Abstract
Background
Within the Coleoptera, the largest order in the animal kingdom, the exclusively herbivorous Chrysomelidae are recognized as one of the most species rich beetle families. The evolutionary processes that have fueled radiation into the more than thirty-five thousand currently recognized leaf beetle species remain partly unresolved. The prominent role of leaf beetles in the insect world, their omnipresence across all terrestrial biomes and their economic importance as common agricultural pest organisms make this family particularly interesting for studying the mechanisms that drive diversification. Here we specifically focus on two ecotypes of the alpine leaf beetle Oreina speciosissima (Scop.), which have been shown to exhibit morphological differences in male genitalia roughly corresponding to the subspecies Oreina speciosissima sensu stricto and Oreina speciosissima troglodytes. In general the two ecotypes segregate along an elevation gradient and by host plants: Oreina speciosissima sensu stricto colonizes high forb vegetation at low altitude and Oreina speciosissima troglodytes is found in stone run vegetation at higher elevations. Both host plants and leaf beetles have a patchy geographical distribution. Through use of gene sequencing and genome fingerprinting (AFLP) we analyzed the genetic structure and habitat use of Oreina speciosissima populations from the Swiss Alps to examine whether the two ecotypes have a genetic basis. By investigating a wide range of altitudes and focusing on the structuring effect of habitat types, we aim to provide answers regarding the factors that drive adaptive radiation in this phytophagous leaf beetle.
Results
While little phylogenetic resolution was observed based on the sequencing of four DNA regions, the topology and clustering resulting from AFLP genotyping grouped specimens according to their habitat, mostly defined by plant associations. A few specimens with intermediate morphologies clustered with one of the two ecotypes or formed separate clusters consistent with habitat differences. These results were discussed in an ecological speciation framework.
Conclusions
The question of whether this case of ecological differentiation occurred in sympatry or allopatry remains open. Still, the observed pattern points towards ongoing divergence between the two ecotypes which is likely driven by a recent shift in host plant use.
Background
The debate about the relative importance of ecological speciation in species diversification spans several decades [1-20]. However, concrete cases based on empirical evidence remain relatively scarce [1,21-25]. In essence, ecological speciation is related to the "ecological species concept", which was defined as follows [26]: "a species is a lineage (or a closely related set of lineages), which occupies an adaptive zone minimally different from that of any other lineage in its range and which evolves separately from all lineages outside its range". The driving force behind ecological speciation is thus divergent natural selection between environments or, in other words, reproductive isolation of populations by means of adaptation to different environments or niches [18,19,21,27,28]. Ecological selection is a consequence of individual-based interactions with the environment. From this interaction follows that divergent selection between ecological niches is a major driving force differentiating lineages until reproductive isolation occurs [17]. Ecologically divergent pairs of populations will show higher levels of reproductive incompatibility and lower levels of gene flow than ecologically more similar population pairs [29]. A resulting corollary is that ecological speciation is more likely to arise in regions with patchworks of contrasting habitats and/or distinct environmental gradients.
The number of taxa within the insect order Coleoptera exceeds that of any known plant or animal group [30]. More than half of the beetles are phytophagous, including the species rich superfamilies Curculionoidea and Chrysomeloidea, of which a majority feeds on angiosperms [31]. The increase in phytophagous beetle diversity was facilitated by the rise of flowering plants [31]. The family Chrysomelidae currently consists of more than thirty-five thousand recognized species including economically important pest species such as the Colorado potato beetle (Leptinotarsa decemlineata), the Northern corn rootworm (Diabrotica virgifera), the Cereal leaf beetle (Oulema melanopus), and the Striped turnip flea beetle (Phyllotreta nemorum). The biological and economic importance of the superfamily Chrysomeloidea make it vital to understand the factors that drive diversification in this group.
Here, we present a case of ecological niche differentiation in the alpine leaf beetle Oreina speciosissima that may represent the early stages of ecological speciation. The genus Oreina currently includes twenty-eight species, of which only seven early-diverging taxa do not exclusively occur in high forbs (i.e. five develop in stone run vegetation and two can be found in both high forbs and stone runs) [32]. According to current knowledge [34], the most parsimonious explanation is that high forbs vegetation is the ancestral niche for the remaining twenty-one Oreina lineages, among which only our focal taxon Oreina speciosissima shows a partial reversal, since it is found both in high forbs and stone run vegetation.
Oreina speciosissima is distributed across nearly the entire range of the genus Oreina (from the Pyrenees in the west to the Carpathian Mountains in the east) through a wide altitudinal gradient (ranging from 800 to 2700 m above sea level). At lower elevations it generally colonizes the very abundant high forbs vegetation whereas at higher elevations it is found in stone run habitats across a small portion of its distribution range [unpublished observations MB, TVN][32]. Kippenberg [32] and personal observations suggest that Oreina speciosissima feeds exclusively on Asteraceae (Achillea, Adenostyles, Cirsium, Doronicum, Petasites, Senecio and Tussilago) and colonizes four distinct habitats represented by well-established plant associations: two occurring in high forbs - Petasition officinalis and Adenostylion - and two in stone run -Androsacion alpinae on siliceous bedrock and Petasition paradoxi on calcareous bedrock - (see Figure 1)[33]. These plant communities are often patchily distributed due to the myriad of spatially proximate microclimates that occur in the Alps, especially sites with calcareous bedrocks which regularly present a mosaic of microhabitats. For instance, sinkholes or dolines, formed through water erosion in so-called karstic areas represent ecological islands inhabited by high forbs vegetation surrounded by areas covered by stone run vegetation [unpublished observation TVN]. Beetles inhabiting the highly divergent habitats have been categorized in two different subspecies, namely Oreina speciosissima sensu stricto (distributed across the whole species range) and Oreina speciosissima troglodytes (restricted to the Swiss and neighbouring Italian Alps), on the basis of differences in elytral coloration and the shape of male genitalia (aedeagus) [32]. Oreina speciosissima sensu stricto beetles are bright metallic green or blue whereas the coloration in Oreina speciosissima troglodytes is generally darker and mat [32]. It is not known whether the morphological differences between ecotypes are in any way adaptive and/or have a genetic basis, although color patterns in another species of Oreina are known to influence natural selection through predation pressure [35]. Like most other members of the genus, Oreina speciosissima sensu stricto can be found feeding or mating throughout the day on, or in the vicinity of its host plants. In contrast, Oreina speciosissima troglodytes is usually found adjacent to its host plants, concealed in crevices and under loose rocks. Previous studies by the authors [36,37] greatly challenged the existence of clear species boundaries within the genus: it is therefore realistic from a biological point of view to refer to these taxonomic entities as ecotypes rather than subspecies. Leaf beetles from the genus Oreina are generally thought to make only limited use of their dispersal capabilities [38,39] even though Kalberer et al. [40] reported an average flight dispersal of approximately one hundred meters for Oreina cacaliae beetles during autumn migration. Rowell-Rahier [35] showed that low vagility in concert with a patchy host plant distribution resulted in a low level of genetic structuring in Oreina speciosissima. Oreina speciosissima sensu stricto beetles inhabit high forbs patches that are generally larger in size, lie closer together and harbor more beetles per unit of surface area than stone run patches inhabited by Oreina speciosissima troglodytes [unpublished observations MB, TVN].
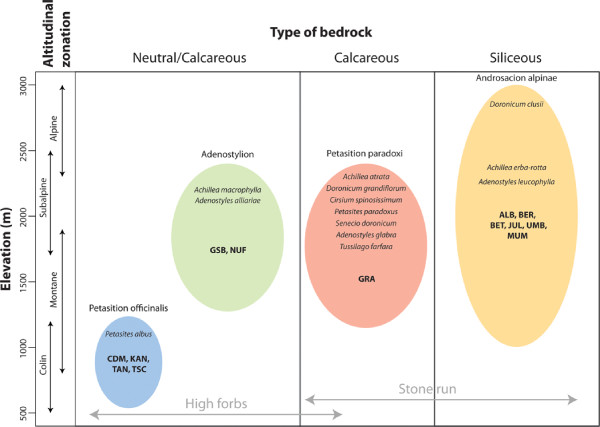
Figure 1.
Host plants of Oreina speciosissima and their altitudual zonation and habitat. According to literature [32] and personal observations of the authors, 13 plant species were referred to as putative host plants for Oreina speciosissima. These species belong to four plant associations, i.e. Petasition officinalis, Adenostylion, Petasition paradoxi and Androsacion alpinae [33], which segregate along altitudinal zonation and bedrock type gradients. Whereas Petasition officinalis and Adenostyles correspond to high forbs habitat, Petasition paradoxi and Androsacion alpinae represent stone run habitats. In particular conditions, the Petasition paradoxi can merge with high forbs (see text). Each association includes bold written codes (see Table 1 for details) of sampled populations. Plant associations are written from left to right as a function of their mean elevation, and also according to a putative scenario of colonization of central Alpine silicious stone run vegetation by specific Oreina speciosissima lineages (high forbs representing the original habitat for most Oreina species). Main host plant species of Oreina speciosissima in each of the four associations are as follows: Petasites albus in Petasition officinalis, Adenostyles alliariae in Adenostylion, Doronicum grandiflorum in Petasition paradoxi, and Doronicum clusii in Androsacion alpinae.
The present work investigates 13 populations representative of the two ecotypes, using sequencing of nuclear (ITS2) and mitochondrial (hereafter mtDNA) (16S, COI and COII) DNA regions as well as AFLP genome fingerprinting in a way to address the following questions:
1. Are ecotypes monophyletic?
2. Is adaptation to different habitats and host plants associated with genetic divergence?
Results
Phylogenetic reconstruction of the DNA sequence data sets
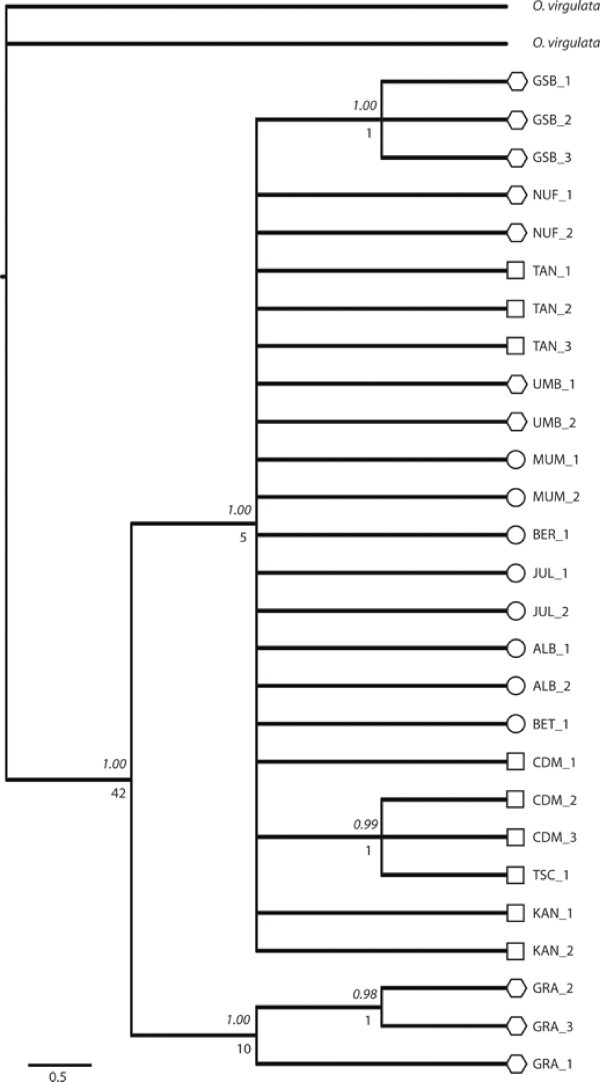
The nuclear ITS2 region showed no variation for Oreina speciosissima and was thus excluded from further analyses. In contrast, the three mtDNA regions were polymorphic with a total alignment length of 1632 bp; 529 bp for 16S, 470 for COI and 633 bp for COII. Excluding the outgroup, 30 characters were potentially parsimony informative (hereafter PPIc) among 37 variable characters. The three mtDNA regions contributed as follows: 16S (3 PPIc among 5 polymorphic sites), COI (13 PPIc among 16 polymorphic sites) and COII (14 PPIc among 16 polymorphic sites). The best substitution models were Hasegawa-Kishino-Yano (HKY) [41] for 16S and Hasegawa-Kishino-Yano plus Gamma (HKY+G) [41,42] for COI and COII. The alignments of the mtDNA markers were combined in a total evidence approach, after pairwise incongruence length difference ILD test [43] revealed no incongruence among the three mtDNA markers (COI and COII, P value = 1.00; COI and 16S, P value = 1.00; COII and 16S, P value = 1.00). The resulting dataset was investigated using maximum parsimony (hereafter MP) and Bayesian phylogenetic inference methods [44]. Both approaches produced highly congruent topologies with the same major nodes. The MP topology with Bremer supports [45] and corresponding Bayesian posterior probabilities from the Bayesian analysis (hereafter bpp) are shown in Figure 2. The ingroup is well supported with a Bremer support of 42 and a bpp of 1.00. The ingroup splits into two groups, a well-supported clade (Bremer support = 10 and bpp = 1.00) containing all individuals from the GRA population and a polytomy (Bremer support = 5 and bpp = 1.00) containing all other individuals. Apart from a clade containing all individuals of GSB and one with two individuals from CDM and one from TSC, there is no resolution within the polytomy. Sample NUF_3 failed to amplify and is therefore not shown in Figure 2. Only samples that rendered both satisfactory DNA sequences and AFLP fingerprints were used for phylogenetic analysis.
Figure 2.
Strict consensus tree of Oreina speciosissima, as revealed by mtDNA regions 16S, COI and COII (maximum parsimony tree). Node supports are given by Bremer supports (decay index) ≥ 1 and Bayesian posterior probabilities (italic). Specimens are labeled according to morphotypes (i.e. square -Oreina speciosissima sensu stricto, circle - Oreina speciosissima troglodytes and polygon - intermediate forms).
AFLP
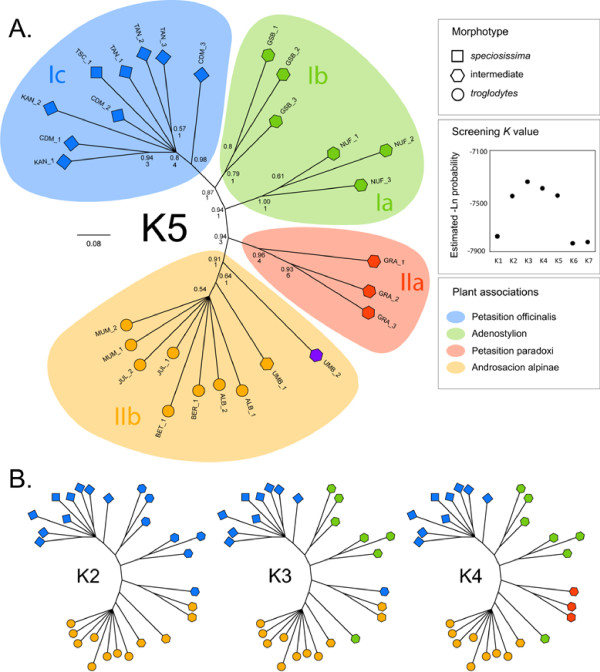
The AFLP analysis produced a total of 530 bands (171, 166 and 173 for EcoRI-ACA/MseI-AGC, EcoRI-ACA/MseI-ACG and EcoRI-ACA/MseI-AAC, respectively) with an average of 254 bands per individual and an average reproducibility rate of 96.1%. Among 510 variable characters, 458 were potentially parsimony informative. Just as the mtDNA data, the AFLP dataset was investigated using MP and Bayesian phylogenetic inference methods. Again, both approaches were highly congruent as the MP and Bayesian trees shared the same major nodes. Consequently, only the Bayesian phylogeny (including the bpp and Bremer supports) is displayed in Figure 3. Due to the lack of an outgroup, we present an unrooted topology (with supports extracted from the corresponding midpoint-rooted topology), which led to a separation of specimens into two well supported clans sensu Wilkinson et al. [46] (clan I and clan II), each with a bpp of 0.94 (Figure 3). Clan I includes three sub-clans supported with bpp values of 1.00 (Ia), 0.79 (Ib) and 0.98 (Ic) respectively. Clan I contains nine specimens with a strict Oreina speciosissima sensu stricto morphology (sub-clan Ic) and six specimens with an intermediate morphology (sub-clans Ia and Ib). Within clan II, two sub-clans were well supported with a bpp of 0.96 (IIa) and 0.91 (IIb) respectively. Clan II contains eight individuals with strict Oreina speciosissima troglodytes morphology (all of sub-clan IIb, except UMB specimens) and five individuals with an intermediate morphology (all of sub-clan IIa and UMB specimens from sub-clan IIb). Notably, specimens with intermediate morphologies were sorted close to the midpoint root of the tree topology. The AFLP dataset was further investigated using a Bayesian (i.e. STRUCTURE see [47,48]) and a distance-based (i.e. K-means; see [49,50]) clustering algorithm. The approaches produced fully congruent relationships and only results of the former are provided here. The STRUCTURE analysis showed highly likely clusters when considering K values ranging between two and five (see box Figure 3). The obtained results were largely congruent across K values (Figure 3) and highly compatible with the phylogenetic relationships. The only incongruence that could be observed when considering all K values, or when comparing STRUCTURE results with the tree topology, involved specimens with an intermediate morphology. Hereafter, we will consider and discuss results based on K = 5, given that they are the most informative. When viewed from a host plant perspective it becomes apparent that all leaf beetle specimens in clan II occur in the same stone run habitat with individuals from sub-clans IIa and IIb developing respectively in alkaline Petasition paradoxi and in acidic Androsacion alpinae habitats (Figures 1 and 3). In contrast all individuals from clan I occurred in high forbs, in Adenostylion or Petasites officinalis habitat, either on alkaline, neutral or slightly acidic soils [33] (see Figures 1 and 3).
Figure 3.
Half-compat consensus tree and clustering of Oreina speciosissima, as revealed by AFLP data (Bayesian tree and STRUCTURE clustering). a) Specimens are labeled according to morphotypes (i.e. square - Oreina speciosissima sensu stricto, circle - Oreina speciosissima troglodytes and polygon -intermediate forms) and clusters defined using the Bayesian model-based STRUCTURE algorithm applied to AFLP data (i.e. colors of tips according to K = 5 groups; legend center panel - log-likelihood values of the best STRUCTURE runs for K1 to K7 groups, see text for further details). In addition, the corresponding habitat types (translated into plant associations) are displayed as color coded backgrounds. The names of clans (Ia,b,c and IIa,b, based on the Bayesian AFLP tree topology) and the node supports (i.e. above - Bayesian posterior probabilities and below - Bremer supports ≥ 1) are provided. b) Insights from alternative STRUCTURE results (i.e. K2 to K4). The tree and morphotype symbols are as in a).
Discussion
Are ecotypes monophyletic?
The phylogenetic tree based on mtDNA markers provides high support for the monophyly of Oreina speciosissima sensu lato (Figure 2). However, very little polymorphism and genetic structure are revealed within the ingroup, although the mtDNA markers proved variable enough to reconstruct intra-specific phylogenetic relationships in other arthropod systems (e.g [51-53]). Indeed, the resulting topology supports neither geographical nor ecotypic grouping of the beetles, possibly suggesting a recent divergence of Oreina speciosissima lineages, with the exception of specimens from the GRA population, which cluster as the sister lineage to all other specimens. Beetles from this latter population thus form an orphan clade [54], which may correspond to an isolated refugial lineage. AFLP data on the other hand shows a clear-cut differentiation of specimens (Figure 3). However, this pattern does not appear to have a geographical basis (Figure 4) and instead correlates with the beetle ecotypic definition, or in other words, to the plant habitat (Figure 1). In contrast, AFLP genetic structuring only partly correlates with morphotypes (sub-clans Ic and the larger part of IIb, see Figure 3). Ecotypes per se are thus not monophyletic, although there is a strong tendency for specimens and populations to cluster within the boundaries set by plant associations and their intrinsic ecologies.
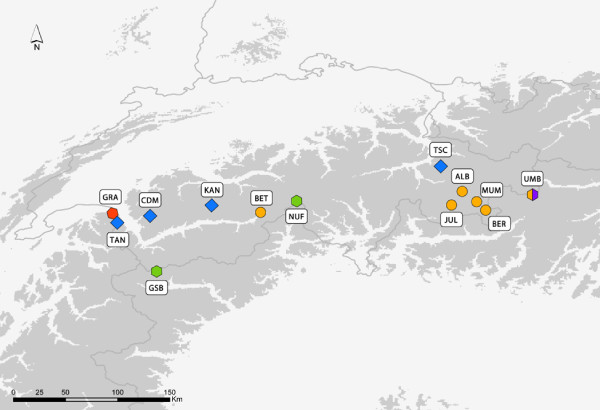
Figure 4.
Geographical distribution of the 13 sampled populations of Oreina speciosissima and results from AFLP Bayesian clustering. Populations are labeled according to morphotype (i.e. square - Oreina speciosissima sensu stricto, circle - Oreina speciosissima troglodytes and polygon - intermediate forms) and clusters defined using the Bayesian model-based STRUCTURE algorithm (i.e. colors of tips according to K = 5 groups). Grey shaded areas represent elevated regions with altitudes above 1000 m. For full names of the populations see Table 1.
Is adaptation to different habitats and host plants associated with genetic divergence?
Our results showed that genetic differentiation among Oreina speciosissima lineages was clearly associated with plant communities (Figure 3). Accordingly, clustering in Oreina speciosissima is well explained by differences in bedrock type and host plants (translated here into different plant associations) (Figures 1 and 3). While specimens feeding in the Petasition paradoxi association (in which the calcicolous Doronicum grandiflorum is the main host plant for Oreina speciosissima [unpublished observations MB, TVN]) cluster in sub-clan IIa, specimens developing in the Androsacion alpinae association (in which the silicicolous Doronicum clusii is largely dominant as a host plant for Oreina speciosissima [unpublished observations MB, TVN]) are restricted to sub-clan IIb. The effect of soil acidity is less striking in clan I, probably because the Adenostylion and Petasition officinalis associations, which are characteristic of all specimens within this clan, are defined by intermediate soil pHs. These two plant communities include species showing an intermediate tolerance to acidic-alkaline variation, such as Achillea macrophylla, Adenostyles alliarae and Petasites albus [55]. Whereas the latter two represent the main host plant species of Oreina speciosissima in high forbs habitat [unpublished observations MB, TVN], other species (particularly in the Petasition paradoxi association) could play or have been playing the role of subalpine bridge species between the montane high forbs and alpine stone runs (see below). We are confident that these results are robust to potential shortcomings inherent to our limited sampling size (see [56] for a review). First, specimens were collected throughout the common geographical range of both ecotypes, a strategy that maximized both the phylogeographic and ecological representativity of our sampling. Second, robust and consistent results were obtained using both phylogenetic and clustering algorithms.
Towards a scenario of ecological speciation in Alpine Oreina speciosissima
Although our data does not allow for divergence time estimates between Oreina speciosissima ecotypes, it seems likely that they diverged relatively recently. Indeed, the current distribution of Oreina populations suggest that the ecotype divergence might have arisen after one of the last glacial maxima, given that populations were probably not able to survive cold periods at high altitudes due to the presence of ice caps (with the possible exception of the GRA population; see above). This hypothesis is consistent with the low level of genetic variation observed in nuclear sequences and the low resolution in the mtDNA topology, as well as with a preliminary dating of the Oreina genus, in which the origin of Oreina speciosissima is estimated at circa 0.4 million years ago [36].
Our results suggest that from an ancestral niche associated with high forbs (see above) beetle populations were able to colonize new habitats along an altitudinal gradient (Figure 1) and invaded the acidic siliceous stone run habitat (corresponding to the Androsacion alpinae association), which is typical for Alpine regions in Central Europe. We propose that this habitat change could have been associated with host shifting events. Accordingly, the plant communities on which Oreina ecotypes feed appear to be connected by phylogenetically related host species. In a framework of plant-insect coevolution [57,58], adaptation to a given plant species might allow beetles to spread to other similarly-defended congeneric species [59,60]. Accordingly, Doronicum species occur in the Petasition paradoxi and the Androsacion alpinae, Petasites species link the Petasition officinalis to the Petasition paradoxi and finally, Adenostyles species are shared among the Adenostylion, the Petasition paradoxi and the Androsacion alpinae. Assuming host-plant conservatism, the connections described above might represent "shifting" routes that could explain how Oreina speciosissima lineages transited among habitats via host switching. Furthermore, these connections could account for the presence of putatively admixed specimens showing intermediate morphologies (e.g. UMB), thereby outlining a possible ongoing migration of beetles from one habitat to the other.
Conclusions
Our study reveals a genetic structure in Oreina speciosissima as a function of the plant community in which beetles develop. We discussed several possible ecological features that could cause the divergence between ecotypes, among which the habitat and host-plant switches seem key factors. These results could be consistent with an ecological speciation scenario. Still, non-adaptive processes such as genetic drift, founder events and population bottlenecks might also have produced the observed pattern. Hence, further investigation is needed, for instance, fine scale studies relying on genomic approaches and targeting populations from a patchy distribution of the two ecotypes following an approach such as described by [61] could provide a powerful framework for detecting adaptive signatures associated to ecological speciation. Additionally reciprocal transplantation experiments in concert with crossings using local and non-local beetles could possibly reveal performance differences between locally adapted and non-adapted beetles and strengthen our argument for the existence of host races and ongoing or incomplete speciation (cf. [62,63]).
Methods
Sampling
During the summers of 2004, 2005 and 2008, specimens of Oreina speciosissima sensu stricto and Oreina speciosissima troglodytes were collected from 13 populations (Table 1) and stored in pure ethanol at -20°C. All sampled beetles were found on four distinct plant associations, namely Petasition officinalis, Adenostylion, Petasition paradoxi and Androsacion alpinae. The Petasition officinalis (populations CDM, KAN, TAN and TSC) and the Adenostylion (populations GSB and NUF) occurred on neutral to slightly calcareous bedrocks, at low and medium altitudes, respectively. The Petasition paradoxi (population GRA) and the Androsacion alpinae (populations ALB, BER, BET, JUL, UMB and MUM) grew on medium-high altitude calcareous and siliceous bedrocks, respectively (see Figure 1). Three individuals from each population were selected for genetic analysis, using only males to ensure accurate identifications based on genitalia. Following the reasoning of Nosil et al. 2002, 2003 [64,65] a 'population' is defined as all of the insects collected within a homogenous patch of plants belonging to one of the four abovementioned plant communities. 'Parapatric' populations are those in contact with a second population using host plants of a different plant community. If we take the maximum migration distance of Oreina cacaliae as reported in [40] as a proxy for migration ability of Oreina speciosissima, and thus the possibility for gene flow, this study incorporates only one true parapatric pair (TAN - GRA). As a result of this our study is not suitable to test the influence of geographical distance with regard to genetic distance between beetles that use different plant communities as host plants. The dataset was completed with two individuals of Oreina virgulata (i.e. a closely related species) that were used as the outgroup [34].
Table 1.
Sampled populations of Oreina speciosissima
| Code | Population | Altitude | Coordinates | Morphotype | Habitat | Year |
|---|---|---|---|---|---|---|
| KAN | Kandersteg | 1314 m | 46°28'21"N, 07°39'23"E | speciosissima | hf | 2004 |
| TSC | Tschiertschen | 1325 m | 46°48'55"N, 09°36'31"E | speciosissima | hf | 2004 |
| CDM | Col des Mosses | 1716 m | 46°23'26"N, 07°07'30"E | speciosissima | hf | 2005 |
| TAN | Lac Taney | 1389 m | 46°20'38"N, 06°50'01"E | speciosissima | hf | 2008 |
| GRA | Le Grammont | 1974 m | 46°21'15"N, 06°49'04"E | intermediate | sr | 2008 |
| NUF | Nufenenpass | 2172 m | 46°28'41"N, 08°22'36"E | intermediate | hf | 2008 |
| GSB | Grand St. Bernard | 2410 m | 45°52'04"N, 07°10'27"E | intermediate | hf | 2008 |
| BET | Bettmerhorn | 2628 m | 46°24'44"N, 08°04'33"E | troglodytes | sr | 2008 |
| UMB | Umbrailpass | 2647 m | 46°32'53"N, 10°25'43"E | intermediate | sr | 2008 |
| BER | Berninapass | 2315 m | 46°24'37"N, 10°01'36"E | troglodytes | sr | 2008 |
| ALB | Albulapass | 2324 m | 46°34'46"N, 09°50'15"E | troglodytes | sr | 2008 |
| MUM | Muottas Muragl | 2735 m | 46°30'27"N, 09°56'29"E | troglodytes | sr | 2008 |
| JUL | Julierpass | 2373 m | 46°28'02"N, 09°43'35"E | troglodytes | sr | 2008 |
Sample locations with altitude (meters above sea level), coordinates (WGS 84) and year of collection with their codes, morphotype and habitat (hf: high forbs; sr: stone run).
DNA sequence data and phylogenetic analyses
The DNA extraction, amplification and sequencing protocols as well as primers for the nuclear (ITS2) region and the three mtDNA markers (16S, COI, COII) are provided in [36]. The alignments of mtDNA markers (using the Clustal-Wallis algorithm [66]) were combined in a total evidence approach [67] after having performed pairwise incongruence length difference ILD tests [43]. We followed the snowball procedure as implemented in the program mILD[68].
Phylogenetic analyses were performed using the maximum parsimony (MP) and Bayesian Markov chain Monte Carlo (MCMC) criteria. Each partition and the combined data set were analyzed using parsimony ratchet [44] as implemented in PAUPrat[69] and further run in PAUP* 4b10 [70]. Ten independent searches were performed with 200 iterations and 15% of the parsimony informative characters perturbed [44]. The shortest most parsimonious trees were combined to produce a strict consensus tree. Branch supports were calculated using the Bremer support (also known as 'decay index') [45] as implemented in TreeRot[71] and further run in PAUP* 4b10 [70]. The Bremer support measures the number of extra steps in tree length required before a node collapses [45,72]. Model selection for the mtDNA data partitions in the MCMC was carried out with MrModeltest2 v.2.3 [73] based on the 'Akaike information criterion' [74]. Two Metropolis-coupled Markov chains with incremental heating temperature of 0.1 were run in MrBayes 3.1.2 [75] for 30 million generations and sampled every 1000th generation. The simulation was repeated six times, starting from random trees. Convergence of the MCMC was checked using the Potential Scale Reduction Factor (PSRF) [76] implemented in MrBayes 3.1.2 [75] and the effective sample size (ESS) criterion for each parameter as implemented in Tracer 1.4 [77]. To yield a single hypothesis of the phylogeny, the posterior distribution was summarized in a 50% majority rule consensus tree (the "halfcompat consensus tree" from MrBayes) after burn-in (for each analysis 10000 trees were discarded). The combined dataset was analysed using partition specific model parameters [73].
AFLP
Genome fingerprinting was performed using the AFLP protocol described in [78]. The selective amplifications were performed using 5-FAM fluorescently labelled EcoRI primer (i.e. EcoRI + ACA) with one of the following: MseI primer + AXX (AGC, ACG and AAC). All amplifications were run in a Biometra TGradient thermocycler (Biometra, Göttingen, Germany). Samples were randomly displayed on a 96-well PCR plate, with ten individuals being replicated to assess the overall reproducibility of reactions. PCR products were analysed using the GeneScan technology with a capillary sequencer (ABI 3730XL, Applied Biosystems, Foster City, CA; the service was provided by Macrogen Inc. Seoul, South Korea).
Resulting electropherograms were analysed with PeakScanner (ABI, peak detection parameters: default parameters with the addition of a light peak smoothing) in order to detect and calculate the size of AFLP bands. The scoring was performed using an automated scoring R CRAN package, RawGeno 2.0 [79,80]. The library was settled as follows: scoring range = 100 - 250 bp for EcoRI-ACA/MseI-AGC, EcoRI- ACA/MseI-ACG and 100-280 for EcoRI-ACA/MseI-AAC, minimum intensity = 50 rfu, minimum bin width = 0, maximum bin width = 1 bp and closely sized bins (5%) were removed. Finally, the matrices of the three scored primer pairs were concatenated into a single binary matrix where individuals and bands were stored as lines and columns, respectively.
Phylogenetic and clustering analyses of the AFLP data set
Phylogenetic analyses of the AFLP data were performed using the MP and Bayesian MCMC criteria. The MP analysis (including Bremer support analysis) was performed as described above. Parameters for the Bayesian MCMC analysis performed in MrBayes 3.1.2 were set as follows: "datatype = restriction" and "coding = noabsencesites". Four metropolis-coupled Markov chains with incremental heating temperature of 0.1 were run for 5 million generations and sampled every 1000th generation. The simulation was repeated six times, starting from random trees. Convergence of the analysis was checked using the PSRF and ESS criteria (see above for more details). The posterior distribution was summarized in a halfcompat consensus tree (see above) after burn-in (for each analysis 1500 trees were discarded).
Two independent clustering algorithms were used to assign Oreina speciosissima specimens into a user-defined number of groups (hereafter K). First, we used non-hierarchical K-means clustering [81], a distance-based algorithm that proves reliable in an AFLP framework [49,50,82]. A total of 100 000 independent runs was carried out for each value of K clusters assumed (i.e. ranging from two to seven) and only runs yielding the highest inter-cluster variance were considered for further analysis. The optimal K value was determined based on the second derivative of the intercluster inertia, as in [50]. Computations were performed using R CRAN [83] (script available upon request to NAR). Second, we performed a model-based Bayesian inference clustering as implemented in STRUCTURE 2.2 [47,48]. The analysis assumed an admixture model and independent allele frequencies between clusters. Five independent runs were carried out for each value of K (i.e. ranging from one to seven), with parameters and model likelihood estimated over 1 000 000 MCMC generations (following a burn-in period of 200 000 generations). For each K value, only runs that obtained the highest likelihood value were taken into account for further analyses. The majority-rule criterion (>0.5 in the assignment probability) was applied to assign samples to a given cluster as in [50]. Both clustering approaches provided fully congruent insights and therefore only results from STRUCTURE are displayed here.
Authors' contributions
MB and TVN collected the samples, carried out the morphological and genetic analyses, participated in phylogenetic analysis and drafted the manuscript. NAR, SB and NAL designed phylogenetic tools, participated in phylogenetic analysis and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Matthias Borer, Email: matthias.borer@unine.ch.
Tom van Noort, Email: tom.vannoort@unine.ch.
Nils Arrigo, Email: arrigon@email.arizona.edu.
Sven Buerki, Email: s.buerki@kew.org.
Nadir Alvarez, Email: nadir.alvarez@unil.ch.
Acknowledgements and funding
The work was funded by the Swiss National Science Foundation (grants 3100-064864.01 and 3100-AO-118031(TVN) the SNSF National Centre of Competence in Research Plant Survival, and a university doctoral assistantship to MB. NAR and NAL were funded by the Swiss National Science Foundation (grant No. 132747 and an Ambizione fellowship PZ00P3_126624, respectively). Financial support to SB was provided by Marie-Curie Intra-European Fellowship (CRADLE; no 253866). TVN wishes to thank CP for beetle collection at GRA and Carolien Jacobs for useful advice. We thank Pascal Vittoz for sharing his botanical knowledge, Jessica Litman for language editing, and three anonymous referees for their helpful and constructive remarks that led to a substantial improvement of the manuscript.
References
- Berlocher SH, Feder JL. Sympatric speciation in phytophagous insects: Moving beyond controversy? Annual Review of Entomology. 2002;47(1):773–815. doi: 10.1146/annurev.ento.47.091201.145312. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Fitzpatrick BM. Sympatric speciation: Models and empirical evidence. Annual Review of Ecology Evolution and Systematics. 2007;38:459–487. doi: 10.1146/annurev.ecolsys.38.091206.095804. [DOI] [Google Scholar]
- Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400(6742):354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- Drès M, Mallet J. Host races in plant-feeding insects and their importance in sympatric speciation. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 2002;357(1420):471–492. doi: 10.1098/rstb.2002.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma DJ, Peterson SC. Genetic-variation in the use of resources by insects. Annual Review of Entomology. 1985;30:217–238. doi: 10.1146/annurev.en.30.010185.001245. [DOI] [Google Scholar]
- Craig TP, Itami JK, Abrahamson WG, Horner JD. Behavioral Evidence for Host-Race Formation in Eurosta solidaginis. Evolution. 1993;47(6):1696–1710. doi: 10.2307/2410214. [DOI] [PubMed] [Google Scholar]
- Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends in Ecology & Evolution. 2009;24(3):145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nosil P, Sandoval CP, Crespi BJ. The evolution of host preference in allopatric vs. parapatric populations of Timema cristinae walking-sticks. Journal of Evolutionary Biology. 2006;19(3):929–942. doi: 10.1111/j.1420-9101.2005.01035.x. [DOI] [PubMed] [Google Scholar]
- Feder JL, Berlocher SH, Roethele JB, Dambroski H, Smith JJ, Perry WL, Gavrilovic V, Filchak KE, Rull J, Aluja M. Allopatric Genetic Origins for Sympatric Host-Plant Shifts and Race Formation in Rhagoletis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10314–10319. doi: 10.1073/pnas.1730757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Chilcote CA, Bush GL. Genetic differentiation between sympatric host races of the apple maggot fly Rhagoletis pomonella. Nature. 1988;336(6194):61–64. doi: 10.1038/336061a0. [DOI] [Google Scholar]
- Feder JL, Xie X, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M. Mayr, Dobzhansky, and Bush and the Complexities of Sympatric Speciation in Rhagoletis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6573–6580. doi: 10.1073/pnas.0502099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filchak KE, Roethele JB, Feder JL. Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature. 2000;407(6805):739–742. doi: 10.1038/35037578. [DOI] [PubMed] [Google Scholar]
- Egan SP, Nosil P, Funk DJ. Selection and genomic differentiation during ecological speciation: isolating the contributions of host association via a comparative genome scan of Neochlamisus bebbianae leaf beetles. Evolution. 2008;62(5):1162–1181. doi: 10.1111/j.1558-5646.2008.00352.x. [DOI] [PubMed] [Google Scholar]
- Funk DJ. Isolating a Role for Natural Selection in Speciation: Host Adaptation and Sexual Isolation in Neochlamisus bebbianae Leaf Beetles. Evolution. 1998;52(6):1744–1759. doi: 10.2307/2411347. [DOI] [PubMed] [Google Scholar]
- Egan SP, Funk DJ. Ecologically dependent postmating isolation between sympatric host forms of Neochlamisus bebbianae leaf beetles. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(46):19426–19431. doi: 10.1073/pnas.0909424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Mooers AÃ. Testing Hypotheses about Ecological Specialization Using Phylogenetic Trees. Evolution. 2005;59(10):2256–2263. [PubMed] [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8(3):336–352. doi: 10.1111/j.1461-0248.2004.00715.x. [DOI] [Google Scholar]
- Schluter D. Evidence for Ecological Speciation and Its Alternative. Science. 2009;323(5915):737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Via S. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution. 1999;53(5):1446–1457. doi: 10.2307/2640891. [DOI] [PubMed] [Google Scholar]
- Pappers S, van der Velde G, Ouborg J. Host preference and larval performance suggest host race formation in Galerucella nymphaeae. Oecologia. 2002;130(3):433–440. doi: 10.1007/s00442-001-0822-3. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends in Ecology & Evolution. 2001;16(7):381–390. doi: 10.1016/S0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Singer MC, McBride CS. Multitrait, host-associated divergence among sets of butterfly populations: implications for reproductive isolation and ecological speciation. Evolution. 2009;64(4):921–933. doi: 10.1111/j.1558-5646.2009.00866.x. [DOI] [PubMed] [Google Scholar]
- Mopper S. Adaptive genetic structure in phytophagous insect populations. Trends in Ecology & Evolution. 1996;11(6):235–238. doi: 10.1016/0169-5347(96)10036-7. [DOI] [PubMed] [Google Scholar]
- Mopper S, Beck M, Simberloff D, Stiling P. Local Adaptation and Agents of Selection in a Mobile Insect. Evolution. 1995;49(5):810–815. doi: 10.2307/2410404. [DOI] [PubMed] [Google Scholar]
- de Jong PW, Breuker CJ, de Vos H, Vermeer KMCA, Oku K, Verbaarschot P, Nielsen JK, Brakefield PM. Genetic differentiation between resistance phenotypes in the phytophagous flea beetle, Phyllotreta nemorum. Journal of Insect Science. 2009;6(69) doi: 10.1673/031.009.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239. doi: 10.2307/1219444. [DOI] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology & Evolution. 2001;16(7):372–380. doi: 10.1016/S0169-5347(01)02198-X. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene Flow in Natural Populations. Annual Review of Ecology and Systematics. 1985;16:393–430. [Google Scholar]
- Stork N. Insect diversity: facts, fiction and speculation*. Biological Journal of the Linnean Society. 1988;35(4):321–337. doi: 10.1111/j.1095-8312.1988.tb00474.x. [DOI] [Google Scholar]
- Farrell BD. "Inordinate Fondness" Explained: Why Are There So Many Beetles? Science. 1998;281(5376):555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- Kippenberg H Lohse GA, Lucht WH. 88. Familie Chrysomelidae Die Käfer Mitteleuropas 19943Supplementband mit Katalogteil. -Bd. 14. Krefeld, Germany: Goecke & Evers Verlag; 65–83.22139176 [Google Scholar]
- Delarze R, Gonseth Y. Guide des milieux naturels de Suisse. 2. Bussigny: Rossolis; 2008. [Google Scholar]
- Hsiao TH, Pasteels JM. In: Advances in Chrysomelidae Biology 1. Cox ML, editor. Leiden, The Netherlands: Backhuys Publishers; 1999. Evolution of host-plant affiliation and chemical defense in Chrysolina-Oreina leaf beetles as revealed by mtDNA phylogenies; pp. 321–342. [Google Scholar]
- Borer M, Van Noort T, Rahier M, Naisbit RE. Positive frequency-dependent selection on warning color in alpine leaf beetles. Evolution. 2010;64(12):3629–3633. doi: 10.1111/j.1558-5646.2010.01137.x. [DOI] [PubMed] [Google Scholar]
- Borer M, Alvarez N, Buerki S, Margraf N, Rahier M, Naisbit RE. The phylogeography of an alpine leaf beetle: Divergence within Oreina elongata spans several ice ages. Molecular Phylogenetics and Evolution. 2010;57(2):703–709. doi: 10.1016/j.ympev.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Triponez Y, Buerki S, Borer M, Naisbit RE, Rahier M, Alvarez N. Discordances between phylogenetic and morphological patterns in alpine leaf beetles attest to an intricate biogeographic history of lineages in postglacial Europe. Molecular Ecology. 2011;20(11):2442–2463. doi: 10.1111/j.1365-294X.2011.05096.x. [DOI] [PubMed] [Google Scholar]
- Rowell-Rahier M. Genetic-structure of leaf-beetles populations -Microgeographic and sexual-differentiation in Oreina cacaliae and Oreina speciosissima. Entomologia Experimentalis Et Applicata. 1992;65(3):247–257. doi: 10.1111/j.1570-7458.1992.tb00678.x. [DOI] [Google Scholar]
- Margraf N, Verdon A, Rahier M, Naisbit RE. Glacial survival and local adaptation in an alpine leaf beetle. Molecular Ecology. 2007;16(11):2333–2343. doi: 10.1111/j.1365-294X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Kalberer NM, Turlings TCJ, Rahier M. An alternative hibernation strategy involving sun-exposed 'hotspots', dispersal by flight, and host plant finding by olfaction in an alpine leaf beetle. Entomologia Experimentalis Et Applicata. 2005;114(3):189–196. doi: 10.1111/j.1570-7458.2005.00244.x. [DOI] [Google Scholar]
- Hasegawa M, Kishino H, Yano Ta. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22(2):160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Yang ZH. Maximum-likelihood phylogenetic estimation from DNA-sequences with variable rates over sites - approximate methods. Journal of Molecular Evolution. 1994;39(3):306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics-the International Journal of the Willi Hennig Society. 1994;10(3):315–319. doi: 10.1111/j.1096-0031.1992.tb00071.x. [DOI] [PubMed] [Google Scholar]
- Nixon KC. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Bremer K. The limits of amino-acid sequence data in angiosperm phylogenetic reconstruction. Evolution. 1988;42(4):795–803. doi: 10.2307/2408870. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, McInerney J, Hirt R, Foster P, Embley M. Of clades and clans: terms for phylogenetic relationships in unrooted trees. Trends in Ecology & Evolution. 2007;22(3):114–115. doi: 10.1016/j.tree.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7(4):574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo N, Felber F, Parisod C, Buerki S, Alvarez N, David J, Guadagnuolo R. Origin and expansion of the allotetraploid Aegilops geniculata, a wild relative of wheat. New Phytologist. 2010;187(4):1170–1180. doi: 10.1111/j.1469-8137.2010.03328.x. [DOI] [PubMed] [Google Scholar]
- Burnier J, Buerki S, Arrigo N, Küpfer P, Alvarez N. Genetic structure and evolution of Alpine polyploid complexes: Ranunculus kuepferi (Ranunculaceae) as a case study. Molecular Ecology. 2009;18(17):3730–3744. doi: 10.1111/j.1365-294X.2009.04281.x. [DOI] [PubMed] [Google Scholar]
- Ribera I, Vogler AP. Speciation of Iberian diving beetles in Pleistocene refugia (Coleoptera, Dytiscidae) Molecular Ecology. 2004;13(1):179–193. doi: 10.1046/j.1365-294X.2003.02035.x. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene-sequences and a compilation of conserved polymerase chain-reaction primers. Annals of the Entomological Society of America. 1994;87(6):651–701. [Google Scholar]
- Zehnder GW, Sandall L, Tisler AM, Powers TO. Mitochondrial DNA Diversity Among 17 Geographic Populations of Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Annals of the Entomological Society of America. 1992;85:234–240. [Google Scholar]
- Emerson BC, Hewitt GM. Phylogeography. Current Biology. 2005;15(10):367–371. doi: 10.1016/j.cub.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Aeschimann D, Lauber K, Moser DM, Theurillat JP. Flora alpina. Bern: Hauptverlag; 2004
- Funk DJ. Molecular systematics of cytochrome oxidase I and 16S from Neochlamisus leaf beetles and the importance of sampling. Molecular Biology and Evolution. 1999;16(1):67–82. doi: 10.1093/oxfordjournals.molbev.a026039. [DOI] [PubMed] [Google Scholar]
- Thompson JN. Specific Hypotheses on the Geographic Mosaic of Coevolution. The American Naturalist. 1999;153:S1–S14. doi: 10.1086/303208. [DOI] [Google Scholar]
- Thompson JN, Cunningham BM. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417(6890):735–738. doi: 10.1038/nature00810. [DOI] [PubMed] [Google Scholar]
- Alvarez N, Romero Napoles J, Anton KW, Benrey B, Hossaert-McKey M. Phylogenetic relationships in the Neotropical bruchid genus Acanthoscelides (Bruchinae, Bruchidae, Coleoptera) Journal of Zoological Systematics and Evolutionary Research. 2006;44(1):63–74. doi: 10.1111/j.1439-0469.2005.00344.x. [DOI] [Google Scholar]
- Kergoat GJ, Alvarez N, Hossaert-Mckey M, Faure N, Silvain JF. Parallels in the evolution of the two largest New and Old World seed-beetle genera (Coleoptera, Bruchidae) Molecular Ecology. 2005;14(13):4003–4021. doi: 10.1111/j.1365-294X.2005.02702.x. [DOI] [PubMed] [Google Scholar]
- Vermeer KMCA, Dicke M, de Jong PW. The potential of a population genomics approach to analyse geographic mosaics of plant-insect coevolution. Evolutionary Ecology. 2011;25:977–992. doi: 10.1007/s10682-010-9452-8. [DOI] [Google Scholar]
- de Jong PW, Nielsen JK. Host plant use of Phyllotreta nemorum: do coadapted gene complexes play a role? Entomologia Experimentalis Et Applicata. 2002;104(1):207–215. doi: 10.1046/j.1570-7458.2002.01008.x. [DOI] [Google Scholar]
- Nosil P. Divergent Host Plant Adaptation and Reproductive Isolation between Ecotypes of Timema cristinae Walking Sticks. The American Naturalist. 2007;169(2):151–162. doi: 10.1086/510634. [DOI] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature. 2002;417(6887):440–443. doi: 10.1038/417440a. [DOI] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP. Reproductive Isolation Driven by the Combined Effects of Ecological Adaptation and Reinforcement. Proceedings of the Royal Society B: Biological Sciences. 2003;270(1527):1911–1918. doi: 10.1098/rspb.2003.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge AG. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes) Systematic Zoology. 1989;38(1):7–25. doi: 10.2307/2992432. [DOI] [Google Scholar]
- Planet PJ, Sarkar IN. mILD: a tool for constructing and analyzing matrices of pairwise phylogenetic character incongruence tests. Bioinformatics. 2005;21(24):4423–4424. doi: 10.1093/bioinformatics/bti744. [DOI] [PubMed] [Google Scholar]
- Sikes DS, Lewis PO. PAUPRat: PAUP implementation of the parsimony ratchet. Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs. 2001.
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Sinauer Associates, Sunderland, Massachusetts; 2002. [Google Scholar]
- Sorenson MD, Franzosa EA. TreeRot, version 3. Boston University, Boston, MA 2007.
- Baker RH, DeSalle R. Multiple sources of character information and the phylogeny of Hawaiian Drosophilids. Systematic Biology. 1997;46(4):654–673. doi: 10.1093/sysbio/46.4.654. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Program distributed by the author Evolutionary Biology Centre, Uppsala University. 2004.
- Akaike H. In: Second International Symposium on Information Theory. Kiado A. Budapest, editor. 1973. Information theory and an extension of the maximum likelihood principle; pp. 267–281. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. Inference from Iterative Simulation Using Multiple Sequences. Statistical Science. 1992;7(4):457–511. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007. http://tree.bio.ed.ac.uk/software/tracer
- Gugerli F, Englisch T, Niklfeld H, Tribsch A, Mirek Z, Ronikier M, Zimmermann NE, Holderegger R, Taberlet P. Relationships among levels of biodiversity and the relevance of intraspecific diversity in conservation - a project synopsis. Perspectives in Plant Ecology, Evolution and Systematics. 2008;10(4):259–281. doi: 10.1016/j.ppees.2008.07.001. [DOI] [Google Scholar]
- Arrigo N, Holderegger R, Alvarez N. In: Population Genomics: Methods and Protocols. Bonin A, Pompanon F., editor. New York, USA: Humana Press; From raw AFLP chromatograms to ready-to-use binary matrices using RawGeno 2.0, an user-friendly interface for automatized and semi-automatized bining and scoring of genotypes in the open-source R environment. in press . [Google Scholar]
- Arrigo N, Tuszynski JW, Ehrich D, Gerdes T, Alvarez N. Evaluating the impact of scoring parameters on the structure of intra-specific genetic variation using RawGeno, an R package for automating AFLP scoring. BMC Bioinformatics. 2009;10(33) doi: 10.1186/1471-2105-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A K-means clustering algorithm. Applied Statistics. 1979;28:100–108. doi: 10.2307/2346830. [DOI] [Google Scholar]
- Arrigo N, Guadagnuolo R, Lappe S, Pasche S, Parisod C, Felber F. Evolutionary Applications. 5. Vol. 4. Gene flow between wheat and wild relatives: empirical evidence from Aegilops geniculata, Ae. neglecta and Ae. triuncialis; pp. 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]