Abstract
The in vitro resistance profile of BI 201335 was evaluated through selection and characterization of variants in genotype 1a (GT 1a) and genotype 1b (GT 1b) replicons. NS3 R155K and D168V were the most frequently observed resistant variants. Phenotypic characterization of the mutants revealed shifts in sensitivity specific to BI 201335 that did not alter susceptibility to alpha interferon. In contrast to macrocyclic and covalent protease inhibitors, changes at V36, T54, F43, and Q80 did not confer resistance to BI 201335.
TEXT
The hepatitis C virus (HCV)-encoded NS3 protease is essential for viral replication and has long been considered an attractive target in drug design efforts (3, 5). NS3 protease inhibitors (PIs) can induce substantial reductions in HCV RNA plasma levels, and several candidates have progressed through clinical development to offer improved treatment options (for a review, see reference 27). Two PIs, boceprevir and telaprevir, were recently approved for use in combination with pegylated interferon (Peg-IFN) and ribavirin (1, 6, 7, 19). The selection of drug-resistant variants is commonly observed in patients experiencing virologic rebound during treatment with PIs (16, 20–22, 24).
BI 201335 is a potent HCV NS3/4A PI (15, 28) currently in phase 3 clinical trials in combination with Peg-IFN and ribavirin as well as phase 2 assessment with other HCV direct acting antivirals in IFN-sparing regimens. BI 201335 exhibited a profound reduction in viral load when administered for 14 days as monotherapy in treatment-naïve patients or for 28 days in combination with Peg-IFN and ribavirin in treatment-experienced patients (16). In these studies, viral breakthrough was observed in most patients on monotherapy, whereas breakthrough was less frequent in patients undergoing combination treatment. Distinct resistant NS3 variants R155K and D168V predominated for genotype 1a and 1b (GT 1a and GT 1b), respectively (8, 16).
This study was designed to evaluate the genotypic and phenotypic profiles of the resistant variants that emerged during in vitro selection in the presence of BI 201335 in the replicon system and to relate these results to clinical observations. Replicons resistant to BI 201335 were selected in GT 1a H77 and GT 1b CON-1 replicon cell lines in the presence of 2 concentrations (100× and 1,000× drug concentration required to reduce HCV RNA or the luciferase reporter levels by 50% [EC50]) of drug for 3 weeks and G-418 as previously described (9). With the lower concentration of BI 201335, resistant variants encoding NS3 changes at residues 155, 156, and 168 were selected with the GT 1b replicon, with D168G as the predominant variant (55%). R155K was the predominant variant (68%) selected with the GT 1a replicon (Table 1) and is consistent with the predominant variant selected in GT 1a HCV-infected patients (16). At the higher concentration of BI 201335, essentially only D168 variants were selected with D168 A and V as the predominant variants in both genotypes.
Table 1.
Frequency of amino acid substitutions identified in the NS3 protein-encoding gene of HCV subgenomic replicons of genotype 1a and 1b selected in vitro in the presence of BI 201335
| Amino acid substitution(s)a | Frequency (%) of amino acid substitutions identified in the NS3 gene of HCV replicons in the presence of the indicated concn of BI 201335 |
|||
|---|---|---|---|---|
| Genotype 1b |
Genotype 1a |
|||
| 0.4 μM (n = 33)b | 4.0 μM (n = 25) | 0.6 μM (n = 58) | 6.0 μM (n = 61) | |
| R155K | 64 | |||
| L106M, R155K/R | 2 | |||
| I48V, R155K | 2 | |||
| R155Q | 6 | |||
| R155W | 3 | |||
| A156V | 12 | |||
| A156V, E176G | 8 | |||
| A156T | 6 | |||
| D168G | 52 | 2 | ||
| D168A | 9 | 28 | 2 | |
| D168H | 3 | |||
| D168I | 2 | 3 | ||
| D168V | 3 | 28 | 10 | 50 |
| D168Y | 3 | 4 | ||
| A150V, D168G | 3 | |||
| T42A, D168A | 2 | |||
| I17V, D168A | 8 | |||
| R24Q, D168A | 4 | |||
| D168A, T177A | 2 | |||
| T38N, D168V | 8 | |||
| P86A, D168V | 7 | |||
| P131S, D168V | 8 | |||
| L106 M, D168V | 5 | 32 | ||
| R109K, D168V | 3 | |||
| T38N, D168I, P194S | 2 | |||
| T72A, Q86R, D168V | 12 | |||
| L106M, R155K/R, D168V | 2 | |||
Amino acid substitutions in boldface type represent those selected in replicon colonies resistant to BI 201335.
The concentrations of BI 201335 represent 100× and 1,000× EC50 values that were used to select GT 1b and GT 1a HCV-resistant replicon cell lines. n is the number of resistant cell colonies analyzed for each condition by NS3-NS4A sequencing.
In order to confirm that the mutations observed in the selected resistant replicons were responsible for the phenotypic changes observed, the single amino acid variants were engineered into the GT 1a and 1b replicon backgrounds by site-directed mutagenesis, and the EC50 was determined as previously described (10, 28). As demonstrated in Table 2, the change in BI 201335 susceptibility of the engineered variants, relative to the wild-type sequence for each subtype, confirmed that the selected resistant mutations confer the reduced susceptibility to BI 201335. Similar shifts in BI 201335 potency were observed with D168V variants in both GT 1a and 1b backgrounds. The R155K mutant had a similar effect on the susceptibility of the GT 1a and 1b replicons with 360- and 350-fold shifts in the EC50 for BI 201335, respectively (Table 2). The in vitro selection of R155K mutants in the GT 1a replicon systems but not in the GT 1b replicon systems is consistent with the differences in the resistance profile between genotype 1 subtypes observed in HCV-infected patients during short-term monotherapy with BI 201335 (16) and other NS3 PIs (22). Most likely, the R155K mutant did not emerge during in vitro selection in GT 1b replicons in this study because the codon would require 2 nucleotide changes, whereas the corresponding codon in GT 1a replicons requires only 1 nucleotide change (12, 18, 24). As further demonstrated in Table 2, analysis of the shift in PI sensitivity by the engineered clones revealed the following. (i) R155 variants resulted in a broad range of cross-resistance to all PIs (12, 14, 23, 30). (ii) Variants at position D168 exhibited cross-resistance to BI 201335 and the macrocyclic class of PIs (10, 12, 13), but not to the covalent inhibitors telaprevir and boceprevir (13, 14, 26). Table 2 includes resistant variants reported for other PIs and shows the following. (i) There was no cross-resistance to BI 201335 for variants at positions V36 and T54, which have been shown to be important in resistance to the covalent inhibitors telaprevir and boceprevir (24, 26, 29). (ii) The natural variants at Q80 (2, 4, 12), which confer resistance to the macrocyclic inhibitor TMC435, remain susceptible to BI 201335. Finally, the potency of mechanistically distinct inhibitors such as alpha interferon (IFN-α) or a HCV polymerase inhibitor, is unaffected by the BI 201335 resistance mutations (Table 2) (10).
Table 2.
In vitro susceptibility of NS3 protease inhibitor-resistant HCV replicon molecular clones to BI 201335 and other HCV inhibitors
| Varianta | Change in susceptibility (EC50 fold change) tob: |
||||||
|---|---|---|---|---|---|---|---|
| BI 201335 | MK-7009 | TMC435 | ITMN-191 | Boceprevir | Telaprevir | IFN-α | |
| WT GT 1a H77 | |||||||
| D168V | 620 ± 180 | 1,000 ± 460 | 780 ± 170 | 86 ± 16 | ND | ND | 0.8 ± 0.2 |
| R155K | 360 ± 90 | 260 ± 22 | 43 ± 8 | 140 ± 41 | 4.2 ± 0.7 | 2.9 ± 1.3 | 0.8 ± 0.2 |
| WT GT 1b CON-1 | |||||||
| A156V | 150 ± 10 | ND | 160 ± 9 | 5.7 ± 2.1 | 5.2 ± 0.4 | >74d | 1.9 ± 0.2 |
| R155Q | 60 ± 9 | 84 ± 62 | 1.6c | 19 ± 2.8 | 1.4c | 2.5c | 0.9 ± 0.2 |
| R155K | 350 ± 14 | 220e | 30c | 447c | 4.0f | 10f | 0.6 ± 0.2 |
| A156T | 270 ± 98 | 76 ± 32 | 44c | 4 ± 1.2 | 85f | >30 | 1.2 ± 0.4 |
| D168G | 80 ± 30 | ND | 4.4c | 8.1c | 0.4c | ND | 0.8 ± 0.1 |
| D168A | 690 ± 332 | 220 ± 12 | 594c | 30 ± 3.2 | 0.7c | 0.4c | 1.5 ± 0.5 |
| D168V | 970 ± 256 | 560 ± 120 | 1,140 ± 570 | 50 ± 10 | 0.7 ± 0.1 | 0.5 ± 0.1 | 0.9 ± 0.4 |
| V36M | 2.1 ± 0.5 | 2.0 ± 0.7 | 2.7 ± 0.5 | 2.1 ± 0.4 | 2.0 ± 0.4 | 7.0d | 0.8 ± 0.2 |
| T54A | 0.9 ± 0.2 | 0.6 ± 0.3 | 0.9 | 0.5 ± 0.2 | 6.0f | 6.3d | 0.9 ± 0.2 |
| T54S | 3.5 ± 0.7 | ND | 1.2c | 1.2 ± 0.5 | 5.0 ± 0.07 | 3.0 | 0.9 ± 0.2 |
| Q80K | 2.2 ± 0.1 | 2.9 ± 0.2 | 14 ± 1.3 | 2.3 ± 0.3 | 1.0 ± 0.1 | 0.9 ± 0.04 | 1.0 ± 0.1 |
| Q80L | 1.2 ± 0.2 | 1.5 ± 0.1 | 1.9 ± 0.2 | 1.3 ± 0.2 | 1.0 ± 0.06 | 1.0 ± 0.04 | 1.0 ± 0.1 |
| Q80N | 0.6 ± 0.1 | 0.8 ± 0.2 | 0.7 ± 0.04 | 0.7 ± 0.2 | ND | 0.5 ± 0.1 | 0.8 ± 0.3 |
| Q80R | 2.6 ± 0.5 | 2.6 ± 0.3 | 15 ± 0.7 | 3.1 ± 0.5 | 1.0 ± 0.2 | 0.9 ± 0.04 | 0.9 ± 0.2 |
WT, wild type.
EC50 values were determined after 72 h of incubation with serial dilution of the indicated inhibitor and quantified by real-time reverse transcription-PCR (RT-PCR) for R155Q, A156T, D168A, and D168G variants (10). All other EC50s were obtained from luciferase reporter replicons in 72-h assays. Fold change was calculated as follows: EC50 for the variant/EC50 for the WT. For each condition, the fold change represents the mean ± standard deviation (SD), which was calculated from at least 3 independent experiments. Replicon variants in boldface type represent those selected in replicon colonies resistant to BI 201335. Values in italic type are from published results. ND, not determined.
Published value obtained from reference 12.
Published value obtained from reference 13.
Published value obtained from reference 25.
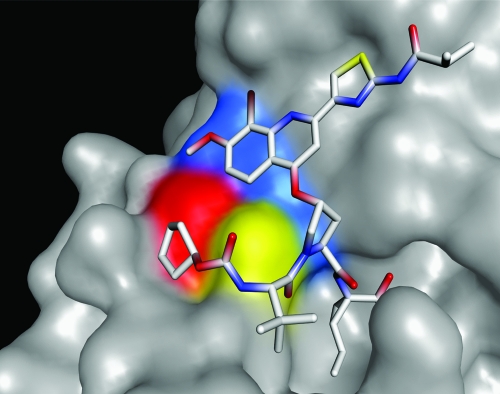
The NS3 protease enzymes containing the variants described in Table 2 were expressed and purified as previously described (17, 25) in order to assess the relative affinity of BI 201335 to these variant enzymes. As demonstrated in Table 3, there is a good correlation between the relative affinity of BI 201335 to the different enzymes and the observed changes in susceptibility of their respective resistant variants to the drug. These results are consistent with structural studies on the interaction of BI 201335 with the active site of the enzyme (Fig. 1) (11).
Table 3.
Relative affinity of genotype 1 NS3-NS4A variant enzymes to BI 201335
| NS3/4A varianta | Change in the relative affinity of the enzyme (fold change in Ki [app]) tob: |
||||
|---|---|---|---|---|---|
| BI 201335 | Telaprevir | Boceprevir | TMC435 | ITMN-191 | |
| GT 1a H77 | |||||
| R155K | 420 ± 30 | 24 ± 6 | 14 ± 6 | ||
| D168V | >690 | 0.34 ± 0.02 | 0.37 ± 0.09 | ||
| V36M/R155K | 530 ± 80 | 264 ± 105 | 126 ± 54 | ||
| GT 1b CON-1 | |||||
| R155K | 210 ± 52 | 20 ± 9 | 6.5 ± 1.6 | 44c | 31 ± 2 |
| R155Q | 430 ± 20 | 13c | 18 ± 1 | ||
| A156T | 710c | >416 | 540c | 21c | 4.0 ± 0.2 |
| A156V | 1100 ± 43 | >398 | >1,041 | 7.3 ± 0.8 | |
| D168V | 830 ± 106 | 0.3 ± 0.1 | 0.6 ± 0.1 | >52 | 5.2 ± 0.2 |
| D168A | 236 ± 27 | >68 | 15 ± 2 | ||
| V36M/A156T | 462 ± 40 | >459 | 1,080 ± 34 | ||
| V36 M | 1c | 18 ± 2 | 6.3 ± 0.5 | 1.6c | |
| Q41R | 2.2 ± 0.1 | 1.5 ± 0.2 | 2.5 ± 0.7 | 1.7 ± 0.2 | |
| T54A | 1.3 ± 0.3 | 9.8 ± 2.9 | 6.6 ± 2.7 | 0.9 ± 0.2 | |
| F43S | 1.3 ± 0.3 | 7.2 ± 0.0 | 4.4 ± 0.5 | 8.0 ± 1.9 | |
| F43V | 1.6 ± 0.6 | 6.9 ± 0.2 | 8.5 ± 0.3 | >34 | |
| Q80K | 1.9 ± 0.1 | 1.0 ± 0.0 | 2.8 ± 0.4 | 1.3 ± 0.1 | |
| Q80R | 1.7 ± 0.1 | 1.1 ± 0.1 | 4.0 ± 0.2 | 1.3 ± 0.2 | |
Replicon variants in boldface type represent those selected in replicon colonies resistant to BI 201335.
Kinetic parameters for NS3-NS4A proteins and inhibition by PIs were determined using the depsipeptide fluorogenic substrate derived from the NS5A-NS5B cleavage site: anthraniloyl-DDIVP-Abu[C(O)-O] AMY(3-NO2)-TW-OH. The assay conditions are similar to those previously described (17, 25). For boceprevir and telaprevir, the compound was preincubated with enzyme for 60 min prior to the addition of substrate. The calculated percentage of inhibition at each inhibitor concentration was used to determine the median effective concentration (IC50) by nonlinear regression routine Symyx Assay Explorer v3.2 SP1 of Symyx Technologies. Ki (apparent [app]) values were calculated from the IC50s using the equation IC50 = Ki (1 + [S]/Km) where S is substrate. Fold change = Ki of the NS3 mutant/Ki for the WT NS3. For most of the data, the fold change value is the mean ± SD, which was calculated from at least 3 experiments.
n < 3 experiments.
Fig 1.
X-ray crystal structure of the NS3 protease domain in complex with BI 201335. The proximity of major amino acid positions that confer resistance are visualized with BI 201335 bound in the active site. Substitutions at residues that confer resistance to BI 201335 are identified by color as follows: Arg155 in blue, Ala156 in yellow, and Asp168 in red. BI 201335 contains a unique C-terminal carboxylic acid that binds noncovalently to the active site, and a bromoquinoline substitution on its proline residue that provides significant potency. R155K and D168V substitutions disrupt the binding of BI 201335, which results in a several hundredfold loss in affinity of the protease inhibitor.
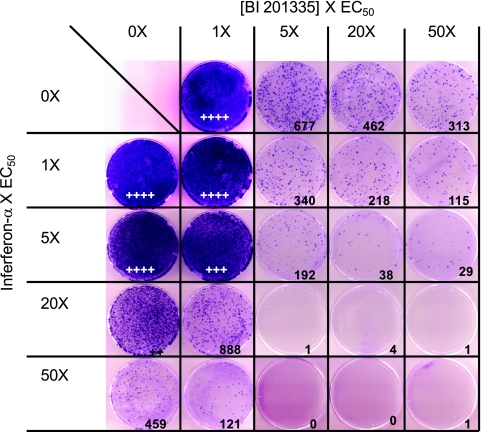
Clinical trials with HCV direct acting antivirals have demonstrated the potential for the selection of drug-resistant viruses when used as monotherapy, while combinations of HCV PIs with interferon and ribavirin have increased response rates. We examined the combined effect of BI 201335 and IFN-α on the emergence of resistant replicons in a 3-week in vitro selection as previously described (9). As demonstrated in Fig. 2, BI 201335 alone moderately suppressed the emergence of resistant colonies, whereas the combination of BI 201335 and IFN-α was extremely effective at reducing the emergence of such resistant colonies. Similar studies of BI 201335 in combination with other HCV direct acting antivirals may provide predictive tools for the design of interferon-free cotherapy regimens for the treatment of chronic HCV infection.
Fig 2.
Effects of combinations of BI 201335 and IFN-α on the frequency of resistant colony formation. Replicon cells were exposed to the indicated concentrations of BI 201335 and/or IFN-α for 3 weeks in the presence of G418. The top row depicts the colonies selected with the indicated concentration (1× to 50×, fold EC50 [5 nM]) of BI 201335 alone. The leftmost column depicts the colonies selected with the indicated concentration (1× to 50×, fold EC50 [0.2 IU/ml]) of IFN-α. The remaining plates depict colonies that were selected with dual combination of BI 201335 and IFN-α at the indicated concentrations derived from the matrix. The value indicated in the bottom right-hand corner of each square represents the number of colonies. If the colony count was too high to be determined accurately, the results of a qualitative evaluation were expressed as a number of + symbols, which was proportional to the intensity of the staining. No cytotoxicity was observed at any of the drug combinations tested (data not shown).
ACKNOWLEDGMENTS
We thank Michael Cordingley, Richard Bethell, and colleagues from Boehringer Ingelheim (Canada) Ltd. R&D for their contributions to the review and preparation of this article.
We are employees of Boehringer Ingelheim (Canada) Ltd. R&D that has discovered and is developing BI 201335.
Footnotes
Published ahead of print 24 October 2011
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Asselah T, Marcellin P. 2011. New direct-acting antivirals' combination for the treatment of chronic hepatitis C. Liver Int. 31(Suppl. 1):68–77 [DOI] [PubMed] [Google Scholar]
- 2. Bae A, et al. 2010. Susceptibility of treatment-naive hepatitis C virus (HCV) clinical isolates to HCV protease inhibitors. Antimicrob. Agents Chemother. 54:5288–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cummings MD, et al. 2010. Induced-fit binding of the macrocyclic noncovalent inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew. Chem. Int. Ed. Engl. 49:1652–1655 [DOI] [PubMed] [Google Scholar]
- 5. De Francesco R, Migliaccio G. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953–960 [DOI] [PubMed] [Google Scholar]
- 6. Fried MW. 2011. The role of triple therapy in HCV genotype 1-experienced patients. Liver Int. 31(Suppl. 1):58–61 [DOI] [PubMed] [Google Scholar]
- 7. Jacobson IM, et al. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 8. Kukolj G, et al. 2009. BI 201335, a potent HCV NS3 protease inhibitor in treatment-naive and experienced chronic HCV genotyope-1 infection: genotypic and phenotypic analysis of the NS3 protease domain. J. Hepatol. 50:S350 [Google Scholar]
- 9. Kukolj G, et al. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280:39260–39267 [DOI] [PubMed] [Google Scholar]
- 10. Lagacé L, et al. 2006. BILN 2061 and beyond: pre-clinical evaluation of HCV subgenomic replicon resistance to a NS3 protease inhibitor, p 263–278 In Schinazi RF, Schiff ER. (ed), Framing the knowledge of viral hepatitis. IHL Press, College Park, GA [Google Scholar]
- 11. Lemke CT, et al. 2011. Combined X-ray, NMR and kinetic analyses reveal uncommon binding characteristics of the HCV NS3-NS4A protease inhibitor BI 201335. J. Biol. Chem. 286:11434–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lenz O, et al. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin C, et al. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 279:17508–17514 [DOI] [PubMed] [Google Scholar]
- 14. Liverton NJ, et al. 2010. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob. Agents Chemother. 54:305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Llinas-Brunet M, et al. 2010. Discovery of a potent and selective noncovalent linear inhibitor of the hepatitis C virus NS3 protease (BI 201335). J. Med. Chem. 53:6466–6476 [DOI] [PubMed] [Google Scholar]
- 16. Manns MP, et al. 2011. Potency, safety, and pharmacokinetics of the NS3/4A protease inhibitor BI 201335 in patients with chronic HCV genotype-1 infection. J. Hepatol. 54:1114–1122 [DOI] [PubMed] [Google Scholar]
- 17. Massariol MJ, Zhao S, Marquis M, Thibeault D, White PW. 2010. Protease and helicase activities of hepatitis C virus genotype 4, 5, and 6 NS3-NS4A proteins. Biochem. Biophys. Res. Commun. 391:692–697 [DOI] [PubMed] [Google Scholar]
- 18. McCown MF, Rajyaguru S, Kular S, Cammack N, Najera I. 2009. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob. Agents Chemother. 53:2129–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poordad F, et al. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reesink HW, et al. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921 [DOI] [PubMed] [Google Scholar]
- 21. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 22. Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462 [DOI] [PubMed] [Google Scholar]
- 23. Shimakami T, et al. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Susser S, et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 25. Thibeault D, et al. 2004. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78:7352–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tong X, et al. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 77:177–185 [DOI] [PubMed] [Google Scholar]
- 27. Vermehren J, Sarrazin C. 2011. New HCV therapies on the horizon. Clin. Microbiol. Infect. 17:122–134 [DOI] [PubMed] [Google Scholar]
- 28. White PW, et al. 2010. Preclinical characterization of BI 201335, a C-terminal carboxylic acid inhibitor of the hepatitis C virus NS3-NS4A protease. Antimicrob. Agents Chemother. 54:4611–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y, et al. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 52:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Y, et al. 2007. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J. Biol. Chem. 282:22619–22628 [DOI] [PubMed] [Google Scholar]