 min at 4°C. The resulting cell pellets were suspended in 6 ml of chloroform-methanol (2:1) and incubated at room temperature for 6 h with constant agitation, and the solvents were evaporated under a stream of N2. The dried cells were suspended in 100 mM ammonium bicarbonate, pH 7.0, and physically disrupted by vigorous mixing with 0.1-mm zirconium beads. The lysate was centrifuged at 18,000 × g for 20 min, and the supernatant was collected. Macromolecular products were removed by passing the supernatant through a Millipore UltraFree centrifugal ultrafiltration device (Millipore, Billerica, MA) with a 5,000-Da-cutoff membrane. The final filtrate was acidified with formic acid to a final concentration of 0.1% and centrifuged at 18,000 × g for 20 min, and an aliquot was subjected to liquid chromatography-mass spectrometry (LC-MS) analysis.
Liquid chromatography-mass spectrometry analysis of urine samples.
The creatinine concentrations of urine specimens were determined by the alkaline picrate method using a creatinine assay kit from Oxford Biomedical Research (Oxford, MI). Urine samples were clarified by centrifugation and subjected to LC-MS. An aliquot of urine containing 13 μg of creatinine was applied to an X-Bridge or Atlantis T3 reversed-phase C18 3.5-μm column (2.1 by 150 mm; Waters Corp., Milford, MA) that was connected to an Agilent 1200 series high-performance liquid chromatography (HPLC) system (Agilent Technologies, Palo Alto, CA). The metabolites were eluted with a 0 to 90% nonlinear gradient of methanol in 0.1% formic acid at a flow rate of 250 μl/min. The eluent was introduced directly into an Agilent 6220 Accurate-Mass TOF (time of flight) or Agilent 6250 quadrapole time of flight (Q-TOF) mass spectrometer equipped with an Agilent multimode source that was operated in simultaneous electrospray ionization and atmospheric pressure chemical ionization modes. The parameters for the mass spectrometers were as follows: gas temperature, 300°C; vaporizer temperature, 200°C; drying gas at 8 liters/min; nebulizer at 45 lb/in2; charging voltage, 2,000 V; capillary voltage, 2,000 V; corona, 2 μA; fragmentation energy, 120 V; skimmer, 60 V; and octapole RF setting, 250 V (TOF) or 750 V (Q-TOF). The positive-ion MS data for the mass range of 104 to 1,500 Da were acquired at a rate of 1.02 spectra/s and 9,700 transients/spectrum. Data were collected in both centroid and profile modes in 4-GHz high-resolution mode. Positive-ion reference masses of 121.050873 m/z and 922.009798 m/z were introduced to ensure mass accuracy. The urine samples collected at day 0, week 2, and week 8 from 15 patients were analyzed. Some of the week 2 urine samples were subjected to LC-tandem MS (MS/MS) analysis to aid structural analysis of the compounds. MS data were processed with the molecular feature extractor (MFE) algorithm in Agilent MassHunter Qualitative Analysis software to identify molecular features (compounds with a defined exact mass and retention time) present in each sample, with a minimum abundance of at least 400 counts. Data from different sample groups (i.e., treatment time points) were compared using Agilent Mass Profiler Professional software. The relative abundances of the filtered molecular features were compared across different treatment time points, and features that increased significantly (at least 10-fold) in relative abundance in the week 2 and week 8 treatment groups compared to the day 0 group were further analyzed to identify the metabolite structure.
Urine specimens collected from mice were also analyzed by LC-MS as described above. The samples collected at day 0, day 2, day 5, and day 7 from infected and uninfected mice receiving INH treatment through drinking water were analyzed by LC-MS. The day 0, day 2, day 5, and day 23 samples from uninfected mice receiving INH through i.p. injection and the day 0, day 2, and day 23 samples from moxifloxacin-treated mice that received INH through gavage were analyzed by LC-MS. The resulting data were analyzed by the Agilent MassHunter Qualitative Analysis software to determine the presence and relative abundances of INH metabolites.
Synthesis of INH-NAD adducts.
Manganese(III) pyrophosphate was synthesized with equal volumes of 50 mM Mn(III) acetate dihydrate (Sigma-Aldrich) and 250 mM Na-pyrophosphate at room temperature as described by Trouwborst et al. (34). INH activation and INH-NAD+ adduct formation were performed in the presence of manganese(III) pyrophosphate as described by Nguyen et al. (26). The reaction mixture was allowed to stand at room temperature for 1 h and clarified by centrifugation. The supernatant was diluted 50 times with HPLC grade water, and a 10-μl aliquot was analyzed by LC-MS. Similar reactions where NAD+ (Sigma-Aldrich) was replaced with 2 mM nicotinamide (Nam) (Sigma-Aldrich) were also performed.
RESULTS
Analysis of human urine samples.
LC-MS data from analysis of TB patients' urine samples prior to the initiation of a standard four-drug antituberculosis regimen (day 0) and at week 2 and week 8 after the start of treatment were analyzed using Agilent Mass Profiler Professional software. This analysis generated a list of metabolites that were unique to the samples collected after the initiation of treatment (data not shown). An accurate mass database of all known metabolites of INH, RIF, PZA, and EMB was created. The list of molecular features that increased significantly in relative abundance in the treatment group compared to the day 0 group was searched against this database. Most of the known urinary metabolites of INH, RIF, PZA, and EMB were identified and were unique to the samples in the treatment group. Additionally, a product with an [M+H]+ molecular ion of 228.0768 m/z was identified by the Mass Profiler Professional software as unique to the samples obtained after the start of therapy (Fig. 2A). This molecular ion generally had lower relative abundance than other drug metabolites, such as N-acetyl isoniazid (180.0768 m/z) and isonicotinyl glycine (181.0608 m/z), and the abundance of the product defined by 228.0768 m/z differed between time points and patients, as was observed for the other INH metabolites. A full query of the MS data files for the 228.0768 m/z ion in all patient samples confirmed the presence of this product only after the start of antituberculosis therapy, just as was observed for the 180.0768 m/z ion of N-acetyl isoniazid (Fig. 2B). Due to the strong correlation with the onset of drug treatment and the appearance of this molecular feature in urine, the 228.0768 m/z ion product was speculated to be an unidentified drug metabolite.
Fig 2.
Relative abundances of INH metabolites in the urine of patients undergoing anti-TB therapy. (A) N-Acetyl isoniazid, isonicotinyl glycine, and a product with an [M+H]+ molecular ion of 228.0768 m/z were detected by LC-MS in the urine of eight patients at 2 weeks (W2) and 8 weeks (W8) after the start of therapy. (B) The product with an [M+H]+ molecular ion of 228.0768 m/z was not present in patients' urine prior to the start of anti-TB therapy (day 0) but could be detected at significantly above background level after 2 and 6 months of therapy. Each closed circle represents one patient. All eight patients were infected with INH-sensitive organisms, and sputum cultures from these patients after week 8 of therapy were negative for M. tuberculosis.
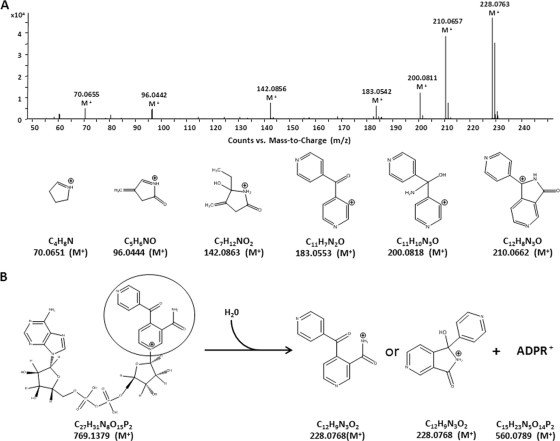
The calculated molecular formula of the 228.0768 m/z ion product was C12H9N3O2, and an interrogation of the molecular formula C12H9N3O2 against potential metabolites of INH, RIF, PZA, or EMB led to the hypothesis that the 228.0768 m/z ion was the result of an INH-nicotinamide adduct, 4-INN. MS/MS fragmentation of the 228.0768 m/z molecular ion resulted in six dominant daughter ions of 210.0657 m/z, 200.0811 m/z, 183.0542 m/z, 142.0856 m/z, 96.0442 m/z, and 70.655 m/z that was consistent with the structure of 4-INN and indicated a mixture of the open and cyclized forms of the molecule (Fig. 3). Although an INH-NAD+ adduct was not observed in the urine of TB patients receiving INH, the 4-INN structure was proposed to result from the hydrolysis of an INH-NAD+ adduct (Fig. 3B) (33).
Fig 3.
Identification of the 228.0768 m/z product. (A) The MS/MS fragmentation pattern of the [M+H]+ molecular ion 228.0768 m/z from urine is consistent with potential products that would arise from fragmentation of an INH-nicotinamide adduct (4-INN). The masses noted for the structures of the fragmentation products are the predicted m/z of the positive ion. All of the observed fragmentation masses were within 6 ppm of the predicted masses. (B) The hypothesized hydrolysis of an INH-NAD+ adduct would lead to an INH-nicotinamide adduct in an open (4-INN) or cyclic {1-hydroxy-1-(pyridin-4-yl)-1H-pyrrolo[3,4-c]pyridin-3(2H)-one} configuration.
Analysis of the origin of 4-INN.
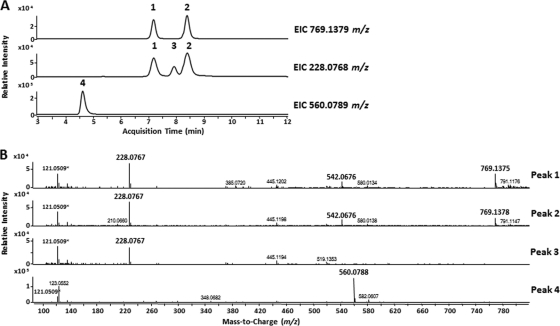
4-INN can be formed by direct conjugation of activated INH and Nam or via the degradation of an oxidized INH-NAD+ adduct. To demonstrate that the 4-INN structure arose from an INH-NAD+ adduct, INH was activated by chemical oxidation with Mn(III) pyrophosphate (26) in the presence of NAD or Nam to synthesize the corresponding adducts and analyzed by LC-MS. Activation of INH in the presence of NAD produced a series of INH-NAD adducts, along with other reaction by-products previously described that were detectable by LC-MS (Fig. 4) (25). The oxidized INH-NAD adduct (INH-NAD+) with a predicted M+ molecular ion of 769.1379 m/z was detected in two separate LC peaks (peaks 1 and 2 in Fig. 4A). The most likely reason for two separate INH-NAD+ LC peaks is the formation of cyclized diastereoisomers in solution (26). The MS spectra of these two peaks also contained an M+ ion of 542.0676 m/z and an [M+H]+ ion of 228.0767 m/z corresponding to the nucleotide and 4-INN fragments of the INH-NAD+ adduct, respectively (Fig. 4B). Finding these two molecular ions in the same chromatographic peak as the 769.1379 m/z ion suggested fragmentation of the INH-NAD+ adduct during ionization. A separate extracted ion chromatogram generated to specifically search for the predicted 228.0768 m/z 4-INN ion resulting from hydrolysis revealed a single LC peak (peak 3) that resolved between the LC peaks (1 and 2) of the INH-NAD+ adduct (Fig. 4A). The MS spectrum of this additional peak demonstrated 228.0767 m/z (4-INN) as the dominant product and no detectable ions corresponding to the INH-NAD+ adducts or the nucleotide fragment. MS/MS analysis confirmed that the 228.0767 m/z ions from the synthetic reaction and the urine samples had identical fragmentation patterns (data not shown). Thus, the presence of a single LC peak (peak 3) containing only 4-INN demonstrated that this product could be generated by nonenzymatic hydrolysis of the INH-NAD+ adduct. Further evidence of nonenzymatic hydrolysis was the presence of LC peak 4 possessing the [M+H]+ molecular ion 560.0788 m/z that corresponded to the adenosine diphosphate ribose (ADPR) (peak 4) (Fig. 4A and B).
Fig 4.
Analysis of the INH-NAD+ adducts after Mn(III)-mediated activation of INH. (A) Extracted ion chromatograms (EIC) for INH-NAD+ adduct (769.1379 m/z) and the 4-INN (228.0768 m/z) and nucleotide fragment of NAD+ (560.0789 m/z) that would be formed via hydrolysis of an INH-NAD+ adduct. (B) MS spectra of the four peaks detected in the extracted ion chromatograms. The molecular ion 121.0509 m/z present in all spectra is the reference ion and is marked with an asterisk. The boldface ions are those assigned to the INH-NAD+ adduct or products from the hydrolysis of the INH-NAD+ adduct.
Analysis of a urine sample from a TB patient at week 2 of antituberculosis therapy that was spiked with synthetic INH-NAD+ and compared to a nonspiked sample revealed that the molecular ion corresponding to the intact INH-NAD+ adduct was present only in the spiked urine sample but that the 4-INN product was present in both (Fig. 5). This suggested that an INH-NAD+ adduct formed in vivo during INH therapy was completely hydrolyzed in the urine. It is also possible, however, that the 4-INN metabolite in urine resulted from direct conjugation of activated INH to Nam. To assess this, INH was activated by chemical oxidation in the presence of Nam. LC-MS analyses of this reaction revealed the presence of 4-INN. However, the yield of 4-INN by direct conjugation to Nam was at least 10-fold lower than that obtained from hydrolysis of the synthetic INH-NAD+ adduct (data not shown).
Fig 5.
Extracted ion chromatograms of LC-MS analyses of unaltered urine from a single TB patient at 2 weeks and the same urine sample spiked with the INH-NAD+ adduct formed in vitro by Mn(III) activation of INH. No INH-NAD+ adducts were detected from the unaltered urine sample, and the 4-INN product in unaltered urine coelutes with that from the hydrolysis of the INH-NAD+ adduct in the spiked sample. Arrows indicate the elution peaks.
Production of 4-INN as a result of bacterial metabolism versus host metabolism.
The activation of INH by M. tuberculosis occurs via oxidation by the katG gene product (17), and this could lead to the generation of the 4-INN metabolite in cultures of M. tuberculosis. Treatment of M. tuberculosis strains H37Rv and CDC1551 and a katG transposon mutant of CDC1551 with 0.5 μg/ml INH and analysis of the metabolites yielded 4-INN at low levels in the wild-type strains but not in the katG mutant (Fig. 6). The molecular ion for the intact INH-NAD+ adduct was also detectable in M. tuberculosis KatG competent strains. The relative abundance of 4-INN, however, was higher than that of the INH-NAD+ adduct. It should be noted that no molecular ion corresponding to the calculated accurate mass of a reduced or oxidized INH-NADP adduct was detected in M. tuberculosis. These data demonstrated that M. tuberculosis can generate the 4-INN metabolite, but only in the presence of KatG activation of INH.
Fig 6.
Extracted ion chromatograms of the LC-MS analysis of metabolites resulting from in vitro exposure of wild-type M. tuberculosis strain CDC1551 (top two chromatograms) and the katG mutant of M. tuberculosis strain CDC1551 (bottom two chromatograms) to INH. Arrows indicate the elution peaks.
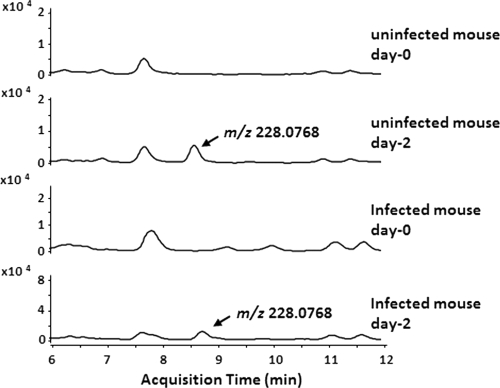
In our analysis of human urine samples, the relative abundances of 4-INN in patients at week 2 and week 8 of antituberculosis therapy were comparable, even though sputum cultures of these patients after week 8 of therapy were negative. This observation, along with previous data indicating that mammalian peroxidases may also participate in the activation of INH (16, 31), led us to hypothesize that the 4-INN metabolite, and presumably activation of INH and its conjugation to NAD+, could occur in the absence of an active M. tuberculosis infection. Urine samples from five M. tuberculosis-infected and five uninfected mice receiving INH were tested for the presence of 4-INN by LC-MS. Whether M. tuberculosis infection was present or not, all mice had 4-INN in their urine following INH administration (Fig. 7). There was also no obvious difference in the relative abundances of 4-INN in samples from M. tuberculosis-infected and uninfected mice (data not shown). The abundance of 4-INN in mouse urine varied between 5.8 × 106 and 2.2 × 106 (peak area). It was further noted that, as with the human samples, the INH-NAD+ adduct was not observed in the urine of mice. The route of INH administration (via drinking water, gavage, or i.p. injection) had no effect on the presence of 4-INN, indicating that metabolism in the gut was not responsible for the generation of 4-INN. Pretreatment of mice with the broad-spectrum fluoroquinolone moxifloxacin for 7 days to reduce normal gut flora that can activate INH did not affect 4-INN formation. These data demonstrate that 4-INN can be a mammalian metabolite of INH treatment and that host mechanisms will conjugate INH with NAD+ in the absence of M. tuberculosis infection.
Fig 7.
Formation of 4-INN in an uninfected mouse treated with INH. Urine from two mice was collected before the start of INH treatment (day 0) and after 2 days of INH treatment (day 2) and analyzed by LC-MS. Extracted ion chromatograms of 228.0768 m/z were used to assess the formation of 4-INN from INH treatment in the absence of M. tuberculosis infection.
DISCUSSION
This study demonstrates the utility of metabolomics to dissect serial changes in complex human biological samples and to gain insight into biological systems that can alter the activity of anti-TB drugs. INH is a key first-line anti-TB drug, and this study provides new knowledge and raises important questions about the host and pathogen metabolism of INH that could impact its activity and toxicity. The mechanism by which INH is activated and inhibits its primary targets has been well described, along with the formation of an INH-NAD+ adduct required for the inhibition of M. tuberculosis (35). However, such a structure has not been detected in humans being treated with INH. The elimination of INH and its metabolites mainly occurs via the urine (11, 12), and our analysis of urine from human TB patients allowed the identification of a novel metabolite (4-INN) that, based on in vitro experiments, presumably resulted from INH-NAD+ adduct formation. The data demonstrated a requirement for INH activation to allow the formation of 4-INN. However, adduct formation subsequent to INH activation could occur with NAD+, NADP+, or Nam. Our conclusion that 4-INN resulted from the hydrolysis of an INH-NAD+ adduct is based on the observation that 4-INN formation in the presence of NAD+ was at least 60-fold more efficient than the direct coupling of activated INH and Nam. The increased efficiency of INH-NAD+ adduct formation must also be considered, with the understanding that the intracellular concentration of Nam is significantly lower than that of NAD+. Likewise, the relative abundance of NAD in cells is generally higher than that of NADP (22, 29), and no evidence of in vivo INH-NADP(+) adduct formation was obtained from our experiments. This collective information supports the preferred formation of INH-NAD+ as a precursor of 4-INN.
The data obtained also provided direct evidence that INH-NAD+ adduct formation by M. tuberculosis cells was dependent on a functional KatG. However, we also observed that 4-INN was present in human urine after 2 months of anti-TB treatment when patients were culture negative for M. tuberculosis and in the urine of uninfected mice after INH administration. Mammalian peroxidases, such as lactoperoxidase, were previously proposed to be involved in the activation of INH in vivo (16, 31), and this explains the formation of 4-INN in the absence of an infection. Although host peroxidases activate INH, the use of INH is poorly effective in vivo against katG mutants of M. tuberculosis. This paradox can be explained by the potential reactivity of activated INH to other host products and raises the premise that activation of INH must occur within the envelope of M. tuberculosis or in close proximity to the pathogen for bactericidal activity.
We observed that hydrolysis of the INH-NAD+ adduct occurred spontaneously under the in vitro INH activation conditions used. However, enzymatic hydrolysis of the INH-NAD+ adduct in vivo is also possible. Several NAD-degrading enzymes are present in mammals and produce Nam from NAD. The NAD glycohydrolase enzyme is abundant in lung tissue and is a potential candidate (10). Other enzymes, such as ADP ribose transferases and protein lysine deacetylases, like sirtuins (Sir2), can also generate Nam from NAD (3, 13). Similar enzymatic activities in M. tuberculosis (14, 15) could also lead to degradation of the INH-NAD+ adduct.
The pharmacological and toxicological implications of the INH-NAD+ adduct and the pathway leading to the 4-INN structure open the path for future studies. Truncated INH-NAD+ adducts with some structural similarity to 4-INN reportedly have antimycobacterial activity (9). Thus, the potential antibacterial activity of the 4-INN product should be investigated. Pellagra is an infrequent side effect of long-term INH therapy that is believed to be due to competitive inhibition of NAD and NADP by INH (6, 8, 24). Based on the data presented, we can postulate that significant amounts of nicotinic acid can be lost through the excretion of 4-INN, resulting in nicotinic acid deficiency and the development of pellagra. Thus, 4-INN formation could be a mechanism for INH-induced pellagra. In fact, individuals who develop INH-associated pellagra despite supplementation with pyridoxine respond to Nam, suggesting niacin deficiency in these individuals (4, 5, 7, 8). Information about INH metabolism can be useful in developing strategies to minimize and manage its toxicity.
ACKNOWLEDGMENTS
This project has been funded by the TBRU, established with federal funds from the National Institute of Allergy and Infectious Diseases and the National Institutes of Health under contract no. HHSN266200700022C/NO1-AI-70022.
Footnotes
This research was part of the Tuberculosis Research Unit (Case Western Reserve University) Surrogate Microbial Marker Clinical Study (TBRU-03).
REFERENCES
- 1. Argyrou A, Vetting MW, Aladegbami B, Blanchard JS. 2006. Mycobacterium tuberculosis dihydrofolate reductase is a target for isoniazid. Nat. Struct. Mol. Biol. 13:408–413 [DOI] [PubMed] [Google Scholar]
- 2. Banerjee A, et al. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230 [DOI] [PubMed] [Google Scholar]
- 3. Belenky P, Bogan KL, Brenner C. 2007. NAD+ metabolism in health and disease. Trends Biochem. Sci. 32:12–19 [DOI] [PubMed] [Google Scholar]
- 4. Bender DA, Russell-Jones R. 1979. Isoniazid-induced pellagra despite vitamin-B6 supplementation. Lancet ii:1125–1126 [DOI] [PubMed] [Google Scholar]
- 5. Burke GJ, Hlangabeza T. 1977. Isoniazid-induced pellagra in a patient on vitamin B supplement. S. Afr. Med. J. 51:719. [PubMed] [Google Scholar]
- 6. Comaish JS, Cooper M. 1977. Isoniazid-induced pellagra. Arch. Dermatol. 113:986–987 [PubMed] [Google Scholar]
- 7. Comaish JS, Felix RH, McGrath H. 1976. Topically applied niacinamide in isoniazid-induced pellagra. Arch. Dermatol. 112:70–72 [PubMed] [Google Scholar]
- 8. Darvay A, Basarab T, McGregor JM, Russell-Jones R. 1999. Isoniazid induced pellagra despite pyridoxine supplementation. Clin. Exp. Dermatol. 24:167–169 [DOI] [PubMed] [Google Scholar]
- 9. Delaine T, et al. 2010. Development of isoniazid-NAD truncated adducts embedding a lipophilic fragment as potential bi-substrate InhA inhibitors and antimycobacterial agents. Eur. J. Med. Chem. 45:4554–4561 [DOI] [PubMed] [Google Scholar]
- 10. DiAugustine RP. 1976. Formation in vitro and in vivo of the isonicotinic acid hydrazide analogue of nicotinamide adenine dinucleotide by lung nicotinamide adenine dinucleotide glycohydrolase. Mol. Pharmacol. 12:291–298 [PubMed] [Google Scholar]
- 11. Ellard GA, Gammon PT. 1976. Pharmacokinetics of isoniazid metabolism in man. J. Pharmacokinet. Biopharm. 4:83–113 [DOI] [PubMed] [Google Scholar]
- 12. Ellard GA, Gammon PT, Wallace SM. 1972. The determination of isoniazid and its metabolites acetylisoniazid, monoacetylhydrazine, diacetylhydrazine, isonicotinic acid and isonicotinylglycine in serum and urine. Biochem. J. 126:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, Brenner C. 2009. Microbial NAD metabolism: lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73:529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gopinathan KP, Sirsi M, Vaidyanathan CS. 1964. Nicotinamide-adenine dinucleotide glycohydrolase of Mycobacterium tuberculosis H37Rv. Biochem. J. 91:277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gu J, et al. 2009. Cloning and characterization of NAD-dependent protein deacetylase (Rv1151c) from Mycobacterium tuberculosis. Biochemistry 74:743–748 [DOI] [PubMed] [Google Scholar]
- 16. Hillar A, Loewen PC. 1995. Comparison of isoniazid oxidation catalyzed by bacterial catalase-peroxidases and horseradish peroxidase. Arch. Biochem. Biophys. 323:438–446 [DOI] [PubMed] [Google Scholar]
- 17. Johnsson K, Schultz PG. 1994. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 116:7425–7426 [Google Scholar]
- 18. Joloba ML, et al. 2000. Quantitative sputum bacillary load during rifampin-containing short course chemotherapy in human immunodeficiency virus-infected and non-infected adults with pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 4:528–536 [PubMed] [Google Scholar]
- 19. Lamichhane G, et al. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100:7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lei BF, Wei CJ, Tu SC. 2000. Action mechanism of antitubercular isoniazid—activation by Mycobacterium tuberculosis KatG, isolation, and characterization of InhA inhibitor. J. Biol. Chem. 275:2520–2526 [DOI] [PubMed] [Google Scholar]
- 21. Lenaerts AJ, et al. 2005. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob. Agents Chemother. 49:2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McLean P, Greenbaum AL, Brown J, Greenslade KR. 1967. Influence of hormones on the nicotinamide nucleotide coenzymes of adipose tissue. Biochem. J. 105:1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mdluli K, et al. 1996. Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J. Infect. Dis. 174:1085–1090 [DOI] [PubMed] [Google Scholar]
- 24. Meyrick Thomas RH, Rowland Payne CM, Black MM. 1981. Isoniazid-induced pellagra. Br. Med. J. 283:287–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen M, Claparols C, Bernadou J, Meunier B. 2001. A fast and efficient metal-mediated oxidation of isoniazid and identification of isoniazid-NAD(H) adducts. Chembiochem 2:877–883 [DOI] [PubMed] [Google Scholar]
- 26. Nguyen M, Quemard A, Broussy S, Bernadou J, Meunier B. 2002. Mn(III) pyrophosphate as an efficient tool for studying the mode of action of isoniazid on the InhA protein of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Preziosi P. 2007. Isoniazid: metabolic aspects and toxicological correlates. Curr. Drug Metab. 8:839–851 [DOI] [PubMed] [Google Scholar]
- 28. Rawat R, Whitty A, Tonge PJ. 2003. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 100:13881–13886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reiss PD, Zuurendonk PF, Veech RL. 1984. Measurement of tissue purine, pyrimidine, and other nucleotides by radial compression high-performance liquid chromatography. Anal. Biochem. 140:162–171 [DOI] [PubMed] [Google Scholar]
- 30. Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Jr., Sacchettini JC. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98–102 [DOI] [PubMed] [Google Scholar]
- 31. Singh AK, et al. 2010. Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution. J. Biol. Chem. 285:1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snider DE, Jr., Farer LS. 1978. Preventive therapy with isoniazid for “inactive” tuberculosis. Chest 73:4–6 [DOI] [PubMed] [Google Scholar]
- 33. Stigliani JL, et al. 2008. Binding of the tautomeric forms of isoniazid-NAD adducts to the active site of the Mycobacterium tuberculosis enoyl-ACP reductase (InhA): a theoretical approach. J. Mol. Graph. Model. 27:536–545 [DOI] [PubMed] [Google Scholar]
- 34. Trouwborst RE, Clement BG, Tebo BM, Glazer BT, Luther GW., III 2006. Soluble Mn(III) in suboxic zones. Science 313:1955–1957 [DOI] [PubMed] [Google Scholar]
- 35. Vilcheze C, Jacobs WR., Jr 2007. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu. Rev. Microbiol. 61:35–50 [DOI] [PubMed] [Google Scholar]
- 36. Wang F, et al. 2010. Mycobacterium tuberculosis dihydrofolate reductase is not a target relevant to the antitubercular activity of isoniazid. Antimicrob. Agents Chemother. 54:3776–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. World Health Organization 2010. Global tuberculosis report 2010 (WHO/HTM/TB/2010.7), p 1–2 WHO, Geneva, Switzerland [Google Scholar]