Abstract
The intimate connection between telomerase regulation and human disease is now well established. The molecular basis for telomerase regulation is highly complex and entails multiple layers of control. While the major target of enzyme regulation is the catalytic subunit TERT, the RNA subunit of telomerase is also implicated in telomerase control. In addition, alterations in gene dosage and alternative isoforms of core telomerase components have been described. Finally, telomerase localization, recruitment to the telomere and enzymology at the chromosome terminus are all subject to modulation. In this review we summarize recent advances in understanding fundamental mechanisms of telomerase regulation.
Keywords: telomere, TERT, TR, ribonucleoprotein, repeat addition processivity, TERRA
1. Introduction
The ends of eukaryotic chromosomes are defined by a tract of simple G-rich repeats and associated proteins that constitute the functional unit termed the telomere. The length of the telomeric DNA tract is highly dynamic and subjected to forces that both shorten and extend the repeat array. Telomeres must be long enough to assemble a protective “cap” that can distinguish the terminus from a double-strand break. Dysfunctional telomeres trigger cell cycle arrest, genome instability and in humans, replicative cell senescence and apoptosis [1, 2]. On the other hand, telomeric DNA loss through incomplete DNA replication or nucleolytic processing suppresses tumorigenesis by limiting the proliferative potential of normal somatic cells. At the heart of this balancing act is telomerase, a ribonucleoprotein reverse transcriptase that consists of two core components: a catalytic reverse transcriptase subunit (TERT), and an RNA subunit (TR or TER), which serves as a template for telomeric DNA addition by TERT.
Telomerase is a highly regulated enzyme and in normal individuals its activity is confined to cells with extended proliferation potential: the germline, embryonic tissues and self-renewing stem cell populations of the hematopoetic system and skin. In other tissues, telomerase is inactivated during gestation, thereby restricting the proliferation program [3]. Mis-regulation of telomerase has dire consequences. As discussed elsewhere in this volume, reactivation of telomerase is associated with approximately 90% of human cancers [4], while insufficient telomerase activity is linked to a litany of stem cell disorders including dyskeratosis congenita, aplastic anemia and idiopathic pulmonary fibrosis [5, 6].
The molecular basis for telomerase regulation is highly complex and entails multiple levels of control. A major determinant of enzyme activity is transcriptional regulation of the catalytic subunit TERT. However, emerging data indicate that TERT is subjected to both post-transcriptional and post-translational control. In addition, transcriptional regulation of TR has also been reported. In some instances the number of genes encoding TERT and TR is expanded, increasing enzyme activity or, over evolutionary time, giving rise to alternative ribonucleoprotein complexes. Finally, telomerase recruitment and enzyme activity at the chromosome terminus are modulated by telomere-associated proteins and by telomeric RNA transcripts. Here we summarize some of the recent advances in understanding telomerase regulation.
2. Transcriptional regulation of TERT
TERT gene expression parallels telomerase activity in many multicellular organisms. For example, in the model plant Arabidopsis, TERT mRNA peaks in flowers and suspension cell culture where telomerase activity is most abundant, but can barely be detected in leaves where telomerase is strongly repressed [4]. Similarly, human TERT is expressed during early development, but with the exception of proliferating cells or renewal tissues, it is absent in most normal somatic cells [7, 8]. Transient transfection of an hTERT promoter-luciferase reporter reveals an expression pattern that mirrors the telomerase activity profile [9]. These and other observations argue that the TERT promoter is a major target of enzyme regulation.
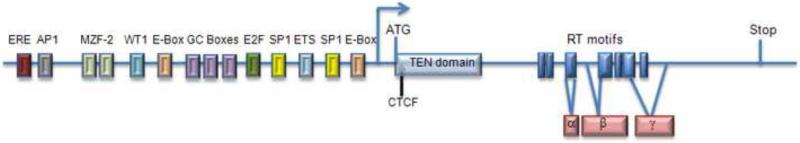
The hTERT promoter has been extensively studied [reviewed in [10, 11]]. A plethora of transcription factor binding sites allow for nuanced hTERT expression (Figure 1). For example, TERT transcription is influenced by Sp1 [11], a general transcription factor that interacts with the TATA binding protein. Notably, TATA boxes are not found in the hTERT promoter, and yet mutations in Sp1 binding sites completely abolish hTERT promoter activity [9, 11]. Telomerase expression is also controlled by oncogenes (i.e. c-Myc) as well as tumor suppressors (i.e. WT1). c-Myc influences telomerase expression by binding the two E-boxes present in the hTERT promoter. Increased expression of c-Myc, which is observed in cancer cells, results in increased telomerase activity [2, 11-13]. Binding of WT1 (Wilm's tumor suppressor) to the hTERT promoter negatively regulates hTERT expression [14]. On the other hand, inactivation of WT1 is associated with telomerase activation during tumorigenesis [15].
Figure 1.
Simplified diagram of regulatory and functional elements associated with the hTERT gene. Rightward arrow depicts start of transcription. Start and stop of translation are indicated. A subset of hTERT promoter elements as well as the binding site for CTCF, a chromatin insulator element, are indicated. Within the TERT coding region, dark blue boxes denote reverse transcriptase motifs, while the light blue box shows the TEN domain. The three major splicing isoforms of TERT (α, β and γ) are shown in pink. Drawing is not to scale. See text for details.
Other factors repress hTERT expression. These include MZF-2, a zinc finger transcription factor [16], and member of the E2F family of transcriptional regulators involved in cell-cycle progression [17]. Repression of hTERT transcription may also be accomplished by inhibiting potential activators like c-Myc, via the TGFβ/Smad signaling pathway [18] or BRCA1, a tumor suppressor for hereditary breast and ovarian cancers that is involved in DNA repair [19, 20]. Finally, it has been suggested that p53 may be involved in the negative regulation of hTERT expression, because most cancers are deficient in p53 [21, 22].
Several lines of evidence indicate that silencing of hTERT is under epigenetic control [23]. The TERT promoter lies within a highly condensed chromatin domain [24] and is associated with hypoacetyled core histones [25, 26]. In addition, both histone and CpG methylation are implicated in hTERT regulation [27, 28], although in the latter case there is controversy as to whether methylation contributes to positive or negative regulation of hTERT expression in cancer cells [29, 30]. CTCF, an insulator that organizes chromatin domains to modulate transcription [31], binds the first exon of hTERT (Figure 1) in cells that repress hTERT expression [32]. Finally, activation of the hTERT promoter is correlated with dramatic changes in the surrounding chromatin environment [23].
3. Post-translational regulation of TERT
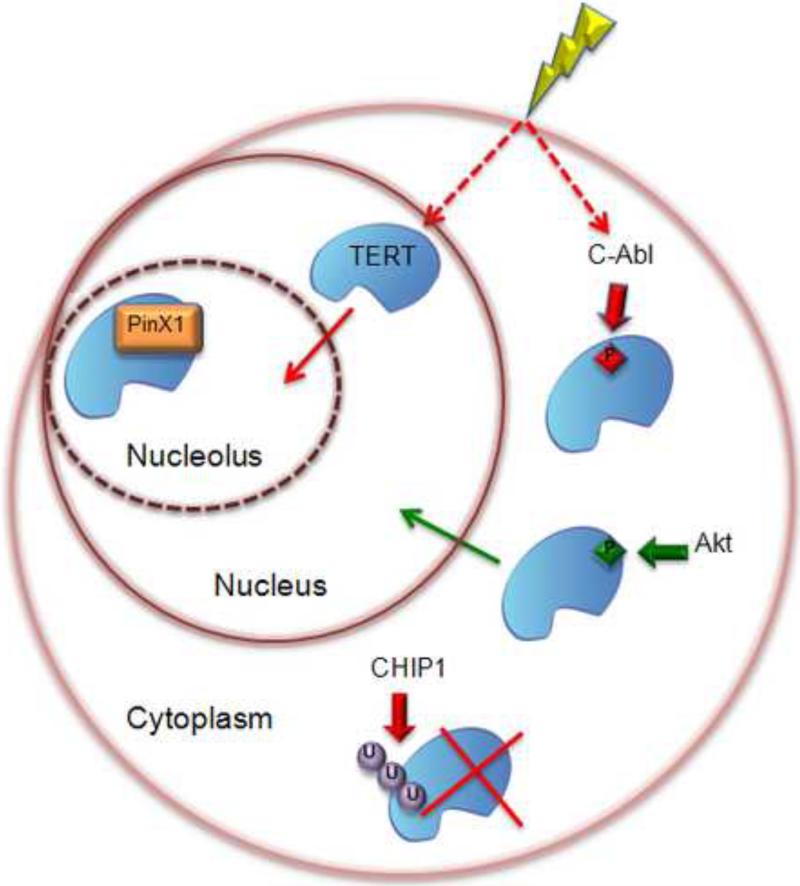
Post-translational regulation of telomerase is supported by the observation that TERT mRNA levels do not always correlate with telomerase enzyme activity [33-35]. Moreover, not all cells with active telomerase are capable of maintaining telomere tracts [36]. The biogenesis and assembly of the telomerase RNP represent other avenues of enzyme regulation and are discussed in detail elsewhere in this issue [37]. Figure 2 depicts some of the post-translational modifications of TERT and how they affect protein stability, subcellular localization and ultimately, enzyme activity. A number of studies indicate telomerase activity is modulated by phosphorylation [reviewed in [38]. Putative phosphorylation sites are present in the TERT sequences from mammals [39] and plants [35]. At least two kinases are implicated in hTERT phosphorylation. In response to ionizing radiation, hTERT is phosphorylated by c-Abl leading to a three-fold reduction in telomerase activity [40]. Mice lacking c-Abl display increased telomerase activity and telomere elongation [40]. Thus, c-Abl negatively regulates telomerase activity. In contrast, phosphorylation of hTERT by Akt correlates with increased telomerase activity, presumably resulting from hTERT translocation from the cytoplasm to the nucleus [39, 41].
Figure 2.
Summary of post-translational modifications of hTERT and their consequences for telomerase activity. Red arrows indicate negative regulation, green arrows positive regulation. See text for details.
Ubiquitination may also influence telomerase activity. The MKRN1 ubiquitin ligase (E3) interacts with hTERT in a yeast-two hybrid assay [42]. Over-expression of MKRN1 leads to degradation of TERT, resulting in decreased telomerase activity and shortened telomeres. This finding suggests that telomerase activity is modulated by TERT stability. The half-life of human telomerase activity is approximately 24 hrs [43], while the half-life of TER is extraordinarily long, five days [44]. These observations support the conclusion that hTERT stability contributes to telomerase regulation. A recent study shows that CHIP (C terminus of Hsc70-interacting protein), a co-chaperone with E3 ubiquitin ligase, controls hTERT stability in the cytoplasm [45]. CHIP interaction with hTERT leads to polyubiquitination, blocking hTERT entry into the nucleus and culminating in proteolytic degradation (Figure 2). Intriguingly, the interaction of CHIP with hTERT peaks in G2/M, and is diminished in S phase when telomerase acts on telomeres. Thus, CHIP may regulate telomerase activity during the cell cycle by controlling the intracellular trafficking and consequently the stability of hTERT [45].
The subnuclear localization of telomerase is also dynamically controlled during the cell cycle and contributes to enzyme regulation [46-48]. Emerging data argue that delivery of enzymatically active telomerase to the chromosome terminus first requires the passage of TR through Cajal bodies via a telomerase-specific Cajal body protein termed TCAB1 [49]. Specifically, Cajal bodies are proposed to act as a type of processing center, where TR and possibly associated proteins are modified, in some fashion before the enzyme can become fully competent for telomere elongation [50, 51] (see below).
Negative regulation of telomerase activity can be achieved by sequestration of the enzyme in the nucleolus. Following DNA damage, hTERT transiently moves from the nucleoplasm to the nucleolus (Figure 2). This re-localization is hypothesized to reduce the probability of de novo telomere formation at sites of DNA damage [46]. PinX1, an interaction partner for the human shelterin component TRF1, is also proposed to regulate telomerase by sequestration [52] (Figure 2). PinX1 directly binds hTERT [53, 54] and hTR [53] and inhibits telomerase activity in vitro [54]. As in human cells, the interaction of PinX1 with Est2 (TERT) leads to sequestration in the yeast nucleolus [55].
4. Transcriptional and post-transcriptional regulation of TR
Although hTR expression is detected in some tissues where hTERT is not, hTR abundance increases in tumors relative to normal cells [56, 57], arguing that hTR abundance contributes to telomerase regulation. Transcription of hTR is activated by Sp1 and HIF-1 and repressed by Sp3, which integrates cues from the MAPK signaling cascade to silence the hTR promoter [reviewed in [10]. Furthermore, like hTERT, hTR transcription appears to be subjected to epigenetic control as repression of hTR expression is associated with decreased levels of H3 and H4 acetylation [27]. Finally, at least six sites in hTR are subjected to post-transcriptional modification by pseudouridylation [58]. Intriguingly, two of these sites lie in a highly conserved domain essential for telomerase catalytic activity. In vitro telomerase reconstitution assays with model pseudouridylated hTR result in modest alterations in enzyme activity and processivity. Whether hTR modification regulates telomerase activity in vivo is still an open question, although recent findings suggest that this is a distinct possibility (see below).
5. Gene dosage and alternative TERT and TR isoforms
TERT and TR exist as single copy genes in most organisms studied and a null mutation in TERT or TR is ultimately lethal. In mice, both TERT [59] and TR [60, 61] are haploinsufficient for maintaining telomere tracts. Indeed, the etiology underlying a growing list of stem cell diseases is linked to hemizygosity of core telomerase subunits [62]. Conversely, amplification of chromosomal loci encoding TERT or TR is correlated with tumor formation [63-66]. Thus, gene dosage plays a critical role in telomerase regulation.
Alternative splicing of TERT is widespread and has been described for a number of multicellular eukaryotes including a variety of mammals [28, 67, 68], birds [69], fish [70] and plants [71]. Ten different splice variants have been reported for human TERT [28, 72-76]. Three of the major products are depicted in Figure 2. Alternative splicing of hTERT results in both nucleotide deletion and mutation. Several splice variants have been correlated with changes in telomerase activity [77-79]. The most well-studied is hTERTα, which encodes a 183 bp deletion with an accompanying nonsense mutation. Expression of TERTα correlates with decreased telomerase activity, and hence this isoform is proposed to act as dominant negative inhibitor of telomerase activity [74, 75].
Variant isoforms of telomerase subunits have also emerged as a consequence of gene duplication. Three different TERT genes are found the ciliated protozoan Euplotes crassus [80]. These genes encode proteins that display 83-87% identity and are differentially expressed during the sexual stage of the life cycle [80]. Expression of the EcTERT-2 gene is limited to macronuclear development, a period when telomeres form de novo on thousands of newly generated mini-chromosomes. In contrast, EcTERT-1 and EcTERT-3 are expressed during vegetative growth when telomerase performs its canonical function of maintaining pre-existing telomere tracts. Remarkably, the EcTERT-2 gene is destroyed by programmed DNA elimination following new telomere formation, presumably to control promiscuous telomere addition at sites of spontaneous DNA damage [80]. Like most other model organisms, E. crassus encodes only a single telomerase RNA. Thus, the E. crassus TER assembles with different TERT subunits into alternative RNP complexes to facilitate a developmentally programmed switch from de novo telomere formation to telomere maintenance.
A second example of alternative telomerase subunits is found in Arabidopsis thaliana [81, 82]. A. thaliana encodes two different template RNA components, TER1 and TER2. The two RNAs assemble with the single TERT isoform into alternative telomerase particles. TER1 is a typical telomerase template critical for telomere maintenance [81]. TER2, on the other hand, is a novel negative regulator of enzyme activity [82]. Why an alternative RNP evolved to negatively regulate the plant telomerase is unclear since, unlike mammals, plants do not face the threat of metastatic cancer as a consequence of unbridled telomerase activity. These findings suggest that additional modes of restraining telomerase could be elucidated in mammals where inappropriate expression is much more deleterious.
6. Regulation of telomerase recruitment to the telomere
Once an active telomerase RNP particle is formed, it must engage the chromosome terminus to facilitate the incorporation of telomere repeats. Here we briefly consider how crosstalk between telomerase RNP components and telomere capping proteins influences the recruitment of telomerase to the telomere. Mechanisms to regulate the length of the telomere tract are discussed elsewhere in this volume [83].
The interaction of telomerase with the telomere is best understood in budding yeast. Lundblad and colleagues established that Est1, a non-catalytic telomerase holoenzyme component, physically links the RNP to the telomere through interactions with the telomerase RNA (Tlc1) and Cdc13, a member of the CST telomere capping complex [84]. The telomerase-telomere interaction is regulated during the cell cycle, peaking during S phase [85]. In addition, the interaction of Est1 with Tlc1 is controlled by cell cycle regulated proteolytic degradation [86]. Moreover, a number of studies suggest that Est1-Cdc13 association is controlled via phosphorylation of Cdc13 by Cdk1 (Cdc28) [87] or by Tel1 (ATM) [88-91]. However, recent analyses of Tel1 consensus phosphorylation sites on Cdc13 do not support this model [92]. In conjunction with its role recruiting telomerase to the telomere, Est1 influences the interaction of telomerase with its DNA primer in vitro to stimulate elongation [93-95].
The Ku heterodimer is also implicated in telomerase recruitment. In budding yeast Ku directly interacts with a stem loop in Tlc1 [96]. Cells lacking Ku exhibit defects in Tlc1 nuclear localization and have shorter telomeres with long G-overhangs [97, 98]. The current view is that Ku assists in positioning telomerase at the telomere in G1 to promote telomere synthesis by the enzyme in S phase [99].
The mechanism of telomerase recruitment is less clear in multicellular organisms. Telomerase associates with components of shelterin, the telomere capping complex in human cells [98]. One of these proteins, TPP1, is implicated in telomerase recruitment [100, 101]. TPP1 forms a subcomplex POT1, another shelterin component, simulating interaction of POT1 with the single-strand 3’ overhang on the chromosome end [101]. TPP1 also interacts with the telomerase RNP, binding the TEN domain of TERT [102]. Intriguingly, the TEN domain promotes repeat addition processivity of the core enzyme [103, 104]. Whether interaction of TPP1 with this region telomerase alters enzyme activity is unknown. Furthermore, unraveling precisely how TPP1 influences telomerase function in vivo is hindered by the fact that depletion of TPP1 also dislodges POT1 from telomeres, activating an immediate DNA damage response and cell cycle arrest, not the ever-shorter-telomere phenotype expected for defect in telomerase recruitment [100, 105].
7. Control of telomerase processivity at the chromosome terminus
Once telomerase engages the single-strand overhang on the telomere, telomere repeat incorporation is facilitated by two enzyme modes: a processive reaction in which multiple telomere repeats are added in a single DNA binding event, and a non-processive or distributive mode in which only one or two repeats are incorporated. The TEN1 domain that promotes repeat addition processivity (RAP) of the core enzyme [103, 104, 106]. Collins and colleagues discovered a new accessory factor, p82, for the Tetrahymena telomerase that strongly stimulates RAP of the catalytic core [107]. In addition, RAP of human telomerase is influenced by the putative human telomerase recruitment factor TPP1 in vitro [108, 109].
Analysis of telomerase dynamics in vivo reveals a striking correlation between RAP and the length of the telomere tract telomerase acts upon. Telomerase is preferentially recruited to shorter telomere tracts [110-112] and in yeast the enzyme does not extend every telomere in every cell cycle [110]. However, telomerase RAP is increased at critically shortened telomeres relative to telomeres in the wild type size range [113]. Thus, modulation of telomerase processivity plays a direct role in establishing telomere length homeostasis.
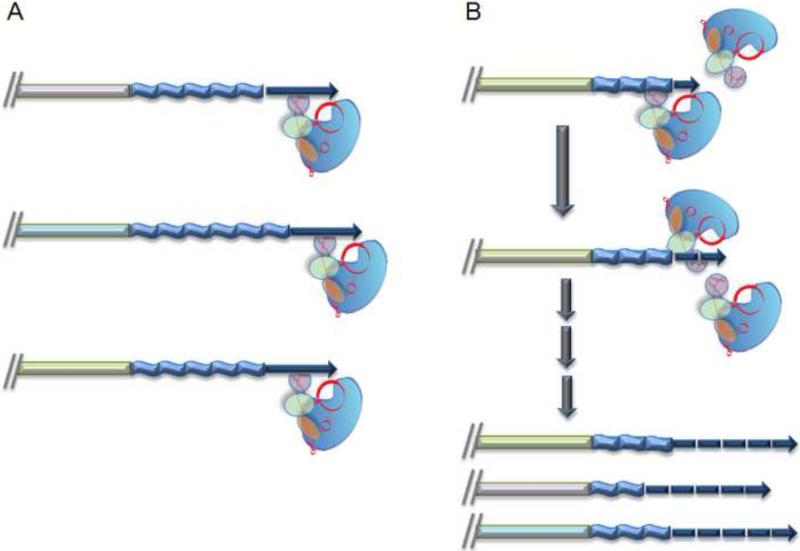
Recent studies reveal that processivity of human telomerase is highly regulated in cancer cells (Figure 3). Unlike yeast, human telomerase extends every telomere end every cell cycle [114]. However, RAP is altered depending on whether the enzyme is establishing or maintaining telomere length homeostasis [51]. On shortened telomere tracts recovering from telomerase inhibition, telomerase acts in a distributive manner. Multiple telomerase enzymes sequentially engage the same chromosome end to rapidly extend the telomere. In contrast, when telomerase maintaining telomere length homeostasis, RAP is strongly stimulated and only a single telomerase enzyme associates with each telomere. Approximately 10 repeats are added before the enzyme dissociates. This remarkable switch from a distributive to a processive mode for telomere synthesis requires trafficking of hTR through Cajal bodies [51]. Hence, post-transcriptional modification of hTR or an auxillary factor may modulates telomerase processivity in vivo.
Figure 3.
A model for the regulation of repeat addition processivity (RAP) of telomerase is regulated in human cancer cells. Diagram summarizes results from [51]. A) Telomerase displays high RAP in cells that maintain telomere length. In this setting, a single telomerase enzyme binds each chromosome end, adding multiple telomere repeats (extended blue arrows) before being released from the DNA. B) Telomerase RAP is decreased following artificial telomere shortening. Under these conditions, telomerase is less processive (fragmented blue arrows), adding fewer telomeric repeats before dissociation (low RAP). However, multiple enzymes bind each chromosome end sequentially to rapidly extend telomere tracts and thereby reestablish telomere length homeostasis.
In budding yeast, telomerase processivity is negatively regulated by the Pif1 helicase. Pif1 unwinds telomeric DNA from the telomerase RNA template, dislodging telomerase from the chromosome terminus [115]. In cells over-expressing Pif1, the interaction of telomerase with telomeres is reduced leading to telomere shortening. Conversely, Pif1 depletion results in telomere elongation. In conjunction with its action at native chromosome ends, Pif1 also promotes genome stability by ejecting telomerase from non-telomeric DNA substrates [116]. Pif1 is phosphorylated in response to DNA damage, which stimulates its helicase activity, decreasing the opportunity for telomerase to form incorporate telomere repeats at sites of DNA damage [117]. Although mammalian PIF1 physically associates with telomerase [118, 119], it is not required for telomere length regulation [115, 118, 119]. However, it is currently unclear whether human PIF1 regulates telomerase activity at double-strand breaks.
8. Telomerase regulation by TERRA
One of the defining features of telomeres is that they are heterochromatic. Consequently, it was surprising when telomere transcripts termed Telomeric Repeat containing RNA (TERRA) were discovered. TERRA molecules are long non-coding RNAs transcribed by RNA polymerase II from subtelomeric and telomeric DNA. Telomere transcription is reported for a number of eukaryotes including mammals, fish and yeast [120-123]. Arabidopsis appears to be unusual in that it transcribes both strands of the telomere, generating TERRA as well as TERRA antisense transcripts, ARRET [123]. Notably, Arabidopsis TERRA and ARRET are not derived exclusively from telomeres; they are also transcribed from centromere-proximal telomeric sequences.
TERRA varies in size ranging from 100nt to 9000nt in mammals and ~380nt in yeast [120]. TERRA molecules are capped by 7-methylguanosine (m7G) [122] and at least a subset of them bear a 3’ poly(A) tail [121, 122]. Intriguingly, only poly(A) minus TERRA is detected in chromatin [124, 125], suggesting that TERRA may have different functions. TERRA interacts with numerous RNA binding proteins [125, 126]. One of these is hnRNPA1, a single-strand nucleic acid binding protein that recognizes RNA as well as telomeric DNA [127, 128]. A recent study reveals that the interaction of TERRA with hnRNPA1 plays a pivotal role in facilitating the exchange of single-strand binding proteins at the chromosome terminus [127]. Following chromosomal replication, the 3’ overhang on the telomere is initially bound by RPA, which is subsequently replaced by hnRPA1. hnRNPA1 is then dislodged from the DNA through its interaction with TERRA, allowing the POT1/TPP1 components of the shelterin complex to bind and thus establish a functional telomere cap.
In addition to this newly discovered role in promoting changes in telomere protein composition, TERRA is also postulated to reinforce the heterochromatic nature of the chromosome terminus [120-122]. Moreover, because TERRA is complementary to the template domain of TR, it has the potential to negatively regulate telomerase. Indeed several studies support this prediction [121, 122, 126]. Human TERRA physically associates with telomerase in nuclear extracts, and as predicted, base pairs with the complementary template region of TR [126]. TERRA may also interact with TERT independently of TR. Further evidence for telomerase regulation by TERRA has been obtained in yeast rat-1 mutants, where TERRA levels increase as telomeres shorten [121]. It is unknown whether TERRA is released from the telomere to inhibit telomerase in trans, or acts in cis on the chromosome terminus to block telomerase action. The latter model is appealing as long telomeres correlate with increased levels of TERRA. Thus, negative regulation of telomerase by TERRA in cis could provide an elegant feedback mechanism to promote telomere length regulation.
9. Conclusions
Although the initial studies of telomerase regulation focused on transcriptional control of core subunits, it is now apparent that the telomerase RNP is subjected to a highly sophisticated network of regulatory pathways that modulate subunit abundance, intracellular trafficking and the interaction with and activity on the chromosome terminus. The necessity of governing telomerase activity is underscored by the remarkable conservation of factors, both protein and RNA based, that control enzyme behavior. Our understanding of telomerase and its pivotal role in safeguarding the genome will undoubtedly mature as new links between enzyme regulation and fundamental aspects of cellular physiology continue to be revealed.
Acknowledgements
Research in the Shippen lab is supported by grants from NSF (MCB-0843399 and MCB-1052018) and NIH (GM065383) to D.E.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare they have no conflicts of interest.
References
- 1.Greider CW. Telomeres. Telomerase and Senescence. BioEssays. 1990;12:363–369. doi: 10.1002/bies.950120803. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg RA. Telomeres, crisis and cancer. Curr. Mol. Med. 2005;5:213–218. doi: 10.2174/1566524053586590. [DOI] [PubMed] [Google Scholar]
- 3.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Devel. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Prescott J, Wentzensen IM, Savage SA, De Vivo I. Epidemiologic evidence for a role of telomere dysfunction in cancer etiology. Mutat. Res. 2011 doi: 10.1016/j.mrfmmm.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson ND, Bertuch AA. Dyskeratosis congenita as a disorder of telomere maintenance. Mutat. Res. 2011 doi: 10.1016/j.mrfmmm.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armanios M. Beyond dyskeratosis congenita: pulmonary phenotypes as a manifestation of telomerase insufficiency. Mutat. Res. 2011 In press. [Google Scholar]
- 7.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 8.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat. Rev. Drug. Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 9.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairney CJ, Keith WN. Telomerase redifined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochemie. 2008;90:13–23. doi: 10.1016/j.biochi.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell. Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 13.Grandori C, Eisenman RN. Myc target genes. Trends Biochem. Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 14.Oh S, Song Y, Yim J, Kim TK. The Wilms’ tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J. Biol. Chem. 1999;274:37473–37478. doi: 10.1074/jbc.274.52.37473. [DOI] [PubMed] [Google Scholar]
- 15.Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167–1176. doi: 10.1093/carcin/bgg085. [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto K, Kyo S, Takakura M, Kanaya T, Kitagawa Y, Itoh H, Takahashi M, Inoue M. Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (hTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nucleic Acids Res. 2000;28:2557–2562. doi: 10.1093/nar/28.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso MM, Fueyo J, Yung WK, Gomez-Manzano C. E2F1 and telomerase: alliance in the dark side. Cell Cycle. 2006;5:930–935. doi: 10.4161/cc.5.9.2698. [DOI] [PubMed] [Google Scholar]
- 18.Lacerte A, Korah J, Roy M, Yang XJ, Lemay S, Lebrun JJ. Transforming growth factor-beta inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal. 2008;20:50–59. doi: 10.1016/j.cellsig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Ballal RD, Saha T, Fan S, Haddad BR, Rosen EM. BRCA1 localization to the telomere and its loss from the telomere in response to DNA damage. J. Biol. Chem. 2009;284:36083–36098. doi: 10.1074/jbc.M109.025825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 21.Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, Rotter V. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 22.Xu D, Wang Q, Gruber A, Bjorkholm M, Chen Z, Zaid A, Selivanova G, Peterson C, Wiman KG, Pisa P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Zhao Y, Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2010;1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Zhu J. Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. J. Biol. Chem. 2003;278:18842–18850. doi: 10.1074/jbc.M209544200. [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol. Biol. Cell. 2007;18:669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atkinson SP, Hoare SF, Glasspool RM, Keith WN. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65:7585–7590. doi: 10.1158/0008-5472.CAN-05-1715. [DOI] [PubMed] [Google Scholar]
- 28.Wick M, Zubov D, Hagen G. Genomic organization and promoter chracterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 29.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 30.Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Exp. Cell. Res. 2003;289:326–334. doi: 10.1016/s0014-4827(03)00281-7. [DOI] [PubMed] [Google Scholar]
- 31.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 32.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850–6860. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR. Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium, International journal of cancer. J. Intl. Cancer. 2000;85:330–335. [PubMed] [Google Scholar]
- 34.Rohde V, Sattler HP, Bund T, Bonkhoff H, Fixemer T, Bachmann C, Lensch R, Unteregger G, Stoeckle M, Wullich B. Expression of the human telomerase reverse transcriptase is not related to telomerase activity in normal and malignant renal tissue. Clin. Cancer Res. 2000;6:4803–4809. [PubMed] [Google Scholar]
- 35.Oguchi K, Tamura K, Takahashi H. Characterization of Oryza sativa telomerase reverse transcriptase and possible role of its phosphorylation in the control of telomerase activity. Gene. 2004;342:57–66. doi: 10.1016/j.gene.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J. Telomerase structure and biogenesis. Mutat Res. 2011 doi: 10.1016/j.mrfmmm.2011.11.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aisner DL, Wright WE, Shay JW. Telomerase regulation: not just flipping the switch. Curr. Opin. Genet. Dev. 2002;12:80–85. doi: 10.1016/s0959-437x(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 39.Kang SS, Kwon T, Kwon DY, Do SI. Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J. Biol. Chem. 1999;274:13085–13090. doi: 10.1074/jbc.274.19.13085. [DOI] [PubMed] [Google Scholar]
- 40.Kharbanda S, Kumar V, Dhar S, Pandey P, Chen C, Majumder P, Yuan ZM, Whang Y, Strauss W, Pandita TK, Weaver D, Kufe D. Regulation of the hTERT telomerase catalytic subunit by the c-Abl tyrosine kinase. Curr. Biol. : CB. 2000;10:568–575. doi: 10.1016/s0960-9822(00)00483-8. [DOI] [PubMed] [Google Scholar]
- 41.Minamino T, Kourembanas S. Mechanisms of telomerase induction during vascular smooth muscle cell proliferation. Circ. Res. 2001;89:237–243. doi: 10.1161/hh1501.094267. [DOI] [PubMed] [Google Scholar]
- 42.Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, Chung IK. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005;19:776–781. doi: 10.1101/gad.1289405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holt SE, Aisner DL, Shay JW, Wright WE. Lack of cell cycle regulation of telomerase activity in human cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10687–10692. doi: 10.1073/pnas.94.20.10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and post-transcriptional mechanisms regulate human telomerase template RNA (hTR) levels. Mol. Cell. Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH, Khadka P, Baek SH, Chung IK. CHIP promotes human telomerase reverse transcriptase degradation and negatively regulates telomerase activity. J. Biol. Chem. 2010;285:42033–42045. doi: 10.1074/jbc.M110.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong JM, Kusdra L, Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 2002;4:731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 47.Yang SW, Jin E, Chung IK, Kim WT. Cell cycle-dependent regulation of telomerase activity by auxin, abscisic acid and protein phosphorylation in tobacco BY-2 suspension culture cells. Plant J. 2002;29:617–626. doi: 10.1046/j.0960-7412.2001.01244.x. [DOI] [PubMed] [Google Scholar]
- 48.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venteicher AS, Artandi SE. TCAB1: driving telomerase to Cajal bodies. Cell Cycle. 2009;8:1329–1331. doi: 10.4161/cc.8.9.8288. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol. Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J, Jin R, Zhang B, Yang PX, Chen H, Bai YX, Xie Y, Huang C, Huang J. Characterization of a novel effect of hPinX1 on hTERT nucleolar localization. Biochem. Bioph. Res. Comm. 2007;353:946–952. doi: 10.1016/j.bbrc.2006.12.123. [DOI] [PubMed] [Google Scholar]
- 53.Banik SS, Counter CM. Characterization of interactions between PinX1 and human telomerase subunits hTERT and hTR. J Biol Chem. 2004;279:51745–51748. doi: 10.1074/jbc.M408131200. [DOI] [PubMed] [Google Scholar]
- 54.Zhou XZ, Lu KP. The Pin2/TRF1-interacting protein PinX1 is an potent telomerase inhibitor. Cell. 2001;107:347–359. doi: 10.1016/s0092-8674(01)00538-4. [DOI] [PubMed] [Google Scholar]
- 55.Lin J, Blackburn EH. Nucleolar protein PinX1p regulates telomerase by sequestering its protein catalytic subunit in an inactive complex lacking telomerase RNA. Genes Dev. 2004;18:387–396. doi: 10.1101/gad.1171804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng J, Funk WD, Wang S-S, Weinrich SL, Avilion AA, Chiu C-P, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 57.Soder AI, Going JJ, Kaye SB, Keith WN. Tumour specific regulation of telomerase RNA gene expression visualized by in situ hybridization. Oncogene. 1998;16:979–983. doi: 10.1038/sj.onc.1201620. [DOI] [PubMed] [Google Scholar]
- 58.Kim NK, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 2010;38:6746–6756. doi: 10.1093/nar/gkq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, Harrington L. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. : CB. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 60.Chiang YJ, Hemann MT, Hathcock KS, Tessarollo L, Feigenbaum L, Hahn WC, Hodes RJ. Expression of telomerase RNA template, but not telomerase reverse transcriptase, is limiting for telomere length maintenance in vivo. Mol. Cell. Biol. 2004;24:7024–7031. doi: 10.1128/MCB.24.16.7024-7031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hathcock KS, Hemann MT, Opperman KK, Strong MA, Greider CW, Hodes RJ. Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrington L. Making the most of a little: dosage effects in eukaryotic telomere length maintenance. Chromosome Res. 2005;13:493–504. doi: 10.1007/s10577-005-0994-5. [DOI] [PubMed] [Google Scholar]
- 63.Soder AI, Hoare SF, Muir S, Going JJ, Parkinson EK, Keith WN. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- 64.Bryce LA, Morrison N, Hoare SF, Muir S, Keith WN. Mapping of the gene for the human telomerase reverse transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in situ hybridization. Neoplasia. 2000;2:197–201. doi: 10.1038/sj.neo.7900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saretzki G, Petersen S, Petersen I, Kolble K, von Zglinicki T. hTERT gene dosage correlates with telomerase activity in human lung cancer cell lines. Cancer Lett. 2002;176:81–91. doi: 10.1016/s0304-3835(01)00644-9. [DOI] [PubMed] [Google Scholar]
- 66.Yokoi S, Yasui K, Iizasa T, Imoto I, Fujisawa T, Inazawa J. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin. Cancer Res. 2003;9:4705–4713. [PubMed] [Google Scholar]
- 67.Angelopoulou K, Zavlaris M, Papaioannou N, Vlemmas I. Canis familiaris telomerase reverse transcriptase undergoes alternative splicing. Mamm. Genome. 2008;19:647–653. doi: 10.1007/s00335-008-9144-7. [DOI] [PubMed] [Google Scholar]
- 68.Guo W, Okamoto M, Park NH, Lee YM. Cloning and expression of hamster telomerase catalytic subunit cDNA. Int. J. Mol. Med. 2001;8:73–78. doi: 10.3892/ijmm.8.1.73. [DOI] [PubMed] [Google Scholar]
- 69.Chang H, Delany ME. Complicated RNA splicing of chicken telomerase reverse transcriptase revealed by profiling cells both positive and negative for telomerase activity. Gene. 2006;379:33–39. doi: 10.1016/j.gene.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 70.Rao F, Wang T, Li M, Li Z, Hong N, Zhao H, Yan Y, Lu W, Chen T, Wang W, Lim M, Yuan Y, Liu L, Zeng L, Wei Q, Guan G, Li C, Hong Y. Medaka tert produces multiple variants with differential expression during differentiation in vitro and in vivo. Int. J. Biol. Sci. 2011;7:426–439. doi: 10.7150/ijbs.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heller-Uszynska K, Schnippenkoetter W, Kilian A. Cloning and characterization of rice (Oryza sativa L) telomerase reverse transcriptase, which reveals complex splicing patterns. Plant J. 2002;31:75–86. doi: 10.1046/j.1365-313x.2001.01337.x. [DOI] [PubMed] [Google Scholar]
- 72.Kilian A, Bowtell DDL, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cells. Human Mol. Gen. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 73.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 74.Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO, Reddel RR. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia. 2000;2:426–432. doi: 10.1038/sj.neo.7900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saeboe-Larssen S, Fossberg E, Gaudernack G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol. Biol. 2006;7:26. doi: 10.1186/1471-2199-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amor S, Remy S, Dambrine G, Le Vern Y, Rasschaert D, Laurent S. Alternative splicing and nonsense-mediated decay regulate telomerase reverse transcriptase (TERT) expression during virus-induced lymphomagenesis in vivo. BMC Cancer. 2010;10:571. doi: 10.1186/1471-2407-10-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rha SY, Jeung HC, Park KH, Kim JJ, Chung HC. Changes of telomerase activity by alternative splicing of full-length and beta variants of hTERT in breast cancer patients. Oncol. Res. 2009;18:213–220. doi: 10.3727/096504009x12596189659123. [DOI] [PubMed] [Google Scholar]
- 79.Lincz LF, Mudge LM, Scorgie FE, Sakoff JA, Hamilton CS, Seldon M. Quantification of hTERT splice variants in melanoma by SYBR green real-time polymerase chain reaction indicates a negative regulatory role for the beta deletion variant. Neoplasia. 2008;10:1131–1137. doi: 10.1593/neo.08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karamysheva Z, Wang L, Shrode T, Bednenko J, Hurley LA, Shippen DE. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell. 2003;113:565–576. doi: 10.1016/s0092-8674(03)00363-5. [DOI] [PubMed] [Google Scholar]
- 81.Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. U. S. A. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Cifuentes-Rojas C, Nelson AD, Kannan K, Shippen DE. A novel regulatory network for Arabidopsis telomerase fueled by TER duplication and processing and assembly of alternative RNP particles. In preparation.
- 83.Price C. Regulation of telomere length. Mutat Res. 2011 In Press. [Google Scholar]
- 84.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 85.Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- 86.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. & Mol. Bio. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 87.Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6065–6074. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tseng SF, Lin JJ, Teng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic. Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 92.Gao H, Toro TB, Paschini M, Braunstein-Ballew B, Cervantes RB, Lundblad V. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics. 2010;186:1147–1159. doi: 10.1534/genetics.110.122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeZwaan DC, Freeman BC. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci U S A. 2009;106:17337–17342. doi: 10.1073/pnas.0905703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 95.Singh SM, Lue NF. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5718–5723. doi: 10.1073/pnas.1036868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peterson SE, Stellwagen AE, S.J. D, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Gen. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 97.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;14:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromsomal DNA structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 99.Fisher TS, Taggart AK, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 100.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 102.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat. Str. & Mol. Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 106.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell. Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 109.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 110.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 111.Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 116.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. Jan 14. 1994;76(1):145–55. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 117.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat. Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell. Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mateyak MK, Zakian VA. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;5:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 120.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 121.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 122.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell. Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 123.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 125.Lopez de Silanes I, Stagno d'Alcontres M, Blasco MA. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010;1:33. doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 126.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]