Summary
The etiological agent of Lyme disease, Borrelia burgdorferi, is transmitted by ticks of the Ixodes genus and, if untreated, can cause significant morbidity in affected individuals. Recent reports have shown that polyunsaturated fatty acids in the B. burgdorferi cell envelope are potential targets for oxidative damage, which can be lethal. How B. burgdorferi responds to this assault is not known. Herein we report evidence that bb0646 codes for a lipase that is located within the bosR operon and that has specificity for both saturated and polyunsaturated fatty acids. Specifically, strains harboring mutated copies of the lipase, either in the form of an insertionally inactivated construct or site directed mutations within the active site, demonstrated attenuated lipolytic and hemolytic phenotypes when compared to the isogenic parent and trans-complements. In vivo analysis showed that while the bb0646 mutant remains infectious, the spirochetal load is significantly lower than both the isogenic parent and the complemented mutant strains. Taken together, these data demonstrate that BB0646 is a broad substrate specific lipase that contributes to lipolytic and hemolytic activity in vitro and is required for optimal B. burgdorferi infection.
Introduction
Lyme disease is caused by the spirochete Borrelia burgdorferi and is transmitted by ticks of the Ixodes genus (Schmid, 1985; Anderson, 1989, 1991; Burgdorfer, 1989; Xu et al., 2003). It is the leading arthropod-borne illness in the United States with 38,468 cases reported to the Center for Disease Control in 2009, which represents a 48% increase in the numbers of cases reported since 2004. The disease that results from B. burgdorferi infection is multiphasic and can be characterized by three stages: early localized, early disseminated and chronic disease (Nadelman and Wormser, 1998; Steere, 2001; Steere et al., 2004). Diagnosis during early-localized disease provides patients the opportunity to be treated with antibiotics and clear the infection. However, if untreated, the infection will progress into a chronic stage that is characterized by arthritis in North American patients, resulting in a significant amount of morbidity (Nadelman and Wormser, 1998; Steere et al., 2004). This, combined with the absence of a commercially available vaccine, makes the initial prevention of B. burgdorferi transmission crucial (Klempner et al., 2001; Steere, 2002; Steere et al., 2004).
B. burgdorferi has limited de novo metabolic capabilities and is largely dependent on the living host or in vitro cultivation media as a nutrient source. Of particular interest to this study, B. burgdorferi lacks the machinery to synthesize fatty acids and, as a direct result, scavenges them from the environment (Barbour and Hayes, 1986; Fraser et al., 1997; Boylan et al., 2008). This scavenging behavior results in a significant amount of polyunsaturated fatty acid incorporation into the cellular envelope, an arrangement that is unusual for prokaryotic organisms. Furthermore, the polyunsaturated fatty acids present in the borrelial membrane represent a unique target for reactive oxygen species (ROS) (Gutteridge and Halliwell, 1990; Boylan et al., 2008). As B. burgdorferi is transmitted to the mammalian host it faces a variety of assaults that it must overcome in order to establish infection, including increased respiration within the arthropod vector and ROS-producing innate immune cells in the infected mammal. Significant advances have been made towards characterizing how B. burgdorferi senses, responds to, and adapts to oxidative stress (Katona et al., 2004; Seshu et al., 2004a, 2004b; Tokarz et al., 2004; Boylan et al., 2006, 2008; Hyde et al., 2006, 2009, 2010; Li et al., 2007); however, some aspects of this response remain unknown. Lipids in the cell envelope of B. burgdorferi, specifically those containing polyunsaturated fatty acid side chains, are targets for ROS (Boylan et al., 2008). Thus, deciphering how the spirochete assimilates lipids, protects the membrane, and responds to lipid oxidation is important for understanding how B. burgdorferi survives during the infectious process.
Herein we report that bb0646, which is genetically linked to and part of an operon with the bosR regulatory locus, encodes a lipase with substrate specificity for both saturated and polyunsaturated fatty acids. Furthermore, we show that bb0646 mutants exhibit reduced hemolytic activity in vitro and have an attenuated in vivo infectivity phenotype when evaluated both qualitatively and quantitatively, specifically at a low inoculum dose. Due to the genetic linkage bb0646 shares with bosR and the known roles BosR plays in regulating the response to oxidative stress and the expression of virulence determinants essential for borrelial pathogenesis (Boylan et al., 2003, 2006; Katona et al., 2004; Seshu et al., 2004b; Hyde et al., 2009, 2010; Ouyang et al., 2009), we hypothesized that BB0646 may also be linked to these processes. The data reported herein demonstrates that BB0646 is needed for optimal infectivity during initial colonization. As such, these observations suggest that the presence of this lipase contributes to the pathogenic potential and/or physiologic fitness of Borrelia burgdorferi.
Results
Isolation of a bb0646 mutant strain in B. burgdorferi
BB0646 encodes a protein with a predicted α/β-hydrolase fold, a conserved folding pattern that has been described for numerous prokaryotic and eukaryotic hydrolytic enzymes (Jaeger et al., 1994; Holmquist, 2000; Gupta et al. 2004). Additionally, BB0646 contains a GXSXG motif that is commonly observed in esterases and lipases (Jaeger et al., 1994; Fraser et al., 1997; Gupta et al. 2004). Within Borrelia spp., homologues to BB0646 are found with 97%, 90%, and 68% identity between B. burgdorferi sensu stricto isolates, B. burgdorferi sensu lato isolates, and relapsing fever Borrelia, respectively. Following this, there is large drop off in identity to other bacterial systems (data not shown). The conservation of a BB0646 homologue in all Borrelia spp. sequenced to date suggests an important role for this protein in borrelial biology.
To assess the role of BB0646 in B. burgdorferi sensu stricto strain B31 (referred to as B. burgdorferi for the remainder of this report), a bb0646 mutant containing an insertionally inactivated copy of bb0646 with the gentR allele was constructed and designated DS102 (Fig 1A). Three separate isolates from independent transformations were obtained and evaluated by PCR, Southern blot, and Western blot analysis (Fig. 1). There was no statistical difference in the growth rate when DS102 was compared with its parent ML23, suggesting that BB0646 is not required for in vitro growth (Hyde et al., 2010). To confirm the presence of the bb0646::gentR allele, oligonucleotide primers specific to the 5’ and 3’ ends of bb0646 were used to PCR amplify a fragment from all DS102 isolates; the resulting PCR product was a 2001 bp fragment. This is consistent with the predicted increase of 1017 bp due to the presence of the gentR cassette relative to the 984 bp fragment amplified from the isogenic parent (Fig. 1B). Additionally, a forward primer flanking the 5’ end of bb0646 and a reverse primer that sits within the gentR produced a 1152 bp fragment and, likewise, a reverse primer flanking the 3’ end of bb0646 and a forward primer that sits within the gentamicin resistance cassette produced an 889 bp confirming the presence and orientation of the gentR cassette (Fig. 1B). The borrelial plasmid composition from all isolates was assessed by PCR to ensure that all expected plasmids were present (data not shown).
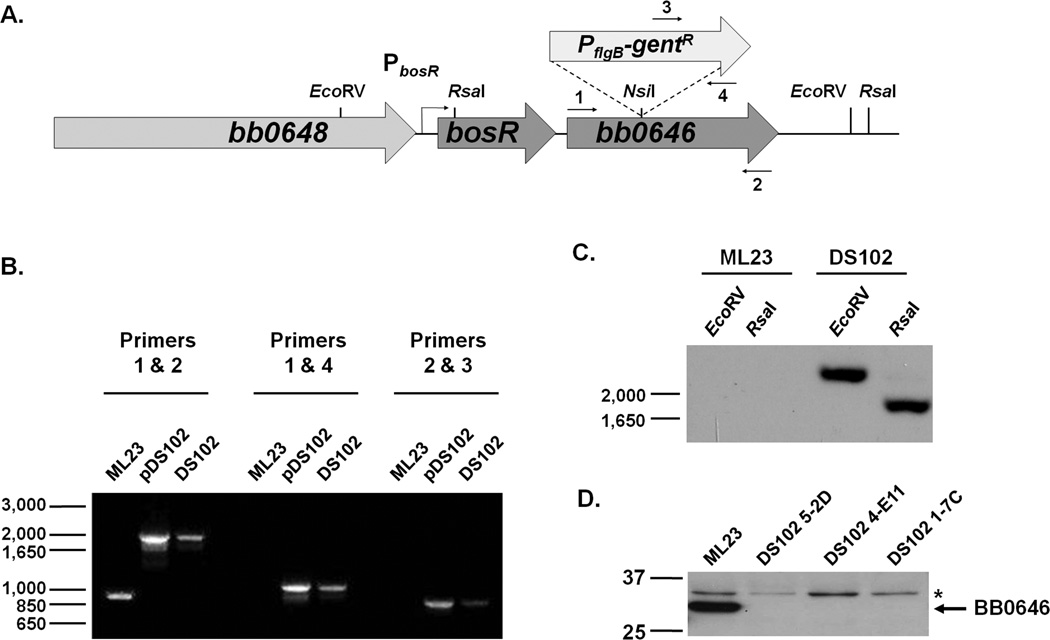
Figure 1.
Isolation and confirmation of bb0646::gentR mutants in B. burgdorferi B31 derivative, ML23. (A) Schematic diagram of the bb0646 insertional mutation of DS102. bb0646 was interrupted using a PflgB-gentR cassette at a unique NsiI restriction site within the gene. Constructs were transformed into ML23 and putative positive clones were screened by PCR using the indicated primers, 1–4, in panel B. (B) Putative transformants were screened with 3 sets of primers: DS111F/R (1 and 2), DS111-NdeI-F/gent_int- R (1 and 4), and DS111-NdeI-R/gent_int-F (2 and 3). (see Table 2 for primers used). Values shown on the left represent markers (in bp). (C) Southern blot confirmed the presence of the bb0646::gentR allele via hybridization with a gentR probe. DNA from B. burgdorferi strains ML23 and DS102 was digested with EcoRV and RsaI as indicated (restriction enzyme sites shown in panel A). Values shown on the left represent markers (in bp). (D) Western blot analysis demonstrated that putative transformants made no detectable BB0646 protein compared to the isogenic parent strain ML23 pBBE22 when B. burgdorferi lysates were analyzed by immunoblot analysis with polyclonal antibody specific for BB0646. An asterisk (*) denotes a non-specific, cross-reactive band that anti-BB0646 recognizes in all isolates. Values shown on the left represent markers (in kDa).
The bb0646 mutant was further verified by Southern and Western blot analyses. A restriction digestion of ML23 and DS102 genomic DNA with EcoRV or RsaI revealed bands consistent with the predicted sizes of 2263 and 1848 bp, respectively, exclusively in the mutant by the binding of the gentR-specific probe (Fig. 1C). Western blotting analysis, using polyclonal antibodies raised against the 33 kDa purified recombinant BB0646, demonstrated that a protein with the predicted molecular mass of BB0646 was present in the parent strain, ML23, but was absent from all three DS102 isolates tested, confirming that the all mutants tested contained the bb0646::gentR allele and were devoid of BB0646 protein (Fig. 1D).
Complementation of the bb0646 mutant
Two schemes were used for the genetic complementation of the bb0646 mutants. The first has bb0646 fused to its native promoter, which is also the promoter for bosR (bb0647), as the two genes are transcribed together (Ouyang et al., 2009; Hyde and Skare, unpublished observation). This construct was designated pDS113 (Fig 2A). The cloning to the native promoter was used to restore any potential transcriptional regulation associated with natively expressed bb0646. The second construct, designated pDS126, placed bb0646 under control of the strong constitutive flagellar promoter, PflgB (Fig 2A). Following transformation of DS102 with both of the complement constructs, isolates were expanded and screened by PCR to verify the presence and orientation of bb0646 on the shuttle vectors (Fig. 2B). Specifically, an oligonucleotide that recognizes pncA (bbe22) on pBBE22, together with an oligonucleotide primer specific for bb0646, amplified a 521 bp or a 926 bp PCR product for DS102 pDS113 or DS102 pDS126, respectively, whereas the negative controls tested, either ML23 pBBE22 genomic DNA or DS102 pBBE22 genomic DNA, did not amplify any products (Fig 2B). Complemented DS102 clones were also PCR screened to confirm the presence of the same borrelial plasmid profile as the parent (data not shown). Isolates that contained the bb0646 complementation constructs were analyzed by Western blot to determine if they produced BB0646 protein. Strains with bb0646 fused to the native promoter showed detectable, but reduced BB0646 protein production relative to the parent, ML23 (Fig 2C). As expected, DS102 pDS126 produced more BB0646 than both ML23 pBBE22 and DS102 pDS113.
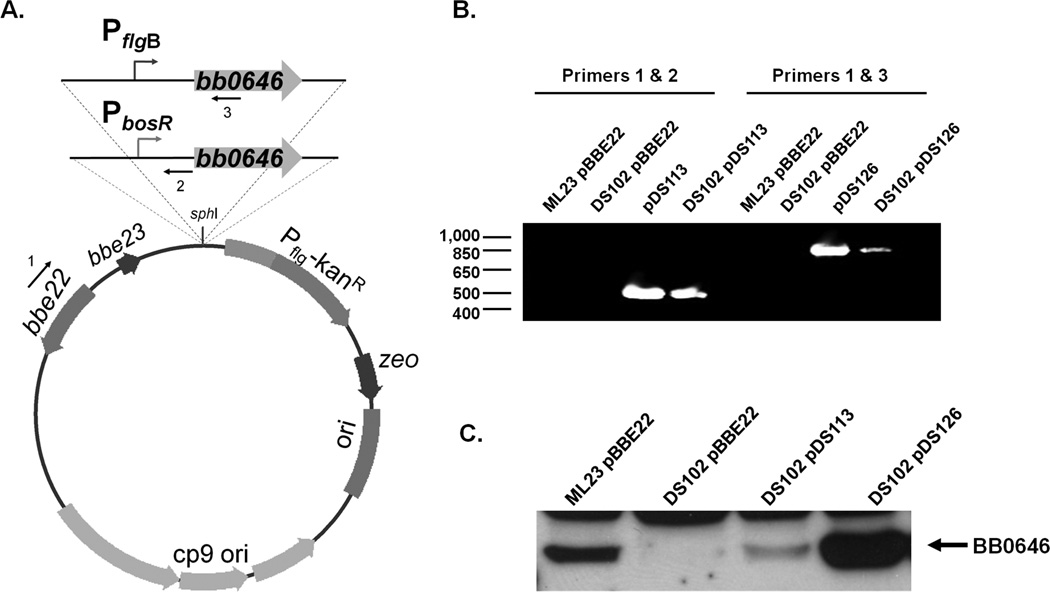
Figure 2.
Complementation of the bb0646::gentR insertional mutant, DS102, with the intact bb0646 gene in trans. (A) Transcriptional fusions of bb0646 to either its native promoter (PbosR; pDS113) or the strong constitutive flagellar promoter (PflgB; pDS126) were cloned into the borrelial shuttle vector pBBE22Gate. (B) Primer pair pncA_nested-F/pDS113_R-confirm (1 and 2; Table 2) were used to confirm the presence of pDS113 in DS102 isolates by PCR. Likewise, primer pair pncA_nested-F/pDS126_R-confirm (1 and 3; Table 2) was used to validate whether pDS126 was present in DS102. Values shown on the left represent markers (in bp). (C) Putative transformants were screened for the presence of BB0646 by Western immunoblot analysis using polyclonal anti-BB0646.
BB0646 has lipase activity with specificity for saturated and polyunsaturated substrates
While we were able to overproduce recombinant BB0646 in Escherichia coli (used for the immunization and subsequent production of polyclonal rabbit antibodies against BB0646), we were unable to obtain functional protein. To assess whether native BB0646 was associated with lipase activity, whole cell lysates from the B. burgdorferi parent, the bb0646 mutant, and the genetic complements were screened for lipase activity. Initially, the whole cell lysates from ML23, the bb0646 mutant, and the genetic complements were tested against a saturated fatty acid substrate, p-nitrophenyl palmitate. The results indicated that B. burgdorferi and genetic complements have lipase activity against saturated fatty acids and that the bb0646 mutant, DS102, is significantly less efficient at hydrolyzing a palmitic acid-containing substrate (Fig 3A). It is important to note that both rabbit serum and BSA alone yielded very low activity in our assay (data not shown) and, because our lysates originated from extensively washed samples, these major components of BSK-II media did not contribute to the substrate turnover observed.
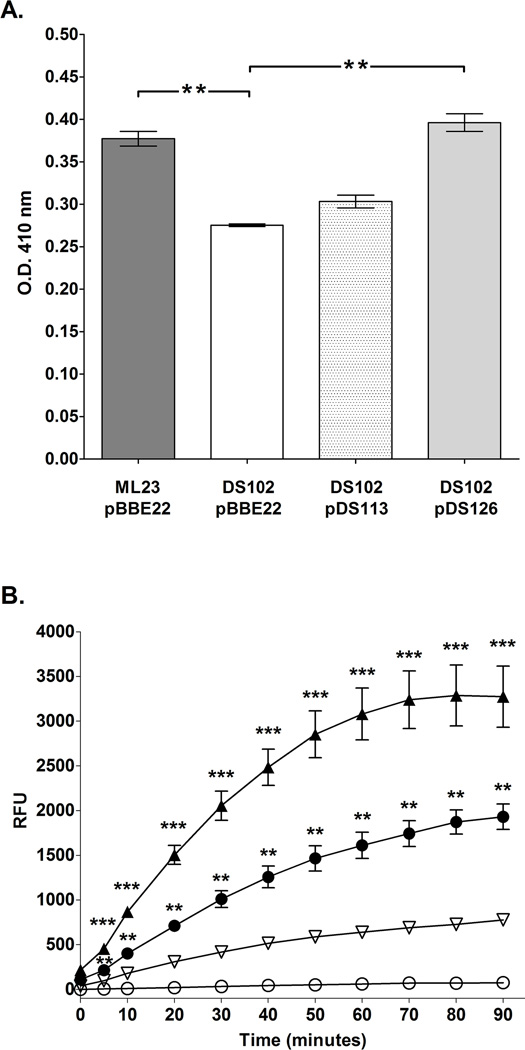
Figure 3.
BB0646 functions as a lipase with specificity for both saturated and polyunsaturated fatty acid substrates. (A) Whole cell lysates from ML23/pBBE22, DS102/pBBE22, DS102/pDS113 and DS102/pDS126 were assayed using p-nitrophenyl palmitic acid as a substrate. Note the statistically significant decrease in lipase activity for DS102/pBBE22 relative to the isogenic parent and the complement DS102/pDS126. The dual asterisks (**) denote a P value of < 0.01. (B) Whole cell lysates of either ML23/pBBE22, DS102/pBBE22, DS102/pDS113 or DS102/pDS126 were incubated with the fluorogenic substrate, 7-hydroxycoumarinyl linolenic acid over a period of 90 min. and read at 10 min. intervals. Note that the bb0646::gentR mutant (DS102/pBBE22; open circles) was nearly devoid of activity whereas the parent (ML23/pBBE22; closed circles) both complements (DS102/pDS113; inverted open triangles and DS102/pDS126; closed triangles) showed restoration of lipase activity with highest activity seen for DS102/pDS126. Bars indicate standard error. Time points from each strain were tested for significance; **, and *** denote P values < 0.001, and 0.0001 respectively.
Although the p-nitrophenyl palmitate assay showed that B. burgdorferi did, indeed, have lipase activity, the overall activity for the parent strain was relatively low (Fig 3A). Given that B. burgdorferi incorporates polyunsaturated fatty acids into the cell envelope (Boylan et al., 2008), it is possible that BB0646 may be involved in their assimilation and thus have a preference for polyunsaturated substrates. To test this hypothesis, whole cell lysates from the parent, mutant, and genetic complements were incubated with the 7-hydroxycoumarin linolenic acid, a fluorogenic polyunsaturated substrate, and monitored over time. In this analysis, potent lipase activity was seen for cell lysates from ML23 and both genetic complements (DS102/pDS113 and DS102/pDS126). In stark contrast, the samples from the bb0646::gentR mutant (DS102) showed a negligible level of activity (Fig. 3B). It is important to note that the complement with the native promoter, DS102/pDS113, demonstrated partial restoration of lipase activity and the complement strain with bb0646 under control of the strong borrelial flgB promoter, DS102/pDS126, showed increased activity when compared to the parent, ML23. This is consistent with an increase in BB0646 produced in DS102/pDS126 relative to DS102 pDS113 and the parent (Fig 2C and 3B). These results indicate that BB0646 also recognizes polyunsaturated fatty acid substrates.
Site-directed mutagenesis of the predicted active site in BB0646
The deduced amino acid sequence of B. burgdorferi strain B31 BB0646 contains the conserved active site motif, GXSXG, which is found in nearly all esterases and lipases (Jaeger et al., 1994; Fraser et al., 1997; Gupta et al., 2004). The catalytic triad that makes up the active site of lipases is composed of a histidine, a serine (which is located in the center of the GXSXG motif), and either an aspartic acid or glutamic acid (Jaeger et al., 1994). To further support the idea that BB0646 is a borrelial lipase and not simply associated with lipase activity, two point mutation constructs were created using PflgB-bb0646 (pDS126) plasmid DNA as template. The point mutation schemes targeted the serine at the center of the GXSXG motif (specifically, GTSNG in residues 150 to 154 in B. burgdorferi strain B31 with the critical serine located at residue 152) and changed it to either an alanine (DS102/pDS127; encodes the bb0646-S152A allele) or a threonine (DS102/pDS128; encodes the bb0646-S152T allele). The resulting bb0646 mutant alleles were transformed into DS102 and screened by PCR, Western blot, and reconfirmed by dideoxy sequencing. As expected, both DS102/pDS127 and DS102/pDS128 were PCR positive for bb0646 and contained the expected sequence with the mutated active site (data not shown). Importantly, each isolate showed that protein production was restored, with levels comparable to DS102/pDS126, which contains the intact copy of bb0646, when analyzed by Western blot using the antibody specific to BB0646 (Fig 4A).
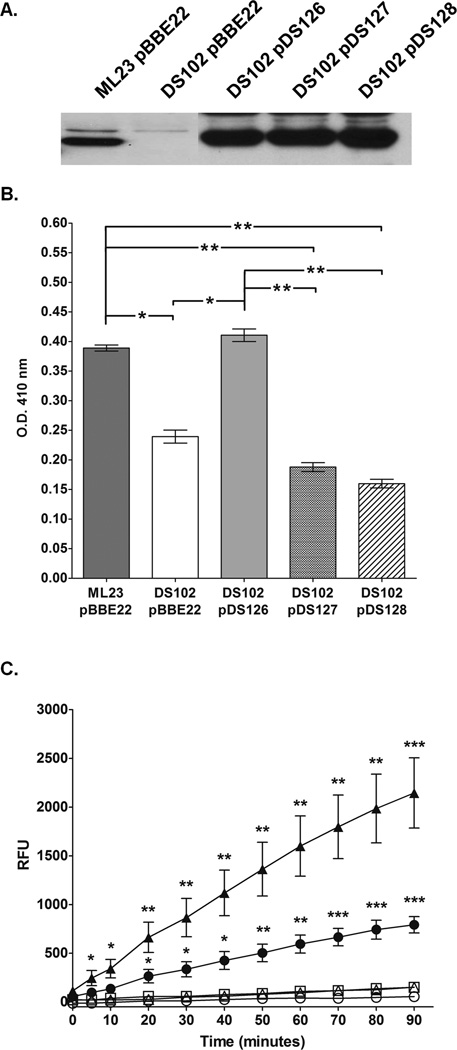
Figure 4.
Mutagenesis of the putative active site serine of BB0646 abrogates lipase activity in B. burgdorferi, but not BB0646 protein production. (A) Production of BB0646 in point mutants. The levels of BB0646 are compared between the parent (ML23/pBBE22), mutant (DS102/pBBE22), complement (DS102/pDS126), and the point mutants within the GXSXG conserved motif (DS102/pDS127; contains the bb0646-S152A allele; and DS102/pDS128; contains the bb0646-S152T allele). Whole cell equivalents were resolved by SDS-PAGE, immunoblotted and probed with antisera to BB0646. (B) Mutagenesis at serine 152 of bb0646 reduces recognition of saturated fatty acid substrate. The B. burgdorferi strains indicated above were tested for lipolytic activity against p-nitrophenyl palmitate as indicated in Fig. 3A. Note the statistically significant decrease in lipase activity for strains carrying the S152A (DS102/pDS127) and S152T (DS102/pDS128) alleles of bb0646 relative to the parent (ML23/pBBE22) and complement (DS102/pDS126). The activity observed for the bb0646 point mutants is commensurate with that observed for the bb0646 mutant (DS102/pBBE22). The single and dual asterisks denote a P value of < 0.05 and 0.01, respectively. (C) bb0646 S152 mutants do not recognize polyunsaturated fatty acid substrates. Whole cell lysates from the B. burgdorferi strains were tested for lipolytic activity against 7-HC linolenate as indicated in Fig. 3B. Samples from the parent (ML23/pBBE22; closed circles), mutant (DS102/pBBE22; open circles), the complement (DS102/pDS126; closed triangles), and the bb0646-S152 mutants (DS102/pDS127 [open triangles] and DS102/pDS128 [open squares]) were tested for their ability to cleave 7-HC linolenate over time. Cleavage of the substrate releases a fluorogenic reporter and the increased fluorescence is plotted as relative fluorescent units (RFU). Significance was measured at each time point between the bb0646 mutant and S152 mutant complements relative to the parent strain and the native complement. Bars indicate standard error. Single, dual, and tri asterisks denote P values of < 0.01, 0.001, and 0.0001 respectively.
Subsequently, lipase assays with whole cell lysates from the parent strain, mutant and complement were compared against borrelial strains carrying the bb0646-S152A and bb0646-S152T alleles (Fig 4B and C). The data indicated that, although the strains carrying the mutant bb0646 alleles make comparable levels of protein relative to the non-mutagenized complement construct, DS102/pDS126 (the most relevant comparator), the lipase activity is significantly reduced for these strains (Fig 4). Taken together, these results indicate that the observed lipase activity seen in the parent and complements, but that is reduced or essentially absent in the bb0646 mutant or point mutation isolates (depending on the assay), is due to functional, wild type BB0646.
BB0646 is required for optimal B. burgdorferi hemolytic activity
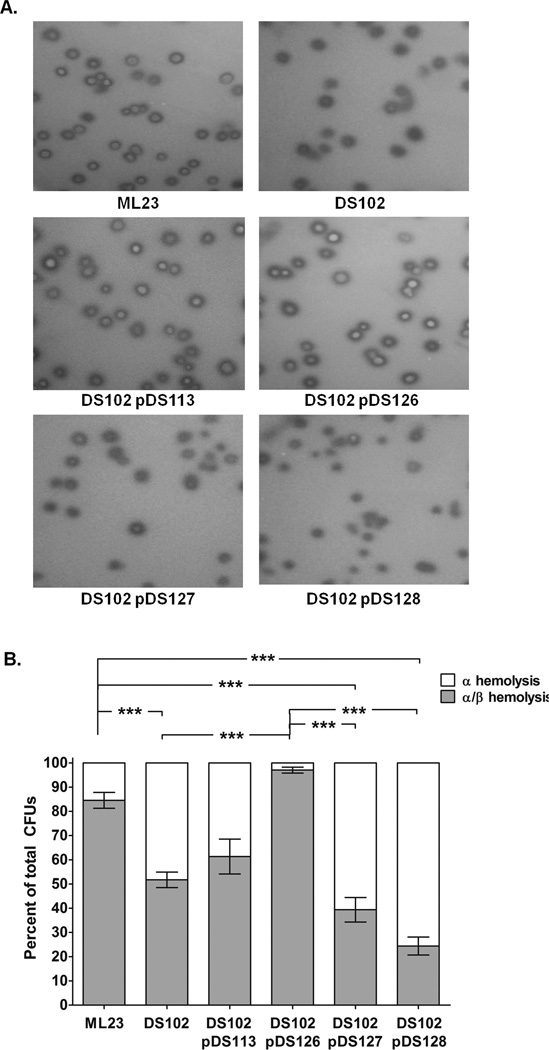
Previously, in vitro hemolytic activity was described for B. burgdorferi when the cells were grown on blood agar plates (Williams and Austin, 1992). BB0646 may have preference for polyunsaturated lipid substrates existing within intact phospholipids, as opposed to triglycerides. Therefore, we tested whether BB0646 could be responsible for the aforementioned borrelial hemolysis. To test this possibility, log phase cultures of ML23, DS102, DS102/pDS113, DS102/pDS126, DS102/pDS127, and DS102/pDS128 were plated on BSK-II agar plates containing 5% horse blood. Colonies from the borrelial parent strain (ML23) showed partial β-hemolytic clearing around them (designated as α/β hemolysis) while the bb0646 mutant (DS102) and the point mutation complement strains (DS102/pDS127 and DS102/pDS128) exhibited α hemolysis and reduced α/β hemolysis (Fig 5A). All of the strains tested exhibited α and α/β hemolysis, with α-hemolysis being defined as no overt lysis, i.e., green/brown in appearance (seen as dark colonies in Fig. 5A) and α/β hemolysis as a zone of clearing where the colony formed with a green/brown border (seen as white colony with a dark border in Fig. 5A). The ratio of these two colony types for each strain tested was then scored (Fig. 5B). DS102 and DS102 transformed with the S152 mutants demonstrated enhanced α-hemolysis and reduced β-hemolysis relative to the parent and the complement, DS102/pDS126. The lone exception was DS102/pDS113, which expresses bb0646 from its native promoter (PbosR) (Ouyang et al., 2009; Hyde and Skare, unpublished observation). DS102/pDS113 showed less β-hemolysis relative to the DS102/pDS126 complement, but slightly more than DS102 alone (Fig. 5B), consistent with the lower amount of BB0646 produced by DS102/pDS113 (Fig. 2C). Overall these observations suggest that the bulk of the hemolytic activity seen is due to functional BB0646 protein.
Figure 5.
BB0646 is required for maximal hemolytic activity in B. burgdorferi. (A) The parent (ML23), bb0646 mutant (DS102), both complements (DS102/pDS113 and DS102/pDS126), and both bb0646 S152 point mutation complements (DS102 carrying the bb0646-S152A and bb0646-S152T alleles, DS102/pDS127 and DS102/pDS128, respectively) were evaluated for hemolytic activity. The bb0646 mutant and point mutation complements exhibited attenuated β–hemolysis while both complement strains restored the deficiency to that observed in the parent strain. (B) Semi-quantitative assessment of borrelial-mediated hemolysis. Colonies from each strain were scored for partial β–hemolysis and α–hemolysis and with values listed as a percentage of the total hemolysis observed. Note that the S152 mutant complements exhibit hemolytic activity indistinguishable from the bb0646 mutant alone (DS102). Both are significantly different from that observed for the parent and PflgB-bb0646 complement (P value < 0.05). Bars indicate standard error. Each strain represents an n of 6.
BB0646 partitions with detergent phase proteins
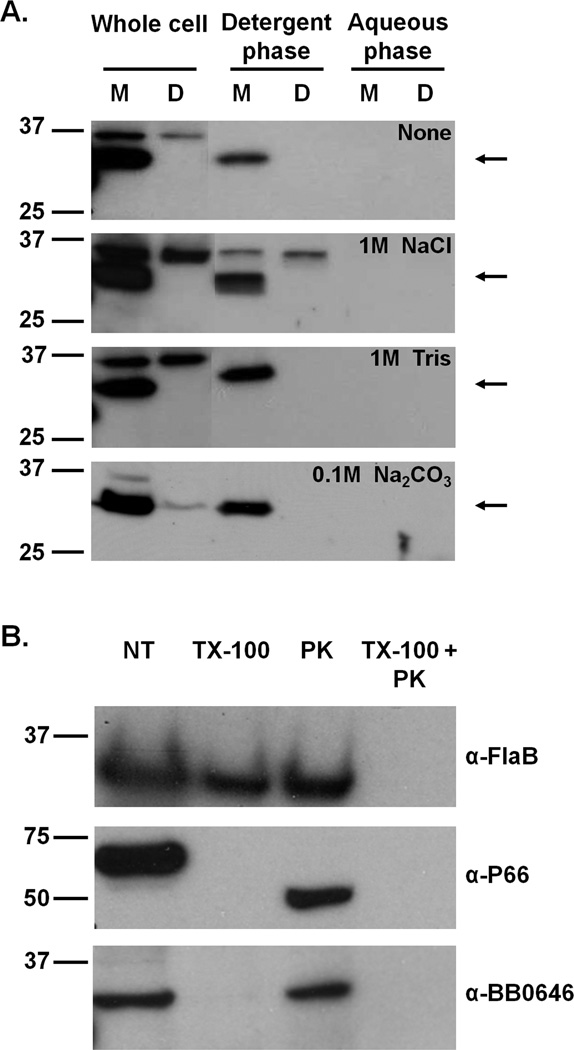
Given that we had difficulty purifying active recombinant BB0646, we reasoned that we could concentrate native BB0646 directly from borrelial protein lysates and subsequently enrich for lipase activity by exploiting the predicted soluble nature of the lipase. To this end, we used Triton X-114 phase partitioning as a means to concentrate BB0646 into the aqueous phase, relative to the membrane protein/lipoprotein containing detergent phase. Although several predictive algorithms indicate that BB0646 has an amino-terminal leader peptide and is thus likely to be an exported, soluble protein (data not shown), the Triton X-114 extraction demonstrated that BB0646 partitioned with the detergent phase (Fig 6A). To determine whether this observation was due to ionic interactions with membrane proteins, a second series of Triton X-114 detergent extractions were performed with high salt concentrations (either 1M NaCl, 1M Tris, or 0.1M sodium carbonate) that would serve to disrupt any putative ionic interactions. In all cases, these conditions did not alter the association of BB0646 with the detergent phase (Fig. 6A), indicating that BB0646 is not peripherally associated with a detergent phase membrane protein or lipoprotein.
Figure 6.
BB0646 is a detergent phase associated protein that is not surface localized. (A) Phase partitioning analysis of BB0646. Whole cell lysates from ML23 and DS102 were subjected to Triton X-114 phase partitioning and following exposure to high salt (i.e., 1M NaCl, 1M Tris, or 0.1 M Na2CO3). Detergent and aqueous phases were compared to whole cell lysates by Western immunoblotting with BB0646 antiserum. Detergent and aqueous phase proteins from 108 whole cell equivalents were loaded into each lane. An arrow indicates the position of BB0646. Numbers on the left refer to the molecular mass of protein makers (in kDa). (B) Localization of BB0646. Strain ML23 was incubated with Proteinase K to assess whether BB0646 was surface exposed. Controls were either incubated in PBS alone (not treated; NT), permeabilized with Triton X-100 (TX-100), treated with Proteinase K alone (PK), or incubated with Proteinase K following exposure to Triton X-100 (TX-100 + PK). Following SDS-PAGE, samples were immunoblotted and probed with antisera against FlaB, P66, and BB0646 (as indicated on the right). Numbers on the left refer to the molecular mass of protein makers (in kDa).
Due to the partitioning of BB0646 with the detergent phase, we next asked whether BB0646 was a surface exposed membrane protein. Following exposure of whole cells to proteinase K, BB0646 was protected from proteolysis when the cells were intact, but was degraded when the cells were permeabilized with Triton X-100 (Fig 6B). This suggests that BB0646 resides in a subsurface location or is not accessible to the protease in its surface exposed locale. As controls, the P66 outer membrane protein was cleaved from the 66 kDa form down to a 50 kDa form, indicating that the proteinase K used was active (Fig 6B). As an additional control, the FlaB endoflagellar protein remained intact in undisturbed cells, demonstrating independently that the cells were structurally intact (Fig 6B).
Taken together, these results suggest that BB0646 is likely a subsurface, periplasmic protein whose association with the Triton X-114 detergent phase may be attributed to unrelated properties exhibited by lipases. Lipases are known to absorb to lipid-water interfaces prior to substrate cleavage (Jaeger et al., 1994; Gupta et al. 2004); due to the lipolytic properties of BB0646, it may absorb to the interface created by the Triton X-114/PBS emulsion and/or recognize the detergent itself as a substrate and anomalously partition with the Triton X-114 detergent phase.
BB0646 is required for optimal B. burgdorferi infectivity
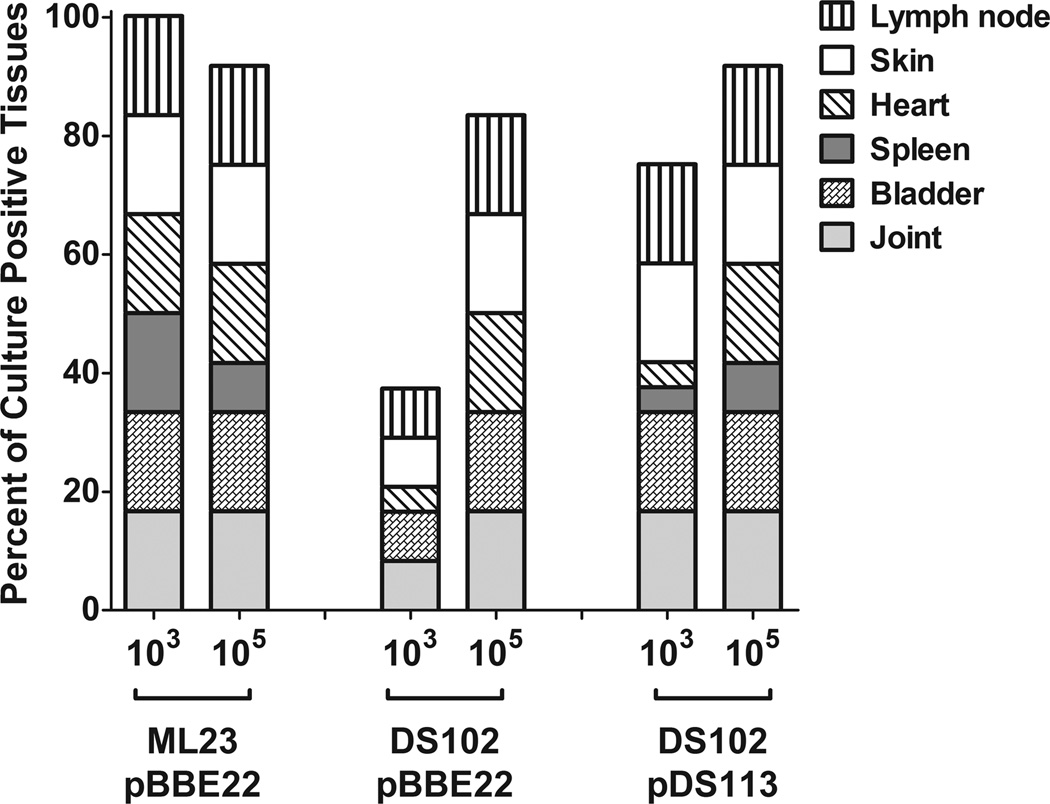
To assess the infectivity potential of the bb0646 mutant relative to its infectious parent and genetic complement, C3H/HeN mice were intradermally inoculated with either 103 or 105 organisms of the B. burgdorferi strains. After 3 weeks, the mice were sacrificed and the presence of B. burgdorferi was qualitatively scored in various organs (Fig 7). The results indicate that the bb0646 mutant (DS102/pBBE22) was attenuated relative to the parent (ML23/pBBE22) and its genetic complement (DS102/pDS113), but only at the 103 inoculum. At the 105 dose, there were no differences observed between the strains tested (Fig 7).
Figure 7.
Loss of BB0646 partially attenuates B. burgdorferi infectivity. C3H/HeN mice were infected with B. burgdorferi strains at either 103 or 105 inoculum doses. Tissues were aseptically removed after 3 weeks of infection and cultivated in BSK-II media. Cultures were evaluated for the presence of B. burgdorferi after 6–14 days. Data is expressed as percentages of culture-positive samples for each strain and dose tested. Each data set represents data obtained from 4–5 mice (depending on the strain).
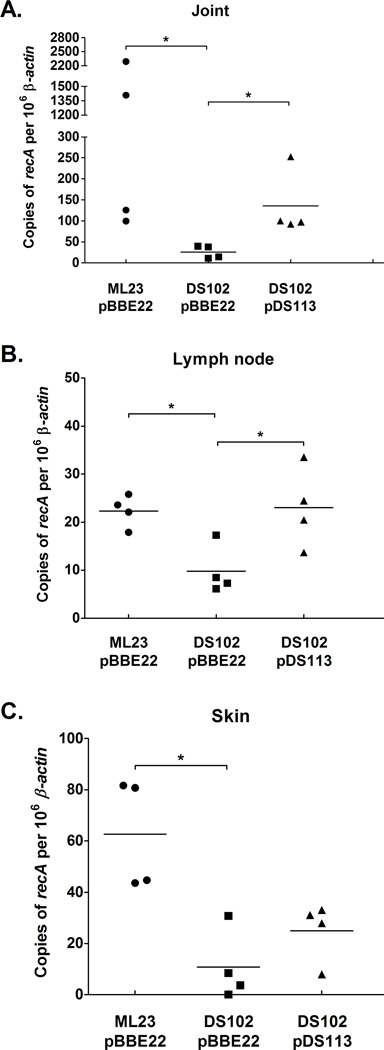
To quantify the borrelial load, quantitative PCR was performed on total DNA extracted from the tibiotarsal joint, lymph node, and skin tissues from mice infected with the parent, bb0646 mutant and genetic complement. Copies of B. burgdorferi genomes per 106 β-actin were compared between different strains from each tissue and inoculums. Fewer borrelial genome copies were quantified from the tissues infected with the bb0646 mutant when compared to the parent and the relevant genetic complement, DS102/pDS113, at an inoculum dose of 103 (Fig 8); no significant difference was observed in borrelial genome equivalents when mice were infected with 105 B. burgdorferi (data not shown). Taken together, these results suggest that presence of BB0646 correlates with bacterial burden in the tissues tested and provides further evidence that BB0646 activity is required for primary colonization of B. burgdorferi following infection.
Figure 8.
Quantitative PCR shows a lowered infectious load for the bb0646 mutant at a low inoculum dose. Quantitative PCR was performed on (A) joint (B) lymph node and (C) skin tissues removed from mice that were infected with 103 organisms to determine the absolute number of B. burgdorferi genomic equivalents in each tissue sample. Copies of recA were quantified and represent the number of total B. burgdorferi genomes. To normalize the data to host tissue, copies of β-actin were also enumerated and data points are expressed as copies of B. burgdorferi genomes per 106 copies of β-actin. Bars indicate average value of the sample tested. The asterisks denote a P value < 0.05.
Discussion
Borrelia burgdorferi has reduced physiological and metabolic capabilities, but maintains the ability to colonize and infect vastly divergent host environments throughout its complex lifecycle. Structure and maintenance of bacterial cell membranes are critical for the survival of bacteria, including pathogens that are subjected to innate and adaptive immune clearance mechanisms. Although B. burgdorferi is organized like a Gram-negative organism, it lacks lipopolysaccharide (LPS) and mimics the fatty acid profile of the environment by assimilating fatty acids, including polyunsaturated fatty acids, into lipids and lipoproteins (Takayama et al., 1987; Boylan et al., 2008; Gherardini et al., 2010). B. burgdorferi’s limited genome does not encode the required genes for fatty acid synthesis (Fraser et al., 1997), which places a premium on B. burgdorferi’s ability to scavenge them from their surroundings. Passive diffusion of free fatty acids into the membrane remains a possibility, but may not be the lone mechanism of acquisition (Cox and Radolf, 2001), particularly if the availability of free fatty acids is not sufficient to support growth. Nevertheless, the precise mechanisms involved in fatty acid acquisition and assimilation remain unknown.
One possible mechanism to circumvent the metabolic limitations of B. burgdorferi in regard to fatty acid biosynthesis, may be via a lipase (or lipases) that are able to obtain fatty acids from host lipids and/or catalyze their incorporation into lipids and lipoprotein species. B. burgdorferi BB0646 encodes a protein of the α/β-hydrolase fold family with a conserved lipase motif (GXSXG) (Fraser et al., 1997) as was indicated in an earlier study (Katona et al., 2004). The inability to synthesize fatty acids requires B. burgdorferi to either rely on the supply of free fatty acids in the environment or to produce enzymes which would catalyze their release from other sources (i.e. other phospholipids and/or triglycerides); BB0646 may be a candidate for this second possibility. Aside from fatty acid acquisition, the BB0646 lipase has the potential to function as a virulence determinant. Several pathogens encode secreted lipases that facilitate invasion by breaking down mucosal linings or escaping from phagosomal or endosomal compartments. Some lipases may also be able to influence the immune response by cleaving phospholipids and releasing diacylglycerol, a secondary lipid messenger, which modulates cellular processes such as neutrophil and macrophage activation (Titball, 1993; Schmiel and Miller, 1999; Istivan and Coloe, 2006). Several of the aforementioned lipase-based activities are associated with obligate intracellular pathogens (i.e., escape from phagosomes); as such, their applicability to B. burgdorferi is not clear.
It is important to note that the putative borrelial lipase is the final locus in a three gene operon whose upstream gene is bosR, which is thought to regulate the borrelial oxidative stress response. In addition to this regulation, BosR is also required for the expression of essential virulence determinants for mammalian infection, including ospC and dbpBA (Hyde et al., 2009, 2010; Ouyang et al., 2009). Genetic linkage often suggests cooperative function and, in this case, it is conceivable that BB0646 may also contribute to pathogenecity and/or the oxidative stress response. Although it is known that the polyunsaturated fatty acids acquired from the host environment and incorporated into the membrane put B. burgdorferi at risk for damage by reactive oxygen species (ROS) (Boylan et al., 2008), how borrelial cells deal with the damage imposed by oxidized lipids and lipoproteins remains unknown. However, for this study, we focused on the ability of the putative borrelial lipase, BB0646, to cleave fatty acids, either saturated or unsaturated, and tested its role in mammalian infection.
To begin characterizing the putative BB0646 lipase, we constructed and isolated an insertional mutation in bb0646. The resulting strain, DS102, exhibited a profound decrease in lipase activity, particularly when a polyunsaturated substrate was tested (Fig. 3B). The known hemolytic activity of B. burgdorferi (Williams and Austin, 1992) was also greatly reduced when bb0646 was inactivated (Fig. 5). To address that the loss of bb0646 was directly linked to the loss of the lipase/hemolytic activity, the mutant strain was genetically complemented in trans with bb0646 expressed from either its native promoter, pDS113, or a strong, constitutive borrelial promoter, (PflgB; Bono et al., 2000), pDS126. (Fig. 2A). Since both of these constructs restored lipase activity to B. burgdorferi, the data strongly suggested that BB0646 was responsible for these activities. The fact that the isogenic parent exhibits partial β-hemolytic activity against erythrocytes (denoted as “α/β hemolysis”) and that the lipase-deficient mutant, DS102, mostly exhibited α-hemolysis (Fig 5), coupled with the decrease in infectivity for DS102 (Fig.’s 7 and 8), suggests that the lipase may be critical at specific points in the borrelial life cycle. For example, this activity would be particularly important under conditions when B. burgdorferi is undergoing host adaptation or rapid expansion. In regard to mammalian infection, it is probable that BB0646 would be enlisted to provide B. burgdorferi with host specific metabolites including fatty acids for lipid and lipoprotein assimilation. Alternatively, on the arthropod side of the infectious life cycle, when an infected nymph starts taking a blood meal, B. burgdorferi undergoes a period of rapid growth (de Silva and Fikrig, 1995) whereby the spirochete would require a sufficient amount of nutrient availability to supplement its limited de novo metabolic capabilities. At this stage, BB0646 may be needed to acquire fatty acids from the incoming blood meal (e.g., from red blood cells) prior to migration to the tick salivary glands.
One limitation of our lipase assays was the reliance on whole cell lysates to track lipase activity from B. burgdorferi. Recombinant BB0646 was purified from E. coli for the immunization of rabbits to obtain a monospecific polyclonal anti-BB0646 reagent (data not shown); however, the recombinant BB0646, even when soluble, demonstrated no lipase activity. To further characterize the lipase activity of BB0646, the serine residue at the center of the conserved lipase motif, GXSXG, was mutagenized and reintroduced into DS102. The serine reside is one of three residues in a conserved catalytic triad that make up the active site found in hydrolases, including esterases and lipases (Jaeger et al., 1994; Gupta et al. 2004). When the conserved active site serine was mutagenized to either an alanine or threonine residue and the resulting whole cell lysates were tested, the lipase activity directed against a polyunsaturated substrate was nearly undetectable (Fig. 4C), despite the observation that the amount of BB0646 produced in the S152 mutant forms was indistinguishable from the most relevant complement construct, DS102/pDS126 (Fig. 4A). This data indicates that the lipase activity observed from borrelial lysates is directly attributable to BB0646 and is not the result of an indirect effect.
One factor that is still unclear is the localization of BB0646. We predicted that the lipase would be a soluble protein and, due to the putative leader peptidase I cleavage site, would be localized to the cellular envelope within the periplasmic space. The Triton X-114, detergent phase partitioning of BB0646 (Fig. 6A) may be due to the affinity of lipases for long hydrocarbon groups that are mimicked by detergents (Jaeger et al., 1994). Localization studies with Proteinase K suggested that BB0646 is not surface exposed, as expected, but is protease accessible when the outer membrane is compromised (Fig. 6B). Because BB0646 is not surface exposed, the observed lipase activity against saturated and polyunsaturated substrates is likely due to the presence of deoxycholate or Triton X-100 in the lipase assay buffers used, respectively, which would effectively release native BB0646 from the B. burgdorferi cells. The hemolytic activity is more difficult to predict, but is presumably due to either protein secretion or minimal localized cell lysis.
The observation that the bb0646 mutant does not exhibit impaired growth (Hyde et al., 2010) suggests that the BB0646 lipase activity is not the exclusive means for obtaining fatty acids, at least during cultivation in BSK-II media. One possible explanation posits that BB0646 works synergistically with passive diffusion to obtain and then enzymatically incorporate fatty acids into the cell envelope (Cox and Radolf, 2001). As such, a growth defect may not be observed for strain DS102 because of the rich, undefined nature of BSK-II media. Yet another possibility is that there is another, uncharacterized, lipase or an esterase encoded in the B. burgdorferi genome. If this is the case, this lipase would presumably exhibit reduced activity towards polyunsaturated substrates, as the bb0646::gentR strain exhibited near null lipase activity when assayed against substrates of this nature (Fig. 3B). The presence of an esterase is plausible given that the bb0646::gentR mutant did exhibit some activity when assayed against the shorter-chained saturated fatty acid, palmitate (Fig. 3A). Similarly, mostly incomplete, or alpha, hemolysis was observed for DS102 colonies on the horse blood BSK-II agar plates (Fig. 5). However, some low level β-like hemolysis was observed in strains lacking intact bb0646 implying, again, that BB0646 may not be the lone hemolytic enzyme encoded by B. burgdorferi.
The lipase and/or hemolytic activities of BB0646 contribute to the ability to infect the mammalian host based on decreased infectivity and bacterial load in cells lacking bb0646 (Fig.’s 7 and 8). In addition, BB0646 may play a role within the tick. It is well established that ticks of various species produce immunoregulatory salivary proteins that are then injected into the host during a blood meal (Bowman et al., 1996; Bowman and Sauer, 2004; Francischetti et al., 2009). These eicosanoids in the saliva of ticks, specifically prostaglandins, suppress the localized immune response and allow a hard-bodied tick to feed for several days (Bowman et al., 1996; Bowman and Sauer, 2004; Francischetti et al., 2009). Eicosanoid production directly correlates with the amount of arachidonic acid liberated from the host cell membranes (Bowman et al., 1996). An additional immunoregulatory role of BB0646 could lie in its ability to recognize and cleave both arachidonic acid (data not shown) and linolenic acid (Fig. 3B), thereby influencing eicosanoid production. It is tempting to speculate that any putative activity of BB0646 during the blood meal could contribute to the localized liberation of fatty acids, which are then converted to prostaglandins, thereby promoting the transmission of the spirochete during tick feeding.
In this study, we have shown the lipase activity of BB0646 can recognize both saturated and polyunsaturated substrates. The ability to liberate saturated fatty acids may serve as a potential nutrient acquisition mechanism for B. burgdorferi, while the more pronounced activity against polyunsaturated fatty acids suggests that it may affect the immunomodulatory response and/or repair of oxidized lipids/lipoproteins. Our preliminary data suggests that the absence of BB0646 does not dramatically alter borrelial sensitivity to oxidized stressors (Shaw and Skare, unpublished), implying that BB0646 is not absolutely needed for the clearance of damaged polyunsaturated lipids. However, BB0646 is required for optimal infectivity as demonstrated by the reduced colonization and bacterial load following 3 weeks of infection (Fig. 7 and 8) seen for the lipase mutant in vivo. Additional experimentation is needed to elucidate the precise role that BB0646 plays during the B. burgdorferi life cycle, particularly in regard to tick transmission and/or membrane envelope homeostasis.
Experimental Procedure
Bacterial strains
Borrelia burgdorferi strain B31 derivatives used in this study are listed in Table 1. All B. burgdorferi strains were grown in BSK-II media supplemented with 6% normal rabbit serum (Pel-Freez, Rogers, AR) lacking gelatin (Zückert, 2007). If required, B. burgdorferi was grown with antibiotics for selective pressure at the following concentrations: gentamicin at 50 µg ml−1; and kanamycin at 300 µg ml−1. The use of infectious B. burgdorferi in this study was reviewed and approved by the Institutional Biosafety Committee at Texas A&M University.
Table 1.
| Strain or plasmid | Genotype and/or characteristics | Source |
|---|---|---|
| E. coli strains | ||
| Rossetta (DE3) pLysS | CamR; F− ompT hsdSB(rB− mB−) gal dcm λ(DE3) pLysSRARE | Novagen |
| Mach-1™-T1R | ϕ80lacZΔM15 ΔlacX74 hsdR (rK−, mk+) ΔrecA1498 endA1 tonA | Invitrogen |
| B. burgdorferi strains | ||
| ML23 | B. burgdorferi strain B31 clonal isolate missing lp25 | (Labandeira-Rey and Skare, 2001) |
| DS102 | ML23, bb0646:gentR | (Hyde et al., 2010) |
| ML23/pBBE22 | Missing lp25 complemented with pBBE22 | (Seshu et al., 2006) |
| DS102/pBBE22 | ML23, bb0646::gentR complemented with pBBE22 | This study |
| DS102/pDS113 | ML23, bb0646::gentR complemented with pBBE22 containing PbosR-bb0646 | This study |
| DS102/pDS126 | ML23, bb0646::gentR complemented with pBBE22 containing PflgB-bb0646 | This study |
| DS102/pDS127 | ML23, bb0646::gentR complemented with pBBE22 containing PflgB-bb0646-S152A | This study |
| DS102/pDS128 | ML23, bb0646::gentR complemented with pBBE22 containing PflgB-bb0646-S152T | This study |
| Plasmids | ||
| pCR8/GW/TOPO | SpecR; Gateway PCR cloning/entry vector | Invitrogen |
| pCR2.1Bactin | KanR; β-actin gene cloned into pCR2.1 vector | (Maruskova et al., 2008) |
| pCR2.1recA | KanR; 1119 bp fragment, containing the recA gene, in pCR2.1 | This study |
| pET15b | CarbR; PT7lac lacI bla His6 coding sequence (5’) pBR322 origin | Novagen |
| pBSV2 | KanR; borrelial shuttle vector | (Stewart et al., 2001) |
| pBSV2G | GentR; borrelial shuttle vector | (Elias et al., 2003) |
| pBBE22 | KanR; borrelial shuttle vector carrying bbe22 | (Purser et al., 2003) |
| pBBE22Gate | CamR, KanR; pBBE22 modified to be a Gateway destination vector containing attL and attR sites | (Weening et al., 2008) |
| pDS200 | SpecR; bb0646 without the coding region of the putative leader peptide (bp 1–63) cloned into pCR8/GW/TOPO | This study |
| pDS201 | CarbR; bb0646 without the coding region of the putative leader peptide (bp 1–63) cloned into pET15b | This study |
| pDS110 | SpecR; 440 bp fragment of the bosR/bb0646 promoter region (PbosR) cloned into pCR/GW/TOPO | This study |
| pDS111 | SpecR; full-length bb0646 cloned into pCR/GW/TOPO | This study |
| pDS112 | SpecR; full-length bb0646 cloned from pDS111 into pDS110 to yield a PbosR-bb0646 transcriptional fusion | This study |
| pDS113 | KanR; pDS112 recombined into pBBE22Gate via the Gateway system | This study |
| pDS120 | SpecR; 535 bp of the flagellar promoter (PflgB) amplified from pBSV2 and cloned into pCR8/GW/TOPO | This study |
| pDS122 | SpecR; full-length bb0646 cloned from pDS111 into pDS120 to yield a PflgB-bb0646 transcriptional fusion | This study |
| pDS126 | KanR; pDS122 recombined into pBBE22Gate via the Gateway system | This study |
| pDS127 | KanR; pDS126 containing the PflgB-bb0646-S152A allele | This study |
| pDS128 | KanR; pDS126 containing the PflgB-bb0646-S152T allele | This study |
Escherichia coli Mach1™-T1R cells were utilized for cloning and Rosetta pLysS E. coli cells were used for the overproduction of recombinant BB0646. The resulting E. coli strain was grown with aeration in LB media and appropriate antibiotic selection: gentamicin at 5 µg ml−1; spectinomycin at 100 µg ml−1; and kanamycin at 50 µg ml−1; chloramphenicol at 25 µg ml−1 and carbenicillin at 100 µg ml−1.
Plasmid constructs
All plasmid constructs used in this study are listed in Table 1. PCR was performed as previously described with Invitrogen’s SuperMix High Fidelity system (Weening et al., 2008) and verified by dideoxy sequencing. An amino-terminal His-tag fusion construct was generated by amplifying a 921bp fragment of bb0646, excluding the type I leader peptide, with primers DS200-NdeI-F and DS200-BamHI-R (Table 2) and cloned into pCR8/GW/TOPO, resulting in pDS200. pDS200 was digested with BamHI and NdeI and the resulting bb0646-containing fragment was cloned into pET15b to obtain pDS201.
Table 2.
| Oligonucleotide | Sequence | Description |
|---|---|---|
| DS200-NdeI-F | CTGGTTCATATGAGGATAAAATTT | Primer pair for amplifying bp 64–984 of bb0646 |
| DS200-BamHI-R | GCTTAGGATCCTTACTTATTAATC | |
| DS_gent-R | ACGCAACAAAAAGATTTGAATTATTATAT | Primer pair for amplifying a 541 bp fragment of the gentamicin resistance cassette for use as a Southern blot probe |
| DS_gent-F | ACGCGAGAGCCACTGCGGGATCGTCACCG | |
| DS110-F | ACGCCTAAAAACAATCCCTTAGGCTACATTAAT | Primer pair for amplifying the bosR promoter (PbosR) |
| DS110-NdeI-R | ACGCCATATGATTATACCTTTTTTGTTTAAAT | |
| DS111-NdeI-F | ACGCCATATGAATATAAAAAATATCATTTTTA | Primer pair for amplifying full-length bb0646 |
| DS111-NdeI-R | ACGCCATATGTTACTTATTAATCTTGTTTATG | |
| gent_int-F | ACGCGCAGCCGCGTAGTGAGATCTAT | Forward primer used for screening positive bb0646::gentR isolates |
| gent_int-R | ACGCATATAGATCTCGTTACGCGGCT | Reverse primer used for screening putative bb0646::gentR isolates |
| PFlgB-F | ACGCTGGCGTTACCCAACTTAATCG | Primer pair for amplifying the flagellar promoter, PflgB, from pBSV2 (Stewart et al., 2001) |
| PFlgB-NdeI-R | ACGCCATATGTATGGAAACCTCCCTCATTTAA A | |
| pncA_nested-F | ACGCGTATACATATATTTTAAATAAAA | Primer that sits within bbe22 and used for confirming borrelial shuttle vector transformants |
| pDS113_R-confirm | ACGCTTGAGAGTAATTTATTACATC | Primer that sits within the promoter region of PbosR and is used for confirming the presence of pDS113 (when used with pDS113_R-confirm). |
| pDS126_R-confirm | ACGCAAAATTCATTTTTAAATTTTATC | Primer that sits within bb0646 and is used for confirming the presence of pDS126 (when used with pDS126_R-confirm). |
| BB0646 ALA quikchange_T454G-F | GTATTAATTGGAACCGCTAATGGGGGCACTG | Primer pair that changes the serine at position 152 to an alanine (TCT →GCT) in bb0646 |
| BB0646 ALA quikchange_T454G-R | CAGTGCCCCCATTAGCGGTTCCAATTAATAC | |
| BB0646 THR quikchange_T454A-F | GTATTAATTGGAACCACTAATGGGGGCACTG | Primer pair that changes the serine at position 152 to an threonine (TCT →ACT) in bb0646 |
| BB0646 THR quikchange_T454A-R | CAGTGCCCCCATTAGTGGTTCCAATTAATAC | |
| recA-F | ACGCAAATTTTCCATATTACTCAGATT | Primer pair used to amplify the 1119 bp fragment containing recA from B. burgdorferi genomic DNA to make pCR2.1recA. |
| recA- R | ACGCAATTTAAGAATGTCAAAGTTAAA | |
| qPCR-Bactin-F | ACGCAGAGGGAAATCGTGCGTGAC | Primer pair used for enumerating copies of mouse β-actin via qPCR (Pal et al., 2008) |
| qPCR-Bactin-R1 | ACGCGGGAGGAAGAGGATGCGGCAGTG | |
| nTM17FrecA | GTGGATCTATTGTATTAGATGAGGCT | Primer pair used for enumerating copies of B. burgdorferi recA via qPCR (Liveris et al., 2002; Weening et al., 2008) |
| nTM17RrecA | GCCAAAGTTCTGCAACATTAACACCT |
The bb0646::gentR insertional mutant, DS102 (Hyde et al., 2010), was complemented with intact bb0646 in trans under control of two different promoters, resulting in the final complementation constructs pDS113 and pDS126, respectively. pDS113 was constructed by making a transcriptional fusion of bb0646 with its native promoter (PbosR), which is located upstream from bosR (bb0647) (Ouyang et al., 2009; Hyde and Skare, unpublished observation). To accomplish this a 440 bp fragment corresponding to the native bosR promoter region was PCR amplified from B. burgdorferi genomic DNA using oligonucleotide primers DS110-F and DS110-NdeI-R (Table 2) and was cloned into pCR8/GW/TOPO (pDS110). A second 984 bp fragment, containing promoterless bb0646, was PCR amplified from B. burgdorferi genomic DNA using oligonucleotide primers DS111-NdeI-F and DS111-NdeI-R (Table 2) and cloned into pCR8/GW/TOPO to generate pDS111. The bb0646 fragment was digested out of pDS111 with NdeI and cloned into pDS110 with the start of bb0646 abutting PbosR; the resulting construct was designated pDS112. The PbosR-bb0646 construct was moved into pBBE22Gate (Weening et al., 2008) using the Gateway based LR recombinase and designated pDS113 (Table 1).
The second complementation construct, pDS126, placed bb0646 under control of the strong, constitutive promoter, PflgB (Bono et al., 2000). To this end, a 535 bp fragment was PCR amplified from pBSV2 (Stewart et al., 2001) using primers pFlgB-F and pFlgB-NdeI-R (Table 2), cloned into pCR8/GW/TOPO, and designated pDS120. The bb0646 fragment from pDS111 was digested with NdeI and cloned into pDS120 with the start of bb0646 transcriptionally fused with PflgB, generating pDS122. The PflgB-bb0646 construct was moved into pBE22Gate, as described above, and was designated pDS126.
To serve as a standard for the enumeration of borrelial genomic equivalents, the B. burgdorferi strain B31 recA gene was PCR amplified using primers recA-F and recA-R (Table 2) and the resulting 1119 bp band was cloned into pCR2.1-TOPO (Invitrogen) generating pCR2.1recA.
Transformation of Borrelia burgdorferi
Borrelia burgdorferi strain DS102 was made competent and was electroporated as described (Samuels, 1995; Seshu et al., 2004b, 2006; Weening et al., 2008; Hyde et al., 2009, 2010). Following recovery overnight, transformants were selected with appropriate antibiotics (i.e., gentamicin at 50 µg ml−1 or kanamycin at 300 µg ml−1) and diluted using the liquid plating method previously described (Yang et al., 2004). Putative transformants were then expanded in BSK-II media and the presence of the desired construct and borrelial plasmid content (as indicated in Labandeira-Rey and Skare, 2001) were confirmed by PCR.
Purification and antibody production of overproduced BB0646
To overproduce recombinant BB0646, Rosetta pLysS pDS201 were grown, pelleted, washed, and frozen at −20°C as previously described (Labandeira-Rey et al., 2001). Recombinant BB0646 was purified from the insoluble fraction that was resolved by SDS-PAGE and stained with E-Zinc® stain (Pierce, Rockford, IL). A protein band corresponding to recombinant BB0646 was excised from the gel, placed in elution buffer (50 mM Tris-HCl, 150 mM NaCl, and 0.1 mM EDTA, pH 7.5), homogenized, and incubated overnight at 30°C. The mixture was separated by centrifugation (10,000 × g) and the supernatant was removed and stored at −20°C. Protein quantification was performed using a BCA assay kit (Pierce, Rockford, IL).
To obtain monospecific polyclonal antisera to BB0646, a New Zealand white rabbit was immunized with approximately 640 µg of purified recombinant BB0646 protein. Three subsequent booster injections with the same amount of protein were administered every 3 weeks over a period of 2.5 months. Serum was collected following the final boost and stored at −80°C. All animal work was performed under the approval of the University Laboratory Animal Care Committee at Texas A&M University.
Southern blotting
Isolation of B. burgdorferi genomic DNA and Southern blotting procedures were performed as previously described (Skare et al., 1999). The probe used in this study, specific for the gentamicin antibiotic resistance cassette, was amplified from pBSV2G plasmid DNA (Elias et al., 2003) using primers DS_gent-R and DS_gent-F (Table 2) and labeled with fluorescein as previously described (Skare et al., 1999).
SDS-PAGE and Western immunoblotting
Protein samples were resolved by SDS-PAGE and either stained with Coomassie Brilliant Blue R-250 (Sigma Aldrich, St. Louis, MO) or transferred to PVDF membranes and immunoblotted as described previously (Seshu et al., 2004b). Primary antibodies were used at the following dilutions: anti-BB0646 at 1:1,000; anti-His-tag (Clontech, Mountain View, CA) at 1:5,000; anti-P66 at 1:5000 (generously provided by Sven Bergström); and anti-FlaB at 1:20,000 (Affinity Bioreagent, Golden, CO). Prior to using the anti-BB0646 for Western immunoblotting, the antisera was adsorbed to DS102 borrelial lysates to reduce the amount of nonspecific antibodies, as previously described (Gruber and Zingales, 1995; Skare et al., 1999). Appropriate secondary antibodies, with horseradish peroxidase (HRP) conjugates (anti-mouse HRP [Invitrogen, Carlsbad, CA] or anti-rabbit HRP [Amersham, Piscataway, NJ], both at 1:4000) were used and developed using the Western Lightning Chemiluminescence Reagent plus (Perkin Elmer, Waltham, MA).
Lipase assays
Chromogenic assays were performed as previously described (Winkler and Stuckmann, 1979; Li et al., 1995) with late log phase B. burgdorferi concentrated to 3 × 108 by centrifugation (9,000 × g for 10 minutes). The final cell pellets were resuspended in 1 ml of lipase assay buffer containing Sorenson Buffer (1.25 mM NaH2PO4, 3.75 mM Na2HPO4, 0.05% CaCl2, pH 8), 4.72 mM sodium deoxycholate sodium salt, 0.9% gum arabic, and 475 µM p-nitrophenyl palmitic acid (Sigma Aldrich, St. Louis, MO). Suspensions were incubated at 37°C for 15 min. and transferred to an ice bath to terminate the reaction. The absorbance of each sample was quantified at 410 nm using clear 96 well plates. Blanks containing buffer and substrate were subtracted from the experimental samples to minimize background and account for nonspecific substrate hydrolysis.
Fluorescent lipase assays were performed with 1 × 108 B. burgdorferi cells that were grown and washed as described above. The final cell pellets were resuspended in 50 µl of lipase assay buffer containing 50 mM Tris-HCl, 1 mM EDTA, 1.1 mM CaCl2, 0.1% Triton X-100 and 10 µM 7-hydroxycoumarinyl linolenate (Cayman Chemicals, Ann Arbor, MI). The samples were read at 10 minute intervals over a period of 2 hours on 96 well black bottom plates at 355 nm excitation and 460 nm emission. Blanks were subtracted from the experimental samples to adjust for non-specific hydrolysis of the substrate.
Site-directed mutagenesis
The active site serine within the conserved lipase motif, GXSXG, was targeted for mutagenesis using the QuikChange I Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). To this end, the serine was converted to either an alanine (TCT →GCT) or a threonine (TCT→ACT) using the primers listed in Table 2 and the PflgB-bb0646 fusion construct (pDS126) as template DNA. Putative point mutants were screened and candidates were verified by sequencing prior to transformation into the bb0646::gentR strain (DS102) resulting in DS102/pDS127 (containing bb0646-S152A) and DS102/pDS128 (containing bb0646-S152T).
Hemolytic assays
BSK-II blood agar plates containing a final concentration of 5% defibrinated horse blood (Cleveland Scientific, Bath, OH) were prepared as previously described (Williams and Austin, 1992). B. burgdorferi cultures were grown to mid log phase and 100 colony forming units were plated per strain assayed. The plates were incubated at 32°C, 1% CO2 until colonies were visible. Alpha hemolysis was defined as green/brown colonies with no overt lysis observed. Partial beta hemolysis, designated as “α/β hemolysis”, was defined as a zone of clearing around a colony with a green/brown border.
Triton X-114 Phase Partitioning
Phase partitioning of B. burgdorferi protein were done as described (Skare et al., 1995).
Protease accessibility assay
Protease accessibility assays were performed as previously described (Barbour et al., 1984; Labandeira-Rey et al., 2001) with the following modifications. Approximately 3 × 108 mid log phase cells were pelleted via centrifugation (9,000 × g for 10 minutes at 4°C) and washed twice with PBS. The cells were resuspended in 50 µl of either: sterile water, proteinase K (to a final concentration of 200 µg/mL), or proteinase K and 0.05% Triton X-100. All samples were incubated at 20°C for 40 min. Reactions were terminated following the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM. Cells were pelleted by centrifugation (9,000 × g for 10 minutes at 4°C), washed twice with PBS containing 1 mM PMSF and resuspended in Laemmli sample buffer (Laemmli, 1970).
Infectivity studies
Infectivity studies were performed as previously described (Hyde et al., 2009). Briefly, 8 week old C3H/HeN mice were infected intradermally with either 103 or 105 inoculums of B. burgdorferi strain ML23/pBBE22, DS102/pBBE22, or DS102/pDS113. For each dose and strain used, 4–5 mice were infected. After 21 days, the mice were sacrificed and lymph node, skin, heart, spleen, bladder, and tibiotarsal joint tissues were aseptically removed. Qualitative analysis of infection was performed by inoculating the aforementioned tissues in BSK-II media with appropriate antibiotics (i.e. kanamycin at 300 µg ml−1 or gentamicin at 50 µg ml−1/ kanamycin at 300 µg ml−1). The presence of B. burgdorferi were scored out to 1 month by dark field microscopy (Labandeira-Rey and Skare, 2001; Labandeira-Rey et al., 2003; Seshu et al., 2006; Gilbert et al., 2007; Weening et al., 2008). All animal studies were performed was approved by the University Laboratory Animal Care Committee at Texas A&M University.
Quantitative PCR
Quantitative analysis of lymph node, skin, and tibiotarsal joint tissues from infected mice was performed by extracting DNA using the Roche High Pure PCR Template preparation kit as described previously (Maruskova et al., 2008; Weening et al., 2008). B. burgdorferi genome copies were enumerated along with mammalian genome copies using the Applied Biosystems ABI 7500 fast real-time PCR system and SYBR green PCR Mastermix (Applied Biosystems Corp., Foster City, CA). Approximately 100 ng of total DNA was used in each reaction with primer set nTM17FRecA and nTM17RRecA for detection of the B. burgdorferi recA gene (Liveris et al., 2002; Weening et al., 2008) or primer set qPCR-Bactin-F and qPCR-Bactin-R1 for detection of mammalian β-actin copies (Pal et al., 2008) (Table 2). Borrelial and mammalian genomic copies were determined separately by comparing the threshold cycle (CT) value of individual tissues to the appropriate standard curve established with known quantities of pCR2.1recA and pCR2.1bactin (Table 1). All samples were assayed in triplicate and presented as copies of B. burgdorferi recA per 106 mouse β-actin copies.
Statistical analysis
For all p-nitrophenyl palmitate lipase assays, a two-tailed Welsh’s t-test was performed between indicated strains. Samples were considered statistically significant if the P value was less than 0.05. For all 7-hydroxycoumarinyl linolenate lipase assays, a one-way analysis of variance was performed between all strains at discrete time points. A Tukey’s post test was used to determine P values between strains. Samples were considered statistically significant if the P value was less than 0.05. A two-tailed Welsh’s t-test was performed with the hemolysis data for indicated strains. Values were considered significantly different if the P value was less than 0.05. For the quantitative PCR analysis, a one-tailed Mann-Whitney’s t-test was performed between the strains indicated. A P value that was less than 0.05 was considered statistically significant.
Acknowledgements
We thank Eric Weening and Michelle McGehee for their valued assistance with the animal experimentation and purification of recombinant BB0646, respectively, and many helpful discussions. We are also grateful to Bob Cluss for helpful discussions and for sharing unpublished information and to Michael Benedik for providing p-nitrophenyl palmitate for the initial lipase assays and for encouraging advice. Thanks also to Patti Rosa for sending pBSV2 and pBSV2G, to Steve Norris for providing pBBE22, and to Sven Bergström for antiserum specific to the borrelial P66 protein. We gratefully acknowledge the technical assistance of Texas A&M University’s Comparative Medicine Program with their assistance in the BB0646 antibody production. This work was supported by Public Health Service grant R01-AI042345 (to J.T.S.).
References
- Anderson JF. Epizootiology of Borrelia in Ixodes tick vectors and reservoir hosts. Rev Infect Dis. 1989;11 Suppl 6:S1451–S1459. doi: 10.1093/clinids/11.supplement_6.s1451. [DOI] [PubMed] [Google Scholar]
- Anderson JF. Epizootiology of Lyme borreliosis. Scand J Infect Dis Suppl. 1991;77:23–34. [PubMed] [Google Scholar]
- Barbour AG, Tessier SL, Hayes SF. Variation in a major surface protein of Lyme disease spirochetes. Infect Immun. 1984;45:94–100. doi: 10.1128/iai.45.1.94-100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Hayes SF. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, Stevenson B, Tilly K, Rosa P. Efficient Targeted Mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Dillwith JW, Sauer JR. Tick salivary prostaglandins: Presence, origin and significance. Parasitol Today. 1996;12:388–396. doi: 10.1016/0169-4758(96)10061-2. [DOI] [PubMed] [Google Scholar]
- Bowman AS, Sauer JR. Tick salivary glands: function, physiology and future. Parasitology. 2004;129 Suppl:S67–S81. doi: 10.1017/s0031182004006468. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: A distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci U S A. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, Crane EJ, Gherardini FC. Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol. 2006;59:475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Lawrence KA, Downey JS, Gherardini FC. Borrelia burgdorferi membranes are the primary targets of reactive oxygen species. Mol Microbiol. 2008;68:786–799. doi: 10.1111/j.1365-2958.2008.06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W. Vector/host relationships of the Lyme disease spirochete, Borrelia burgdorferi. Rheum Dis Clin North Am. 1989;15:775–787. [PubMed] [Google Scholar]
- Cox DL, Radolf JD. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology. 2001;147:1161–1169. doi: 10.1099/00221287-147-5-1161. [DOI] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, 3rd, Stewart PE, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Gherardini FC, Boylan JA, Lawrence KA, Skare JT. Metabolism and Physiology of Borrelia. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk: Caister Academic Press; 2010. pp. 103–138. [Google Scholar]
- Gilbert MA, Morton EA, Bundle SF, Samuels DS. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol Microbiol. 2007;63:1259–1273. doi: 10.1111/j.1365-2958.2007.05593.x. [DOI] [PubMed] [Google Scholar]
- Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. BioTechniques. 1995;19:28. 30. [PubMed] [Google Scholar]
- Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004;64:763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- Gutteridge JM, Halliwell B. The measurement and mechanism of lipid peroxidation in biological systems. Trends Biochem Sci. 1990;15:129–135. doi: 10.1016/0968-0004(90)90206-q. [DOI] [PubMed] [Google Scholar]
- Holmquist M. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Hyde JA, Seshu J, Skare JT. Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative oxidative stress regulon. Microbiology. 2006;152:2599–2609. doi: 10.1099/mic.0.28996-0. [DOI] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R, III, Trzeciakowski JP, Skare JT. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 2009;74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R, Trzeciakowski JP, Skare JT. Characterization of a Conditional bosR Mutant in Borrelia burgdorferi. Infect Immun. 2010;78:265–274. doi: 10.1128/IAI.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istivan TS, Coloe PJ. Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology. 2006;152:1263–1274. doi: 10.1099/mic.0.28609-0. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. The Fur Homologue in Borrelia burgdorferi. J Bacteriol. 2004;186:6443–6456. doi: 10.1128/JB.186.19.6443-6456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempner MS, Hu LT, Evans J, Schmid CH, Johnson GM, Trevino RP, et al. Two controlled trials of antibiotic treatment in patients with persistent symptoms and a history of Lyme disease. N Engl J Med. 2001;345:85–92. doi: 10.1056/NEJM200107123450202. [DOI] [PubMed] [Google Scholar]
- Labandeira-Rey M, Skare JT. Decreased Infectivity in Borrelia burgdorferi Strain B31 Is Associated with Loss of Linear Plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Baker EA, Skare JT. VraA (BBI16) Protein of Borrelia burgdorferi is a Surface-Exposed Antigen with a Repetitive Motif That Confers Partial Protection against Experimental Lyme Borreliosis. Infect Immun. 2001;69:1409–1419. doi: 10.1128/IAI.69.3.1409-1419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Seshu J, Skare JT. The Absence of Linear Plasmid 25 or 28-1 of Borrelia burgdorferi Dramatically Alters the Kinetics of Experimental Infection via Distinct Mechanisms. Infect Immun. 2003;71:4608–4613. doi: 10.1128/IAI.71.8.4608-4613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li X, Tetling S, Winkler UK, Jaeger KE, Benedik MJ. Gene cloning, sequence analysis, purification, and secretion by Escherichia coli of an extracellular lipase from Serratia marcescens. Appl Environ Microbiol. 1995;61:2674–2680. doi: 10.1128/aem.61.7.2674-2680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, et al. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- Liveris D, Wang G, Girao G, Byrne DW, Nowakowski J, McKenna D, et al. Quantitative Detection of Borrelia burgdorferi in 2-Millimeter Skin Samples of Erythema Migrans Lesions: Correlation of Results with Clinical and Laboratory Findings. J Clin Microbiol. 2002;40:1249–1253. doi: 10.1128/JCM.40.4.1249-1253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruskova M, Esteve-Gassent MD, Sexton VL, Seshu J. Role of the BBA64 Locus of Borrelia burgdorferi in Early Stages of Infectivity in a Murine Model of Lyme Disease. Infect Immun. 2008;76:391–402. doi: 10.1128/IAI.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadelman RB, Wormser GP. Lyme borreliosis. Lancet. 1998;352:557–565. doi: 10.1016/S0140-6736(98)01146-5. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol. 2009;74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Wang P, Bao F, Yang X, Samanta S, Schoen S, et al. Borrelia burgdorferi basic membrane proteins A and B participate in the genesis of Lyme arthritis. J Exp Med. 2008;205:133–141. doi: 10.1084/jem.20070962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- Samuels DS. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid GP. The global distribution of Lyme disease. Rev. Infect. Dis. 1985;7:41–50. doi: 10.1093/clinids/7.1.41. [DOI] [PubMed] [Google Scholar]
- Schmiel DH, Miller VL. Bacterial phospholipases and pathogenesis. Microbes Infect. 1999;1:1103–1112. doi: 10.1016/s1286-4579(99)00205-1. [DOI] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved Oxygen Levels Alter Gene Expression and Antigen Profiles in Borrelia burgdorferi. Infect Immun. 2004a;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Hyde JA, Swingle KL, Gherardini FC, Skare JT. A conservative amino acid change alters the function of BosR, the redox regulator of Borrelia burgdorferi. Mol Microbiol. 2004b;54:1352–1363. doi: 10.1111/j.1365-2958.2004.04352.x. [DOI] [PubMed] [Google Scholar]
- Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Höök M, Skare JT. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol. 2006;59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- De Silva AM, Fikrig E. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- Skare JT, Shang ES, Foley DM, Blanco DR, Champion CI, Mirzabekov T, et al. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J Clin Invest. 1995;96:2380–2392. doi: 10.1172/JCI118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare JT, Foley DM, Hernandez SR, Moore DC, Blanco DR, Miller JN, Lovett MA. Cloning and Molecular Characterization of Plasmid-Encoded Antigens of Borrelia burgdorferi. Infect Immun. 1999;67:4407–4417. doi: 10.1128/iai.67.9.4407-4417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC. Lyme disease. N Engl J Med. 2001;345:115–125. doi: 10.1056/NEJM200107123450207. [DOI] [PubMed] [Google Scholar]
- Steere AC. A 58-year-old man with a diagnosis of chronic Lyme disease. JAMA. 2002;288:1002–1010. doi: 10.1001/jama.288.8.1002. [DOI] [PubMed] [Google Scholar]
- Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Takayama K, Rothenberg RJ, Barbour AG. Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1987;55:2311–2313. doi: 10.1128/iai.55.9.2311-2313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball RW. Bacterial phospholipases C. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, Benach JL. Combined Effects of Blood and Temperature Shift on Borrelia burgdorferi Gene Expression as Determined by Whole Genome DNA Array. Infect Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Höök M, Skare JT. Borrelia burgdorferi Lacking DbpBA Exhibits an Early Survival Defect during Experimental Infection. Infect Immun. 2008;76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LR, Austin FE. Hemolytic activity of Borrelia burgdorferi. Infect Immun. 1992;60:3224–3230. doi: 10.1128/iai.60.8.3224-3230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler UK, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fang QQ, Keirans JE, Durden LA. Molecular phylogenetic analyses indicate that Ixodes ricinus complex is a paraphyletic group. J Parasitol. 2003;89:452–457. doi: 10.1645/0022-3395(2003)089[0452:MPAITT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential Role for OspA/B in the Life Cycle of the Lyme Disease Spirochete. J Exp Med. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zückert WR. Laboratory Maintenance of Borrelia burgdorferi. Current Protoc Microbiol. 2007;Chapter 12(Unit 12C.1) doi: 10.1002/9780471729259.mc12c01s4. [DOI] [PubMed] [Google Scholar]