Abstract
We recently demonstrated that colistin resistance in Acinetobacter baumannii can result from mutational inactivation of genes essential for lipid A biosynthesis (Moffatt JH, et al., Antimicrob. Agents Chemother. 54:4971–4977). Consequently, strains harboring these mutations are unable to produce the major Gram-negative bacterial surface component, lipopolysaccharide (LPS). To understand how A. baumannii compensates for the lack of LPS, we compared the transcriptional profile of the A. baumannii type strain ATCC 19606 to that of an isogenic, LPS-deficient, lpxA mutant strain. The analysis of the expression profiles indicated that the LPS-deficient strain showed increased expression of many genes involved in cell envelope and membrane biogenesis. In particular, upregulated genes included those involved in the Lol lipoprotein transport system and the Mla-retrograde phospholipid transport system. In addition, genes involved in the synthesis and transport of poly-β-1,6-N-acetylglucosamine (PNAG) also were upregulated, and a corresponding increase in PNAG production was observed. The LPS-deficient strain also exhibited the reduced expression of genes predicted to encode the fimbrial subunit FimA and a type VI secretion system (T6SS). The reduced expression of genes involved in T6SS correlated with the detection of the T6SS-effector protein AssC in culture supernatants of the A. baumannii wild-type strain but not in the LPS-deficient strain. Taken together, these data show that, in response to total LPS loss, A. baumannii alters the expression of critical transport and biosynthesis systems associated with modulating the composition and structure of the bacterial surface.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative, opportunistic, nosocomial pathogen (18). It can cause infections at most anatomical sites, resulting in outcomes ranging from asymptomatic carriage to fulminant sepsis (15, 18). The treatment of disease is significantly hindered by the propensity of A. baumannii to develop multidrug resistance (MDR); pan-drug-resistant strains have been identified (15, 37). For MDR strains, colistin (polymyxin E) is often the only effective treatment. However, colistin-resistant strains of A. baumannii are being reported increasingly in clinical settings (37).
Colistin is a cationic antibiotic that is composed of a cyclic heptapeptide covalently attached to a fatty acyl chain (50). Critical to the bactericidal action of colistin is its amphiphilic nature that allows interaction with the hydrophobic lipid A component of lipopolysaccharide (LPS) (39). Colistin resistance in A. baumannii can occur by at least two distinct mechanisms, namely, complete LPS loss or modification of lipid A (2, 6, 29). LPS-deficient derivatives of strain ATCC 19606 with mutations in any of the lipid A biosynthesis genes, lpxA, lpxC, or lpxD, can occur spontaneously and be selected for in the presence of a high (10 μg/ml) concentration of colistin (29). Furthermore, we have identified a colistin-resistant clinical isolate which has an lpxD mutation and lacks LPS (29). These colistin-resistant, LPS-deficient A. baumannii strains are the first Gram-negative bacteria reported to spontaneously lose the ability to produce lipid A. It is predicted that A. baumannii lipid A mutants are highly resistant to colistin because the initial charge-based interaction between colistin and lipid A cannot occur.
Lipid A biosynthesis is essential for the viability of E. coli (16) and has been proposed to be essential for the viability of most Gram-negative bacteria (40). However, viable, lipid A-deficient lpxA mutants have been constructed by directed mutagenesis in Neisseria meningitidis and Moraxella catarrhalis (38, 49). Chlamydia trachomatis, treated with various LpxC small-molecule inhibitors, was shown recently to replicate in the reticulate body form while lacking LPS (33). The loss of lipid A, and therefore LPS, in N. meningitidis resulted in the reduced expression of surface-exposed lipoproteins and altered outer membrane (OM) phospholipid composition, with LPS-deficient cells displaying preference for short-chain saturated fatty acids (48, 55). N. meningitidis lpxA mutants displayed significant growth defects in vitro, but lipid A-deficient A. baumannii mutants grow in vitro at the same rate as their parent strains (29). Thus, we hypothesized that A. baumannii lipid A mutants undergo unique changes in gene expression to compensate for the loss of LPS from the OM. How Gram-negative bacteria adapt to survive without LPS is poorly characterized, and for A. baumannii this adaptation may be critical for its ability to become resistant to colistin via LPS loss.
In this paper, we report the results of comparative quantitative transcriptomic analysis using the high-throughput RNA sequencing of the wild-type A. baumannii type strain ATCC 19606 and the isogenic lpxA mutant strain, 19606R. The LPS-deficient strain displayed the increased expression of genes associated with cell envelope and OM biogenesis and multidrug efflux. In particular, genes encoding lipoproteins and components of the Lol lipoprotein transport system were highly upregulated in the LPS-deficient strain, indicating that the alteration of the lipoprotein content of the OM is a critical response to LPS loss. Genes associated with the synthesis and transport of the surface polysaccharide poly-β-1,6-N-acetlyglucosamine (PNAG) also were highly upregulated, and a corresponding increase in the surface expression of PNAG was observed. Finally, we identified a number of genes associated with a type VI secretion system (T6SS) that were downregulated in the LPS-deficient strain. We also showed, using the proteomic analysis of culture supernatants, that the T6SS was active in the A. baumannii wild-type strain ATCC 19606 but was not active in the LPS-deficient mutant. A functional T6SS has not been previously identified in A. baumannii and may constitute a novel virulence factor.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The A. baumannii type strain ATCC 19606 was obtained from the American Type Culture Collection. The lpxA mutant strain 19606R was an LPS-deficient, colistin-resistant derivative of ATCC 19606 described previously (29). A. baumannii cultures were grown on Mueller-Hinton (MH) agar or in cation-adjusted MH broth at 37°C. Colistin sulfate (10 μg/ml) was added to overnight cultures where appropriate.
Total RNA purification.
A. baumannii cultures initially were grown overnight at 37°C in MH broth, with 10 μg/ml colistin sulfate added for the growth of the 19606R strain. Strains then were subcultured 1/50 into fresh MH broth without antibiotic and grown at 37°C with shaking (200 rpm) to an absorbance at 600 nm of 0.5 (mid-exponential phase), equivalent to ∼5 × 108 CFU/ml. The cells were harvested by centrifugation at 9,000 × g at 4°C and resuspended in 1 ml of RNAlater RNA stabilization reagent (Qiagen), followed by incubation for 5 min at room temperature before centrifugation at 5,000 × g. The cell pellet was resuspended in 200 μl of lysis solution (40 μg/μl proteinase K, 2 mg/ml lysozyme, 40 U/μl protector RNase inhibitor [Roche]) and incubated at room temperature for 10 min with intermittent shaking. RNA was purified using the RNeasy Minikit (Qiagen) per the manufacturer's protocol. Contaminating DNA was removed by two treatments with the RNase-free DNase kit (Qiagen). DNase-treated, purified total RNA was utilized for high-throughput RNA sequencing.
High-throughput RNA sequencing.
DNA fragmentation and synthesis of first- and second-strand cDNA was conducted as described by Nagalakshmi et al. (31). Sequencing was conducted on an Illumina GAIIx by the Micromon High-Throughput Sequencing Facility (Monash University). ATCC 19606 and 19606R cDNA samples were multiplexed into a single lane and sequenced using a 75-bp paired-end DNA sequencing protocol per the manufacturer's instructions (Illumina). For each strain, quality-trimmed sequence reads were independently aligned to the draft A. baumannii ATCC 19606 genome sequence (GenBank accession no. ACQB00000000) using SHRiMP 2.0.4 (44). To identify genes that exhibited differential expression, two biological replicates of ATCC 19606 and two of 19606R were sequenced. The differential expression of sequenced RNA was determined using the EdgeR package from Bioconductor, which uses a generalized linear model with a log link function and a negative binomial distribution (42). The negative binomial distribution has a dispersion parameter that must be estimated from the data and was assumed to be equal for all genes. A likelihood ratio test was applied to detect differentially expressed genes, with a false discovery rate of 0.05 and with a further condition that the fold change in expression be greater than 1.5.
Real-time qRT-PCR.
The RNA used for quantitative reverse transcription-PCR (qRT-PCR) was the same as that used for the RNA-seq reactions. Oligonucleotides were designed using Primer-BLAST (NCBI). Reverse transcription and triplicate qRT-PCRs were conducted using gene-specific primers (Table 1) as described by Lo et al. (24) using a Mastercycler Ep Realplex (Eppendorf). The concentration of cDNA in each reaction was determined by comparison to a standard curve constructed using each pair of primers together with genomic DNA. All reactions were normalized against the housekeeping gene gyrB.
Table 1.
Oligonucleotides used for qRT-PCR
| Gene | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| gyrB | CGAGGGTGACTCAGCGGGTG | GCGCACGCTCAACGTTCAGG |
| baeS | CCATTGGCTGTTCTGCAAGCGC | ACTCGAAACTTGTCGCATCATGGCA |
| baeR | GTCTTGGGTCTAAACATGGGGGCA | CGTTCTAAACGGCGTAAAACGGCC |
| mlaC | ACACGATGCGTCCATACAAGGCG | TGCCAACTGGAACGACACAGGA |
Purification of A. baumannii extracellular proteins.
A. baumannii cultures were grown as described for total RNA purification. Culture supernatants were filtered through a Millex GP 0.22-μm syringe filter (Millipore) to obtain cell-free supernatants and then concentrated 16-fold using Amicon Ultra-15 Ultracel 10K centrifugation concentrators (Millipore). Cultures utilized for the detection of PNAG were incubated at 42°C for 1 h to enhance PNAG release and then treated with 100 μg/μl proteinase K at 37°C for 1 h prior to the centrifugation and collection of supernatants.
SDS-PAGE and Western immunoblotting.
SDS-PAGE and Western immunoblotting were conducted by standard methods (4) on supernatants derived from A. baumannii cultures grown to mid-exponential phase. The primary antibody used was generated in goats against a deacetylated glycoform of PNAG conjugated to a diphtheria toxoid carrier (kindly supplied by G. Peir, Channing Laboratory, Harvard Medical School, Boston, MA). Horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin (Chemicon) was used as the secondary antibody. Blots were visualized with ECL Western detection reagents (GE Healthcare) and imaged by autoradiography. Densitometry was performed using ImageJ (http://rsbweb.nih.gov/ij/).
Protein identification.
Bands were excised from SDS-PAGE gels and submitted to the Monash University Biomedical Proteomics Facility for protein identification by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS). Monoisotopic peak data were analyzed using GPS Explorer with the program MASCOT (http://www.matrixscience.com/) and matched against the theoretical tryptic peptides derived from the A. baumannii genomes available in the NCBI nr database.
Transcriptomics data accession number.
The gene expression data in this study have been deposited in the NCBI Gene Expression Omnibus database and are accessible through GEO series accession number GSE31206.
RESULTS AND DISCUSSION
Sequencing of the transcriptomes of A. baumannii ATCC 19606 and the LPS-deficient strain 19606R.
A. baumannii can become resistant to the antibiotic colistin through the complete loss of LPS. To identify the changes in A. baumannii gene expression associated with LPS deficiency, we used high-throughput RNA sequencing (27) to compare the transcriptomes of ATCC 19606 and the LPS-deficient strain 19606R. Overall, a raw combined data set of 21,951,711 reads for ATCC 19606 and 21,576,267 reads for 19606R was generated. Of these, 18,031,171 (ATCC 19606) and 17,192,552 reads (19606R) aligned to the draft genome of A. baumannii ATCC 19606 (GenBank accession no. ACQB00000000), equating to ∼20% unambiguous reads for each combined data set. The aligned 75-bp reads correspond to a total of 1,352,337,750 (ATCC 19606) and 1,289,441,400 (19606R) bases sequenced for each respective strain.
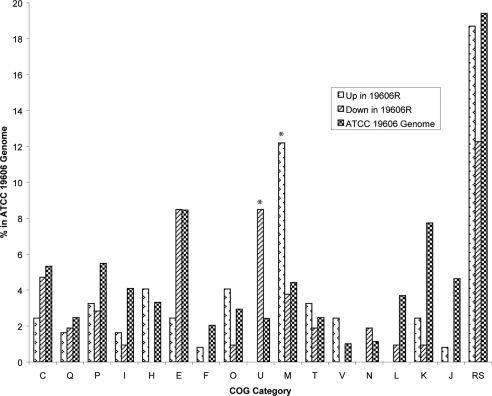
In total, 229 genes displayed altered expression of greater than ±1.5-fold in both replicates at a false discovery rate of ≤0.05. Of these, 123 genes had increased expression in the LPS-deficient strain 19606R (Table 2), whereas the other 106 showed reduced expression (Table 3). Functional categories were assigned on the basis of annotated cluster-of-orthologous-group (COG) categories (Fig. 1) (26). Genes encoding proteins with unknown or poorly characterized functions (categories R and S) made up the largest category of those displaying differential expression (12 to 19%). In the genes upregulated in 19606R, those encoding proteins belonging to COG category M, representing cell wall, membrane, and envelope biogenesis proteins, were the most significantly overrepresented (P < 0.001). The increased expression of genes within this COG category suggests that A. baumannii significantly alters the composition of the OM to compensate for the loss of LPS. Intracellular trafficking, secretion, and vesicle transport-associated genes (COG category U) were significantly overrepresented (P < 0.001) in the set of genes that displayed reduced expression in the LPS-deficient strain 19606R.
Table 2.
Genes with increased expression in LPS-deficient A. baumannii strain 19606R
| Gene identifiera | Gene name | Protein description | Fold change in expression | COG category | Signal peptidec | Predicted locationd |
|---|---|---|---|---|---|---|
| HMPREF0010_03216 | Phosphopantetheinyl transferase | ∞b | H | Cyt | ||
| HMPREF0010_03059 | Transcriptional regulator | ∞ | K | Cyt | ||
| HMPREF0010_00383 | Putative lipase | ∞ | R | |||
| HMPREF0010_00724 | Hypothetical | ∞ | CM | |||
| HMPREF0010_01511 | Hypothetical | ∞ | SpII | |||
| HMPREF0010_03356 | Putative membrane protein | 168.9 | S | SpI | ||
| HMPREF0010_02739 | Hypothetical | 137.2 | SpII | |||
| HMPREF0010_02733 | Putative membrane protein | 119.4 | S | SpI | ||
| HMPREF0010_01945 | Hypothetical | 73.5 | SpII | |||
| HMPREF0010_03654 | Hypothetical | 68.6 | SpII | |||
| HMPREF0010_00185 | Hypothetical | 68.6 | SpI | |||
| HMPREF0010_03296 | Hypothetical | 64.0 | SpI | |||
| HMPREF0010_00181 | pgaC | N-glycosyltransferase | 48.5 | M | CM | |
| HMPREF0010_03182 | Hypothetical | 42.2 | SpII | |||
| HMPREF0010_01712 | macA | Macrolide transporter | 39.4 | M | SpI | CM |
| HMPREF0010_03427 | cobW | Cobalamin synthesis | 29.9 | R | Cyt | |
| HMPREF0010_01944 | Hypothetical | 29.9 | SpII | CM | ||
| HMPREF0010_02579 | Hypothetical | 29.9 | SpI | |||
| HMPREF0010_02249 | Putative secreted protein | 27.9 | S | SpI | ||
| HMPREF0010_02888 | lolA | Outer membrane lipoprotein carrier protein | 27.9 | M | SpI | Per |
| HMPREF0010_01713 | macB | Macrolide transporter ATP-binding protein | 27.9 | V | CM | |
| HMPREF0010_01714 | tolC | Outer membrane efflux protein | 27.9 | M | SpII | OM |
| HMPREF0010_00179 | pgaA | Outer membrane protein | 26.0 | R | SpI | OM |
| HMPREF0010_00186 | Hypothetical | 22.6 | SpI | CM | ||
| HMPREF0010_00069 | Hypothetical | 22.6 | SpII | |||
| HMPREF0010_03655 | Hypothetical | 19.7 | SpII | |||
| HMPREF0010_00180 | pgaB | Outer membrane N-deacetylase | 19.7 | R | SpI | |
| HMPREF0010_02462 | Hypothetical | 18.4 | ||||
| HMPREF0010_03406 | Hypothetical | 17.1 | SpI | |||
| HMPREF0010_00247 | Hypothetical | 17.1 | ||||
| HMPREF0010_01333 | lolB | Outer membrane lipoprotein | 16.0 | M | SpII | |
| HMPREF0010_03425 | Hypothetical | 16.0 | SpII | |||
| HMPREF0010_00182 | pgaD | Putative PNAG biosynthesis | 14.9 | R | ||
| HMPREF0010_02568 | Hypothetical | 14.9 | SpI | |||
| HMPREF0010_03355 | Putative membrane protein | 13.9 | S | SpI | ||
| HMPREF0010_02269 | Hypothetical | 13.9 | SpI | |||
| HMPREF0010_01851 | rpmE2 | 50S ribosomal protein L31 type B | 13.9 | J | ||
| HMPREF0010_03241 | Hypothetical | 13.0 | CM | |||
| HMPREF0010_02727 | Putative glycosyltransferase | 13.0 | R | Cyt | ||
| HMPREF0010_03242 | Ferrichrome outer membrane transporter | 12.1 | P | SpI | OM | |
| HMPREF0010_03516 | Hypothetical | 12.1 | SpI | OM | ||
| HMPREF0010_02675 | Hypothetical | 12.1 | SpI | |||
| HMPREF0010_02288 | Hypothetical | 11.3 | ||||
| HMPREF0010_00814 | Hypothetical | 11.3 | SpII | |||
| HMPREF0010_02730 | TonB-dependent OM receptor | 10.6 | P | SpI | OM | |
| HMPREF0010_00159 | Hypothetical | 9.8 | SpI | |||
| HMPREF0010_02077 | Putative transglycosylase | 9.8 | M | OM | ||
| HMPREF0010_03295 | nadC | Nicotinate-nucleotide pyrophosphorylase | 8.6 | H | Cyt | |
| HMPREF0010_01565 | Putative heat shock protein | 8.6 | O | SpII | ||
| HMPREF0010_02779 | Hypothetical | 8.0 | S | |||
| HMPREF0010_00184 | Putative luciferase protein | 8.0 | C | Cyt | ||
| HMPREF0010_03631 | Hypothetical | 8.0 | ||||
| HMPREF0010_03357 | argA | N-acetylglutamate synthase | 8.0 | E | Cyt | |
| HMPREF0010_02071 | Multidrug efflux transport protein | 7.5 | V | CM | ||
| HMPREF0010_01545 | nlpE | Putative lipoprotein NlpE | 7.5 | SpII | ||
| HMPREF0010_02607 | mlaC | Toluene tolerance, Ttg2D | 7.5 | Q | SpI | |
| HMPREF0010_02608 | mlaB | Anti-anti-sigma factor | 7.0 | T | ||
| HMPREF0010_02815 | Entericidin A | 6.5 | S | SpII | ||
| HMPREF0010_00263 | Mechanosensitive ion channel | 6.5 | M | CM | ||
| HMPREF0010_02487 | Putative sulfate transporter | 6.5 | P | CM | ||
| HMPREF0010_03607 | Hypothetical | 6.5 | ||||
| HMPREF0010_02248 | DNA-binding transcriptional activator LysR | 6.5 | K | Cyt | ||
| HMPREF0010_00610 | Hypothetical | 6.1 | SpII | |||
| HMPREF0010_01177 | Hypothetical | 6.1 | S | |||
| HMPREF0010_01282 | Hypothetical | 6.1 | Cyt | |||
| HMPREF0010_01271 | Hypothetical | 6.1 | ||||
| HMPREF0010_03351 | dsbA | Disulfide isomerase I | 6.1 | SpI | Per | |
| HMPREF0010_00396 | Hypothetical | 6.1 | Cyt | |||
| HMPREF0010_02938 | TonB dependent receptor | 5.7 | P | SpI | OM | |
| HMPREF0010_01939 | Hypothetical | 5.7 | S | SpI | ||
| HMPREF0010_02124 | lolE | Outer membrane lipoprotein transporter | 5.7 | M | CM | |
| HMPREF0010_02166 | Hypothetical | 5.3 | SpI | OM | ||
| HMPREF0010_02606 | mlaD | ABC transporter, substrate-binding protein | 5.3 | Q | ||
| HMPREF0010_00694 | Hypothetical | 5.3 | ||||
| HMPREF0010_01378 | OmpA family lipoprotein | 5.3 | M | SpII | OM | |
| HMPREF0010_02352 | OmpW family protein | 5.3 | SpI | |||
| HMPREF0010_03145 | Peptidase M48 family | 5.3 | O | SpII | ||
| HMPREF0010_03538 | dcpA | Diguanylate cyclase | 5.3 | T | ||
| HMPREF0010_02583 | Hypothetical | 5.3 | SpI | |||
| HMPREF0010_02740 | baeS | Signal transduction histidine-protein kinase | 4.9 | T | ||
| HMPREF0010_02125 | lolD | Lipoprotein transporter | 4.9 | V | CM | |
| HMPREF0010_01748 | Hypothetical | 4.9 | S | SpI | ||
| HMPREF0010_02582 | Hypothetical | 4.9 | SpII | |||
| HMPREF0010_02025 | Putative lysine decarboxylase | 4.6 | R | |||
| HMPREF0010_02797 | Hypothetical | 4.6 | R | SpI | ||
| HMPREF0010_02820 | Putative sulfite reductase | 4.3 | S | CM | ||
| HMPREF0010_02553 | Hypothetical | 4.3 | Cyt | |||
| HMPREF0010_01798 | Hypothetical | 4.3 | S | CM | ||
| HMPREF0010_00045 | mutT | Thiamine monophosphate synthase | 4.3 | H | Cyt | |
| HMPREF0010_02558 | Hypothetical | 4.0 | SpII | |||
| HMPREF0010_01311 | Dethiobiotin synthetase | 4.0 | H | Cyt | ||
| HMPREF0010_03678 | Hypothetical | 4.0 | S | CM | ||
| HMPREF0010_02741 | baeR | DNA-binding transcriptional regulator | 3.7 | K | Cyt | |
| HMPREF0010_02437 | gabD | Succinate-semialdehyde dehydrogenase I | 3.7 | C | Cyt | |
| HMPREF0010_00264 | Hypothetical | 3.5 | Cyt | |||
| HMPREF0010_01938 | htpG | Heat shock protein 90 | 3.5 | O | Cyt | |
| HMPREF0010_02862 | Putative phospholipase | 3.5 | R | SpII | CM | |
| HMPREF0010_03073 | Hypothetical | 3.5 | SpII | |||
| HMPREF0010_02162 | Hypothetical | 3.5 | SpI | |||
| HMPREF0010_00081 | htpX | Heat shock protein HtpX | 3.5 | R | SpI | |
| HMPREF0010_00002 | Hypothetical | 3.2 | ||||
| HMPREF0010_00376 | Response regulator | 3.2 | T | Cyt | ||
| HMPREF0010_01233 | Hypothetical | 3.2 | ||||
| HMPREF0010_00046 | Hypothetical | 3.2 | SpII | |||
| HMPREF0010_02318 | ampC | β-Lactamase | 3.2 | V | SpI | Per |
| HMPREF0010_02681 | Hypothetical | 3.0 | SpI | |||
| HMPREF0010_02882 | adeI | Multidrug efflux system protein | 3.0 | M | SpII | CM |
| HMPREF0010_00792 | Putative 6-pyruvoyl-tetrahydropterin synthase | 2.8 | H | Cyt | ||
| HMPREF0010_02881 | adeJ | Multidrug efflux protein | 2.8 | V | CM | |
| HMPREF0010_02674 | pyrE | Xanthine phosphoribosyltransferase | 2.8 | F | Cyt | |
| HMPREF0010_02200 | Putative lipid binding protein | 2.8 | M | SpI | ||
| HMPREF0010_00334 | fumC | Fumarate hydratase | 2.8 | C | Cyt | |
| HMPREF0010_01332 | ipk | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase | 2.8 | I | Cyt | |
| HMPREF0010_02272 | nlpD | Outer membrane lipoprotein, NlpD | 2.8 | M | SpII | |
| HMPREF0010_01850 | Hypothetical | 2.8 | R | Cyt | ||
| HMPREF0010_00333 | UDP-galactose-4-epimerase | 2.6 | M | Cyt | ||
| HMPREF0010_02068 | groES | Chaperonin, GroES | 2.6 | O | Cyt | |
| HMPREF0010_01234 | metE | Cobalamine-independent methionine synthase | 2.6 | E | Cyt | |
| HMPREF0010_02178 | degP | Serine protease, DegP | 2.6 | O | SpI | Per |
| HMPREF0010_00353 | Lysine family exporter | 2.6 | E | OM | ||
| HMPREF0010_02883 | Putative membrane-associated lipid phosphatase | 2.5 | I | CM | ||
| HMPREF0010_02142 | OmpA-like protein | 2.5 | M | SpII | OM | |
| HMPREF0010_02880 | adeK | Outer membrane protein, AdeK | 2.5 | M | SpII | OM |
Genes were considered to be differentially expressed if they displayed at least a 1.5-fold difference in gene expression at a confidence level of 95%.
The infinity symbol (∞) indicates a gene where replicate samples from ATCC 19606 had zero read counts across the total gene length.
Presence of signal peptide (signal peptidase I [spI] or signal peptidase II [spII] cleavage sites) as predicted by SignalP (7) and LipoP (20).
Cellular localization predicted using PSORTB (17). Localization abbreviations: Cyt, cytoplasmic; CM, cytoplasmic membrane; Per, periplasmic; OM, outer membrane; Ext, extracellular.
Table 3.
Genes with reduced expression in LPS-deficient A. baumannii strain 19606R
| Gene identifiera | Gene name | Protein description | Fold change in expression | COG category | Signal peptidec | Predicted locationd |
|---|---|---|---|---|---|---|
| HMPREF0010_01583 | Hypothetical | −∞b | ||||
| HMPREF0010_01592 | Hypothetical | −∞ | ||||
| HMPREF0010_03584 | Hypothetical | −17.1 | ||||
| HMPREF0010_03084 | Hypothetical | −16.0 | ||||
| HMPREF0010_01874 | Lysine export protein | −13.0 | E | CM | ||
| HMPREF0010_02378 | Hypothetical | −12.1 | ||||
| HMPREF0010_03583 | Hypothetical | −12.1 | ||||
| HMPREF0010_01580 | Hypothetical | −11.3 | Cyt | |||
| HMPREF0010_00379 | Hypothetical | −11.3 | SpII | |||
| HMPREF0010_02377 | Hypothetical | −11.3 | Cyt | |||
| HMPREF0010_02379 | Hypothetical | −10.6 | ||||
| HMPREF0010_00669 | Hypothetical | −9.8 | ||||
| HMPREF0010_01582 | Hypothetical | −9.8 | ||||
| HMPREF0010_03594 | Hypothetical | −9.2 | ||||
| HMPREF0010_03083 | Hypothetical | −9.2 | ||||
| HMPREF0010_03161 | Hypothetical | −8.6 | Cyt | |||
| HMPREF0010_00517 | fhaB | Putative filamentous hemagglutinin | −8.6 | OM | ||
| HMPREF0010_02398 | Hypothetical | −8.6 | Cyt | |||
| HMPREF0010_01993 | pilR | Putative response regulator, PilR | −8.0 | T | Cyt | |
| HMPREF0010_03596 | Fis-like DNA binding protein | −8.0 | K | |||
| HMPREF0010_01126 | Hypothetical | −8.0 | SpI | |||
| HMPREF0010_00670 | Metallo-β-lactamase | −7.0 | R | Cyt | ||
| HMPREF0010_02504 | Sulfate transporter | −7.0 | P | Per | ||
| HMPREF0010_03593 | Hypothetical | −7.0 | Cyt | |||
| HMPREF0010_02381 | Hypothetical | −7.0 | S | Cyt | ||
| HMPREF0010_00518 | Hypothetical | −6.5 | S | Cyt | ||
| HMPREF0010_00006 | antB | Putative antirepressor protein, AntB | −6.5 | K | ||
| HMPREF0010_03013 | Hypothetical | −6.5 | SpII | |||
| HMPREF0010_02376 | Exonuclease | −6.5 | ||||
| HMPREF0010_03014 | Hypothetical | −6.1 | S | SpI | ||
| HMPREF0010_00516 | Haemolysin secretion/activation protein | −6.1 | U | SpI | OM | |
| HMPREF0010_02388 | Hypothetical | −6.1 | ||||
| HMPREF0010_01123 | assC | Type VI secretion effector | −6.1 | U | ||
| HMPREF0010_03431 | hutU | Urocanate hydratase | −6.1 | E | ||
| HMPREF0010_01153 | Hypothetical | −6.1 | CM | |||
| HMPREF0010_00933 | Periplasmic binding protein-dependent ATP binding cassette | −6.1 | P | Cyt | ||
| HMPREF0010_03012 | csgG | Curli assembly/production protein | −6.1 | M | ||
| HMPREF0010_01584 | Hypothetical | −6.1 | SpI | |||
| HMPREF0010_02380 | Hypothetical | −6.1 | ||||
| HMPREF0010_01347 | Putative transporter | −5.7 | M | CM | ||
| HMPREF0010_02923 | Hypothetical | −5.7 | ||||
| HMPREF0010_03591 | Hypothetical | −5.3 | S | |||
| HMPREF0010_01344 | Hypothetical | −5.3 | SpI | OM | ||
| HMPREF0010_00985 | Putative homoserine lactone efflux protein | −5.3 | E | CM | ||
| HMPREF0010_03597 | Hypothetical | −5.3 | Cyt | |||
| HMPREF0010_03595 | DNA-dependent helicase | −4.9 | L | Cyt | ||
| HMPREF0010_00281 | Benzoate membrane transport protein | −4.9 | Q | CM | ||
| HMPREF0010_03087 | Hypothetical | −4.9 | SpI | |||
| HMPREF0010_01125 | assA | Type VI secretion protein | −4.9 | U | Cyt | |
| HMPREF0010_02387 | Hypothetical | −4.9 | Cyt | |||
| HMPREF0010_00598 | fimB | Pili-associated assembly protein | −4.6 | N | SpI | Per |
| HMPREF0010_03432 | hutH | Histidine ammonia-lyase | −4.6 | E | ||
| HMPREF0010_01124 | assB | Type VI secretion protein | −4.6 | U | ||
| HMPREF0010_01122 | assD | Type VI secretion associated lysozyme | −4.6 | U | Cyt | |
| HMPREF0010_02316 | Hypothetical | −4.6 | SpI | |||
| HMPREF0010_03592 | Hypothetical | −4.6 | Cyt | |||
| HMPREF0010_03238 | dadX | Alanine racemase | −4.6 | M | Cyt | |
| HMPREF0010_03635 | actP | ATPase | −4.3 | P | CM | |
| HMPREF0010_03534 | Acetate permease | −4.3 | R | SpI | CM | |
| HMPREF0010_03007 | Hypothetical | −4.3 | S | CM | ||
| HMPREF0010_02929 | Hypothetical | −4.3 | ||||
| HMPREF0010_01346 | Tartrate dehydrogenase | −4.3 | E | Cyt | ||
| HMPREF0010_01404 | ggt | γ-Glutamyltranspeptidase | −4.0 | E | SpII | Per |
| HMPREF0010_01579 | Hypothetical | −4.0 | Cyt | |||
| HMPREF0010_01116 | assJ | Type VI-associated OmpA-like lipoprotein | −4.0 | M | SpI | OM |
| HMPREF0010_02809 | Response regulator receiver protein | −4.0 | T | Cyt | ||
| HMPREF0010_02843 | Hypothetical | −4.0 | ||||
| HMPREF0010_02390 | Hypothetical | −4.0 | R | |||
| HMPREF0010_03598 | Hypothetical | −4.0 | Cyt | |||
| HMPREF0010_00007 | Hypothetical | −4.0 | SpII | |||
| HMPREF0010_03239 | dadA | D-amino acid dehydrogenase | −3.7 | E | SpI | |
| HMPREF0010_01789 | calB | Succinic semialdehyde dehydrogenase | −3.7 | C | Cyt | |
| HMPREF0010_03533 | Hypothetical | −3.7 | S | |||
| HMPREF0010_03604 | Hypothetical | −3.7 | ||||
| HMPREF0010_03298 | Hypothetical | −3.7 | ||||
| HMPREF0010_01111 | assO | Type VI secretion protein | −3.7 | U | ||
| HMPREF0010_02865 | Hypothetical | −3.7 | SpI | |||
| HMPREF0010_03590 | Hypothetical | −3.7 | ||||
| HMPREF0010_03236 | cycA | d-alanine/d-serine/glycine permease | −3.7 | E | CM | |
| HMPREF0010_03763 | Metallo-β-lactamase | −3.5 | R | SpI | ||
| HMPREF0010_01439 | Hypothetical | −3.5 | ||||
| HMPREF0010_01114 | assL | Protein disaggregation chaperone | −3.5 | O | Cyt | |
| HMPREF0010_02375 | Hypothetical | −3.5 | Cyt | |||
| HMPREF0010_03695 | Hypothetical | −3.5 | SpI | |||
| HMPREF0010_01210 | Multicopper oxidase | −3.2 | Q | Per | ||
| HMPREF0010_00597 | fimA | Fimbrial protein | −3.2 | N | SpI | Ext |
| HMPREF0010_00403 | cydB | Cytochrome oxidase subunit II | −3.2 | C | CM | |
| HMPREF0010_01013 | Indolepyruvate ferredoxin oxidoreductase | −3.2 | C | |||
| HMPREF0010_01121 | assE | VasA-like type VI secretion protein | −3.2 | U | Cyt | |
| HMPREF0010_01119 | assG | Hypothetical | −3.2 | CM | ||
| HMPREF0010_01118 | assH | IcmF-like type VI secretion protein | −3.2 | U | CM | |
| HMPREF0010_01428 | Hypothetical | −3.0 | Cyt | |||
| HMPREF0010_03448 | Hypothetical | −3.0 | CM | |||
| HMPREF0010_00402 | Hypothetical | −3.0 | S | SpI | ||
| HMPREF0010_00404 | cydA | Bacterial cytochrome ubiquinol oxidase | −3.0 | C | CM | |
| HMPREF0010_01112 | assN | Type VI secretion protein | −2.8 | U | ||
| HMPREF0010_02666 | Hypothetical | −2.8 | Ext | |||
| HMPREF0010_02523 | Glutamate synthase | −2.8 | E | Cyt | ||
| HMPREF0010_01649 | Hypothetical | −2.6 | SpII | |||
| HMPREF0010_03251 | vgrG | VgrG-like type VI secretion protein | −2.6 | S | OM | |
| HMPREF0010_00401 | Cyd operon protein | −2.6 | S | CM | ||
| HMPREF0010_02596 | Hypothetical | −2.6 | SpII | |||
| HMPREF0010_02999 | hsdM | DNA methylase | −2.5 | V | ||
| HMPREF0010_02847 | Toluene catabolism | −2.5 | I | SpI | OM | |
| HMPREF0010_02245 | Ferredoxin reductase | −2.5 | C | |||
| HMPREF0010_03163 | Hypothetical | −2.5 | Cyt |
Genes were considered to be differentially expressed if they displayed at least a 1.5-fold difference in gene expression at a confidence level of 95%.
The minus infinity symbol (−∞) indicates a gene where replicate samples from 19606R had zero read counts across the total gene length.
Presence of signal peptide (signal peptidase I [spI] or signal peptidase II [spII] cleavage sites) as predicted by SignalP (7) and LipoP (20).
Cellular localization predicted using PSORTB (17). Localization abbreviations: Cyt, cytoplasmic; CM, cytoplasmic membrane; Per, periplasmic; OM, outer membrane; Ext, extracellular.
Fig 1.
Percentage of genes in each COG functional category that were differentially expressed in the LPS-deficient A. baumannii strain 19606R and parent strain ATCC 19606. Groups significantly overrepresented in the up- or downregulated gene sets in comparison to the proportions in the ATCC 19606 genome were determined by χ2 test. *, P < 0.001. The COG functional categories are the following: C, energy production and conversion; Q, secondary metabolite biosynthesis, transport, and catabolism; P, inorganic ion transport and metabolism; I, lipid transport and metabolism; H, coenzyme transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; O, posttranslational modification, protein turnover, and chaperones; U, intracellular trafficking, secretion, and vesicle transport; M, cell wall, membrane, and envelope biogenesis; T, signal transduction; V, defense mechanisms; N, cell motility and secretion; L, replication; K, transcription; J, translation; and RS, poorly characterized, which includes both R (general function prediction) and S (function unknown).
Differentially expressed genes encoding outer membrane proteins and proteins associated with outer membrane biogenesis.
Of the 123 genes with increased expression in the colistin-resistant LPS-deficient 19606R strain, more than 9% were predicted to encode OM proteins or proteins associated with OM biogenesis (Table 2). These included genes encoding components of three transport systems associated with bacterial surface components, namely, the Mla retrograde phospholipid (PL) transport system, the Lol lipoprotein transport system, and the PNAG biosynthesis and transport system.
The Lol lipoprotein transport system.
The expression of four genes that encode components of the Lol lipoprotein transport system (lolA, lolB, lolD, and lolE) increased between 4.9- and 27.9-fold in the LPS-deficient strain 19606R, with the largest increase in expression being observed for lolA (Table 2). Of these genes, only lolD and lolE are contiguous on the genome. In E. coli and P. aeruginosa, the lipoprotein transport system is essential for viability and consists of five proteins, LolA, LolB, LolC, LolD, and LolE, which together form a system that transports lipoproteins to the OM (54). LolC, LolD, and LolE form an inner membrane ABC transport complex, LolA is a periplasmic lipoprotein carrier, and LolB is an OM receptor. It is proposed that lipoproteins are linked to the OM by direct interaction with hydrophobic surface pockets of LolB (53). No clear homolog of LolC was identified in the A. baumannii genome sequence. The increased expression of the Lol system in LPS-deficient A. baumannii indicates that there is a requirement for increased lipoprotein transport to the OM. Indeed, another 25 genes encoding predicted outer membrane lipoproteins (as determined by the SignalP and PSORTb prediction of Signal II peptidase cleavage sites; Table 2) also were upregulated in 19606R. Thus, more than 20% of the genes upregulated in the LPS-deficient A. baumannii encode putative lipoproteins. Interestingly, previous studies on LPS-deficient, lpxA mutants of N. meningitidis and M. catarrhalis did not identify an increase in the expression of the Lol lipoprotein transport system or an increase in lipoprotein composition in the OM (38, 48, 55). Therefore, the compensatory mechanisms induced in A. baumannii as a result of LPS deficiency appear to be different from those described previously.
The PNAG biosynthesis and transport system.
The genes pgaABCD were upregulated between 14.9- and 48.5-fold in LPS-deficient 19606R (Table 2). These genes are involved in the synthesis and transport of the biofilm-associated exopolysaccharide PNAG in A. baumannii (12). PgaA (OM porin) and PgaB (polysaccharide deacetylase) associate to form an OM transport complex that is required for PNAG translocation, while the synthesis of PNAG is predicted to occur through the interaction of PgaC (N-glycosyltransferase) and PgaD (hypothetical protein) (12). Interestingly, studies of E. coli K-12 have shown that the modulation of PNAG expression is associated with perturbations of core LPS biosynthesis genes, suggesting that the PNAG expression in A. baumannii is regulated in a similar fashion (1).
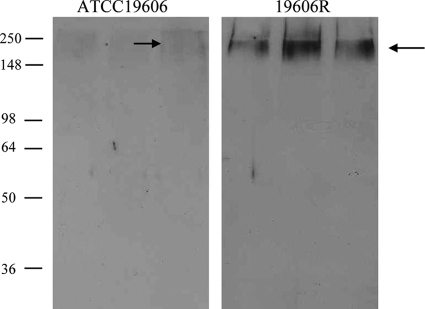
To confirm increased PNAG expression in the LPS-deficient strain 19606R, comparative Western blot analysis was conducted on equivalent concentrations of cell culture supernatants of ATCC 19606 and 19606R. Antiserum raised against PNAG reacted with a single band of approximately 250 kDa in lanes containing supernatant samples derived from either the parent strain, ATCC 19606, or the colistin-resistant, LPS-deficient derivative 19606R (Fig. 2). Densitometric analysis revealed a 7.5 (±4.6)-fold increase in anti-PNAG reactivity across replicate samples of the LPS-deficient strain 19606R, indicating an increase in PNAG expression consistent with the transcriptomic data.
Fig 2.
Lipopolysaccharide-deficient A. baumannii strain 19606R secretes increased poly-β-1,6-N-acetylglucosamine (PNAG). Shown is a Western immunoblot of culture supernatants using PNAG-specific antiserum on three biological replicates of ∼5 × 108 CFU/ml A. baumannii strain ATCC 19606 and the LPS-deficient strain 19606R. Arrows indicate the positions of PNAG-specific reactivity. The positions of molecular mass markers are indicated on the left (in kDa).
PNAG is a major component of the biofilm matrix formed by numerous bacterial species, including Staphylococcus epidermidis (21), S. aureus (13), E. coli (11), and A. baumannii (12). However, despite the increased expression of pgaABCD and increased production of PNAG in 19606R, this strain did not form increased levels of biofilm under in vitro growth conditions (data not shown). We therefore suggest that PNAG acts to stabilize the OM in the absence of LPS. Interestingly, E. coli biofilms containing PNAG show increased tolerance to polymyxin B due to a proposed electrostatic repulsion between the positively charged PNAG and the cationic peptide (1). Therefore, it also is possible that the increased surface PNAG plays a secondary, contributory role in colistin resistance in LPS-deficient strains.
The Mla retrograde phospholipid transport system.
The expression of three genes predicted to be involved in phospholipid transport (mlaBCD) increased between 5.3- and 7.5-fold in the LPS-deficient strain 19606R (Table 2). The increased expression of mlaC was confirmed by qRT-PCR, with 12-fold higher expression observed in 19606R than in ATCC 19606. In E. coli, the genes mlaABCDEF encode components of a transport system proposed to maintain OM PL asymmetry through the removal and transport of PLs from the outer leaflet of the OM to the inner membrane (IM) (25). The system consists of an inner membrane ABC transporter complex (MlaFEDB), a periplasmic substrate binding protein, MlaC, and an OM-associated lipoprotein, MlaA. In A. baumannii ATCC 19606, we identified a single locus containing mlaB (HMPREF0010_02608), mlaD (HMPREF0010_02606), mlaE (HMPREF0010_02605), mlaF (HMPREF0010_02607), and mlaC (HMPREF0010_02607). A gene (HMPREF0010_01630) encoding a protein with 52% similarity to E. coli MlaA was identified elsewhere on the genome. However, the analysis of the encoded protein revealed that HMPREF0010_01630 does not contain an N-terminal lipoprotein signal sequence which was identified as being important for the OM association of MlaA-like lipoproteins (52), suggesting that despite a level of shared amino acid identity, this protein is not the OM component of the A. baumannii Mla system. While the expression of mlaB, mlaC, and mlaD increased significantly in the LPS-deficient strain, there was no significant increase in the expression of the genes encoding the putative ABC transport ATP binding protein (mlaE) or the predicted IM permease (mlaF), which suggests that the levels of these protein products do not determine the rate of PL transport in 19606R.
In Gram-negative bacteria, the OM is an asymmetric bilayer with LPS comprising the majority of the outer leaflet and PLs the entire inner leaflet. This asymmetry is considered critical for the function of the OM as a selective permeability barrier (32, 46). However, under certain conditions, PLs may occur in the outer leaflet of the OM (34), and the Mla PL transport system is predicted to act to retain OM lipid asymmetry by removing PLs from the outer leaflet (25). In the LPS-deficient 19606R strain, we predict that the composition of the OM outer leaflet is significantly altered by the complete absence of LPS and instead must contain very high concentrations of PLs. Thus, we hypothesize that the Mla system is upregulated in response to the complete loss of surface LPS and the concomitant increase in outer membrane PL. Comparative analysis of ATCC 19606 and 19606R using thin-layer chromatography and mass spectrometry may aid in further defining the precise phospholipid composition of the outer leaflet in the absence of LPS.
Induction of the envelope stress response regulators baeS and baeR in LPS-deficient A. baumannii.
The expression of the A. baumannii genes baeR (HMPREF0010_02741) and baeS (HMPREF0010_02740) increased 3.7- and 4.9-fold, respectively, in the LPS-deficient strain 19606R. This increased expression was confirmed by qRT-PCR, with baeR and baeS expression measured as 9.4- and 11-fold higher, respectively, in 19606R compared to that of ATCC 19606. The activity and function of the BaeS/R system in Gram-negative bacteria has not been fully elucidated, but it is suggested to be associated with cellular stress response mechanisms (41). In other bacterial species, alterations to OM structure and composition can result in the induction of cellular stress response mechanisms (41, 47). Thus, we propose that the A. baumannii BaeS/R system responds to the envelope stress associated with LPS loss. In E. coli, the increased expression of BaeS/R also is associated with the increased expression of the multidrug resistance (MDR)-associated efflux proteins MdtABC, ArcD, and TolC (35). This association is further supported by the transcriptional data from LPS-deficient A. baumannii 19606R, which showed the increased expression of the genes encoding the BaeS/R orthologs as well as the increased expression of genes encoding MDR-associated proteins, such as macAB-tolC and adeIJK (Table 2).
RND efflux systems.
The expression of adeIJK and macAB-tolC was upregulated in the LPS-deficient strain 19606R. These genes are predicted to encode components of the AdeIJK and MacAB-TolC resistance nodulation-cell division (RND) family efflux systems, which are associated with the efflux of toxic compounds and antibiotics from Gram-negative bacteria (19). The expression of the adeIJK genes (HMPREF0010_02880, HMPREF0010_02881, and HMPREF0010_02882) was increased approximately 3-fold in 19606R. The genes adeI and adeJ encode the predicted membrane fusion and efflux proteins, respectively, while AdeK is the predicted OM component (14). The MacAB-TolC RND efflux system plays a major role in antibiotic resistance in Salmonella enterica, N. gonorrhoeae, and E. coli (23, 30, 36, 43). However, the role of this system in A. baumannii has not been elucidated. The genes predicted to encode the components of this system in A. baumannii, HMPREF0010_01714 (tolC), HMPREF0010_01713 (macB), and HMPREF0010_01712 (macA), were upregulated between 28- and 39-fold in 19606R.
Interestingly, despite an increase in the expression of genes encoding these predicted efflux systems, 19606R displays increased susceptibility to a number of antibiotics compared to the susceptibility of ATCC 19606 (29). This increased susceptibility to other antibiotics likely results from the significantly increased OM permeability of the 19606R strain resulting from LPS loss (29). Therefore, the induction of macAB-tolC and adeIJK expression in LPS-deficient cells may occur in response to the intracellular accumulation of toxic substances that would result from the increase in membrane permeability. In other bacterial species, the expression of both systems increased in response to the presence of antibiotics and compounds such as sodium dodecyl sulfate and safranin (14, 23, 43). The increased expression of these systems is likely to increase the efflux rate of toxic compounds from the cell, thus helping to compensate for the increased permeability of the LPS-deficient OM.
Reduced expression of genes associated with a T6SS.
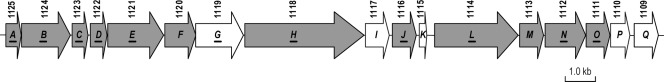
Of the 106 genes downregulated in the LPS-deficient strain 19606R, 11 were located together on the A. baumannii chromosome (HMPREF0010_01111, HMPREF0010_01112, HMPREF0010_01114, HMPREF0010_01116, HMPREF0010_01118, HMPREF0010_01119, HMPREF0010_01121, HMPREF0010_01122, HMPREF0010_01123, HMPREF0010_01124, and HMPREF0010_01125) (Table 3). Bioinformatic analysis of this region identified a putative T6SS locus consisting of 17 genes. We have assigned it the designation ass (for Acinetobacter type VI secretion system) (Fig. 3). Twelve genes within the locus (assABCDEFHJLMNO) encode proteins with homology to core T6SS components characterized in other bacterial species, and the remaining five encode products unique to Acinetobacter (assGIKPQ). Of the 12 genes encoding conserved components, 10 were significantly downregulated in the 19606R strain (assABCDEHJLNO). A number of secreted T6SS effector proteins have been identified in other bacteria, including Hcp and VgrG (8). In the A. baumannii ass locus, the Hcp homolog is encoded by assC, and this gene was significantly downregulated in the LPS-deficient strain 19606R. No proteins with similarity to VgrG effector proteins were identified in the ass locus. However, four genes were identified elsewhere on the A. baumannii ATCC 19606 genome that encoded VgrG homologs. Moreover, one of the genes identified (HMPREF0010_03251) was downregulated in the 19606R strain. Our bioinformatic analysis of other Acinetobacter genomes indicated that the T6SS ass locus is present in all Acinetobacter spp., with the exception of A. lwoffii and A. junii. However, the predicted number and sequence similarity of VgrG effectors differ significantly between strains, suggesting that these effectors are important in determining the precise role of the T6SS in different strains.
Fig 3.
Gene organization of the type VI secretion (ass) locus in A. baumannii. The locus consists of 17 genes (HMPREF0010_01125 to HMPREF0010_01109) designated assABCDEFGHIJKLMNOPQ extending over a 22-kb region. Genes encoding orthologs of the classical T6SS components in Edwardsiella tarda (56) are indicated by gray arrows. White arrows indicate genes unique to the A. baumannii type VI secretion locus. The letter designations of genes downregulated in the LPS-deficient strain 19606R are underlined. The NCBI gene accession numbers (beginning with HMPREF0010_) are shown above the genes.
The A. baumannii T6SS is functional and secretes the effector AssC.
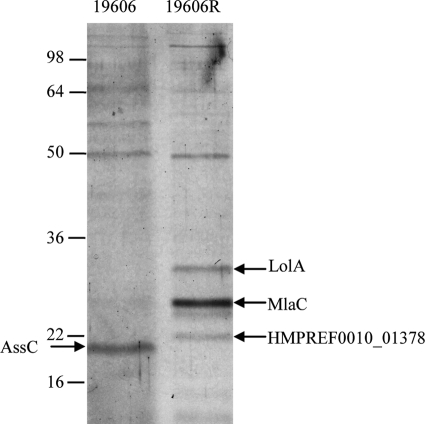
T6SS have been identified as important virulence factors in a number of Gram-negative bacterial species. The T6SS apparatus resembles an inverted bacteriophage tail and functions to secrete effector molecules into host cells (8–10). Hcp proteins are conserved T6SS components that form part of the T6SS needle tip and also are effectors actively secreted by functional T6SS (3, 8, 51, 56). To confirm that the identified A. baumannii T6SS was functional in the wild-type ATCC 19606 strain and to determine if the T6SS was impaired in the LPS-deficient 19606R, we analyzed the proteins present in A. baumannii culture supernatants for the presence of T6SS effector proteins. The analysis of the culture supernatants from ATCC 19606 and the LPS-deficient strain 19606R by SDS-PAGE revealed the presence of an approximately 20-kDa protein present in the ATCC 19606 supernatant sample but not the 19606R supernatant (Fig. 4). MALDI-TOF MS analysis of this protein identified it as the Hcp homolog AssC (7 peptides, 44% sequence coverage), confirming that the T6SS is active in the wild-type A. baumannii strain ATCC 19606 but not in the LPS-deficient strain 19606R. Interestingly, none of the VgrG orthologues was identified in the culture supernatant of the parent strain.
Fig 4.
Differential expression of AssC, LolA, MlaC, and HMPREF0010_01378 in culture supernatants derived from the A. baumannii parent strain ATCC 19606 and the LPS-deficient strain 19606R. Proteins were identified using MALDI-TOF MS. Proteins with increased expression in the LPS-deficient strain 19606R were identified by MALDI-TOF MS as LolA, MlaC, and HMPREF0010_01378, as indicated by arrows on the right. The AssC protein was identified only in the ATCC 19606 supernatant samples and is indicated by the arrow at the left. The positions of molecular mass markers are indicated on the left (in kDa).
Analysis of the culture supernatants also identified three proteins that were detected at higher levels in the supernatants from the 19606R strain than in the parent strain (Fig. 4). These were identified by MALDI-TOF MS analysis as LolA, MlaC, and HMPREF0010_01378 (an OmpA family lipoprotein). The genes encoding each of these proteins also were identified by transcriptomic analyses as being expressed at increased levels in the 19606R strain. Thus, the changes observed in protein production correlate closely with the transcriptional data for these genes.
Reduced expression of genes encoding surface appendages.
Studies of other bacterial species have shown that perturbations in OM integrity can result in the reduced expression of surface-associated adhesins, such as filamentous hemagglutinin (FHA) and pili (5, 22, 28, 45). In the LPS-deficient strain 19606R, the gene encoding an FHA homolog (fhaB) was downregulated by approximately 9-fold. Two genes predicted to be involved in fimbrial biogenesis (fimA and fimB) also were downregulated between 3- and 4.6-fold (Table 3). In addition, a gene predicted to be involved in the synthesis of curli, csgG (HMPREF0010_03012), also was downregulated approximately 6-fold. Interestingly, these genes encode products that are predicted to be required for, or associated with, the formation of channels through the lipid bilayer of the OM. This suggests that the alteration of phospholipid composition and membrane architecture in 19606R inhibits membrane-protein interactions that are important for the synthesis and/or stability of certain membrane-spanning surface structures.
Conclusions.
We have used high-throughput RNA-Seq to compare the global transcriptome of the wild-type A. baumannii ATCC 19606 strain to the LPS-deficient strain 19606R. The LPS-deficient strain showed increased expression of genes involved in membrane biogenesis, lipoprotein transport, and exopolysaccharide production. We propose that increases in lipoprotein and surface polysaccharide expression by the LPS-deficient strain aid in OM stabilization. Furthermore, the reduced expression of membrane-spanning structures, such as the T6SS, may result from a reduced ability to form these structures across an LPS-deficient OM and/or may help to stabilize the LPS-deficient OM. The LPS-deficient strain also displayed the increased expression of several efflux systems, which is likely a response to the elevated levels of toxic compounds inside the cell due to the increased permeability of the LPS-deficient OM. We predict that the observed gene expression changes, and subsequent changes in protein production, enable A. baumannii to retain a functional OM and contribute to the unique ability of A. baumannii to survive and grow normally in the absence of LPS.
ACKNOWLEDGMENTS
We thank Gerry Peir (Channing Laboratory, Harvard Medical School, Boston, MA) for kindly providing the goat antiserum for PNAG expression analysis and Luke Southey for qRT-PCR.
This work was supported by the National Health and Medical Research Council (NHMRC) and the Australian Research Council, Canberra, Australia.
J.L. is an Australian NHMRC Senior Research Fellow. J.H.M. is supported by an Australian Postgraduate Award scholarship.
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Amini S, Goodarzi H, Tavazoie S. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5:e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arroyo LA, et al. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55:3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75:886–899 [DOI] [PubMed] [Google Scholar]
- 4. Ausubel F, et al. (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 5. Baud C, et al. 2009. Role of DegP for two-partner secretion in Bordetella. Mol. Microbiol. 74:315–329 [DOI] [PubMed] [Google Scholar]
- 6. Beceiro A, et al. 2011. Phosphoethanolamine modification of Lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55:3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 8. Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8 [DOI] [PubMed] [Google Scholar]
- 9. Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ. 2010. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect. Immun. 78:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerca N, Jefferson KK. 2008. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 283:36–41 [DOI] [PubMed] [Google Scholar]
- 12. Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191:5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 16. Galloway SM, Raetz CR. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394–6402 [PubMed] [Google Scholar]
- 17. Gardy JL, et al. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 18. Gordon NC, Wareham DW. 2010. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35:219–226 [DOI] [PubMed] [Google Scholar]
- 19. Horiyama T, Yamaguchi A, Nishino K. 2010. TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 65:1372–1376 [DOI] [PubMed] [Google Scholar]
- 20. Juncker AS, et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaplan JB, Jabbouri S, Sadovskaya I. 2011. Extracellular DNA-dependent biofilm formation by Staphylococcus epidermidis RP62A in response to subminimal inhibitory concentrations of antibiotics. Res. Microbiol. 162:535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim SH, Kim YH. 2004. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J. Vet. Sci. 5:119–124 [PubMed] [Google Scholar]
- 23. Kobayashi N, Nishino K, Yamaguchi A. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lo M, et al. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U. S. A. 106:8009–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazar J, Cotter PA. 2006. Topology and maturation of filamentous haemagglutinin suggest a new model for two-partner secretion. Mol. Microbiol. 62:641–654 [DOI] [PubMed] [Google Scholar]
- 29. Moffatt JH, et al. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide. Antimicrob. Agents Chemother. 54:4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morona R, Manning PA, Reeves P. 1983. Identification and characterization of the TolC protein, an outer membrane protein from Escherichia coli. J. Bacteriol. 153:693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagalakshmi U, Waern K, Snyder M. 2010. RNA-Seq: a method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 89:4.11.1–4.11.13 [DOI] [PubMed] [Google Scholar]
- 32. Nagao K, Kimura Y, Mastuo M, Ueda K. 2010. Lipid outward translocation by ABC proteins. FEBS Lett. 584:2717–2723 [DOI] [PubMed] [Google Scholar]
- 33. Nguyen BD, et al. 2011. Lipooligosaccharide is required for the generation of infectious elementary bodies in Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 108:10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikaido H. 2005. Restoring permeability barrier function to outer membrane. Chem. Biol. 12:507–509 [DOI] [PubMed] [Google Scholar]
- 35. Nishino K, Honda T, Yamaguchi A. 2005. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol. 187:1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126–141 [DOI] [PubMed] [Google Scholar]
- 37. Park YK, et al. 2010. A single clone of Acinetobacter baumannii, ST22, is responsible for high antimicrobial resistance rates of Acinetobacter spp. isolates that cause bacteremia and urinary tract infections in Korea. Microb. Drug Resist. 16:143–149 [DOI] [PubMed] [Google Scholar]
- 38. Peng D, Hong W, Choudhury BP, Carlson RW, Gu XX. 2005. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect. Immun. 73:7569–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pristovsek P, Kidric J. 1999. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 42:4604–4613 [DOI] [PubMed] [Google Scholar]
- 40. Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76:295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raffa RG, Raivio TL. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599–1611 [DOI] [PubMed] [Google Scholar]
- 42. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rouquette-Loughlin CE, Balthazar JT, Shafer WM. 2005. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 56:856–860 [DOI] [PubMed] [Google Scholar]
- 44. Rumble SM, et al. 2009. SHRiMP: accurate mapping of short color-space reads. PLoS Comput. Biol. 5:e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ryan KR, Taylor JA, Bowers LM. 2010. The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane ß-barrel proteins in Caulobacter crescentus. Microbiology 156:742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Slamti L, Waldor MK. 2009. Genetic analysis of activation of the Vibrio cholerae Cpx pathway. J. Bacteriol. 191:5044–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steeghs L, et al. 2001. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 20:6937–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steeghs L, et al. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449–450 [DOI] [PubMed] [Google Scholar]
- 50. Storm DR, Rosenthal KS, Swanson PE. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:723–763 [DOI] [PubMed] [Google Scholar]
- 51. Suarez G, Sierra JC, Kirtley ML, Chopra AK. 2010. Role of Hcp, a type 6 secretion system effector, of Aeromonas hydrophila in modulating activation of host immune cells. Microbiology 156:3678–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suzuki T, et al. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31–41 [DOI] [PubMed] [Google Scholar]
- 53. Taniguchi N, Matsuyama S, Tokuda H. 2005. Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed ß-barrel structures. J. Biol. Chem. 280:34481–34488 [DOI] [PubMed] [Google Scholar]
- 54. Tokuda H. 2009. Biogenesis of outer membranes in Gram-negative bacteria. Biosci. Biotechnol. Biochem. 73:465–473 [DOI] [PubMed] [Google Scholar]
- 55. Williams JN, et al. 2007. Proteomic analysis of outer membranes and vesicles from wild-type serogroup B Neisseria meningitidis and a lipopolysaccharide-deficient mutant. Infect. Immun. 75:1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192–1206 [DOI] [PubMed] [Google Scholar]