Abstract
Chromosome end protection is essential to protect genome integrity. Telomeres, tracts of repetitive DNA sequence and associated proteins located at the chromosomal terminus, serve to safeguard the ends from degradation and unwanted double strand break repair. Due to the essential nature of telomeres in protecting the genome, a number of unique proteins have evolved to ensure that telomere length and structure are preserved. The inability to properly maintain telomeres can lead to diseases such as dyskeratosis congenita, pulmonary fibrosis and cancer. In this review, we will discuss the known functions of mammalian telomere-associated proteins, their role in telomere replication and length regulation and how these processes relate to genome instability and human disease.
Keywords: telomere, telomerase, shelterin, replication, CST
1. Introduction
Telomeres can be thought of as protective caps for chromosomes that are composed of repeated DNA sequences bound by a series of specialized telomere proteins (Fig. 1) [1]. The telomere proteins prevent the chromosome terminus from being seen as DNA damage and initiating a DNA damage response. Defects in the protective cap structure lead to cell cycle arrest and/or DNA repair activities that cause end-to-end fusion of chromosomes via non-homologous end joining (NHEJ) [2].
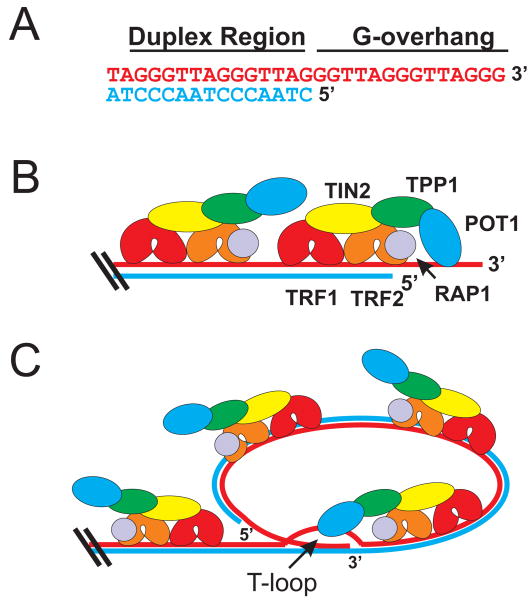
Fig. 1. Telomere Structure.
(A) Telomeric DNA consists of repetitive DNA sequence, a duplex region and a ssDNA G-strand overhang (G-strand, red; C-strand blue). (B) The shelterin complex binds to both the duplex and ssDNA regions through specific protein-DNA interactions. (C) Formation of the t-loop involves strand invasion of the G-overhang to create a displacement-loop (D-loop). The t-loop is proposed to mask the chromosome end from DNA damage sensors. See text for further details.
Telomeres are directly affected by the inability of DNA polymerase to completely replicate the 5′ end of a linear chromosome, a situation known as the end replication problem [3, 4]. Human somatic cells generally lack a mechanism to compensate for the resulting loss of DNA from the chromosome terminus, so progressive rounds of replication lead to gradual telomere shortening. After many rounds of cell division, telomeres become critically short and are sensed as DNA damage [5]. The damage signal stops the cell from further division and it becomes senescent. The ability of telomeres to limit cell proliferation can be thought of as a tumor suppressor mechanism [6, 7]. However under some circumstances, such as embryogenesis or stem cell proliferation, cells must be able to divide without the penalty of progressive telomere shortening. In these situations, a specialized reverse transcriptase, called telomerase, adds DNA to the chromosome terminus to compensate for the sequence lost during DNA replication [8, 9]. Telomerase is an RNA-containing enzyme that uses the catalytic subunit, TERT, to reverse transcribe the template region of the RNA subunit, TR, onto the 3′ end of the telomeric DNA. In some instances telomeres can also be elongated by a recombination based mechanism known as ALT (alternative lengthening of telomeres) [10].
Abnormal telomere shortening or telomere elongation can have adverse effects on human health. For example, undue telomere shortening due to telomerase deficiency in highly proliferative tissue can lead to diseases such as dyskeratosis congenita or pulmonary fibrosis [7, 11](see accompanying articles by M. Armanios [12] and A. Bertuch [13]). Conversely, telomerase upregulation leads to the cellular immortalization that is fundamental to cancer cell growth (see accompanying article by J. Shay [14]). Thus, telomere length regulation is essential for both cellular and organismal well being. Factors that contribute to telomere length regulation include the structure and composition of the telomere, the availability of telomerase and the interplay between telomere proteins, telomerase and the DNA replication machinery. This article describes overall telomere structure in mammalian cells and how mammalian telomere proteins contribute to end protection, telomere replication and length regulation. Telomerase structure, biosynthesis and regulation will be covered in other articles in this issue [15, 16].
2. Telomeric DNA
Telomere length varies between organisms, ranging from several hundred base pairs of DNA in yeast to tens of kilobases (kb) in mammals [1, 17]. In humans, telomeres are generally in the 5-15 kb length range while those of the lab mouse (Mus musculus) are 40-50 kb. Mammalian telomeric DNA consists of short tandem repeats with the 3′ or G-rich strand composed of 5′ TTAGGG repeats and the complementary 5′ or C-rich strand composed of 5′ CCCTAA repeats (Fig. 1A). Structurally, telomeres contain regions of double-stranded (ds) and single-stranded (ss) DNA. The double-stranded region comprises the bulk of the telomere, while the single-stranded region exists as an (∼)100 nucleotide extension at the 3′ end of the G-rich strand [18, 19]. This “G-overhang” can form a structure called a t-loop (telomeric-loop) (Fig. 1C), which is believed to mask the DNA terminus from the double-stranded break (DSB) repair machinery and to limit access to telomerase [17, 20]. To generate the t-loop, the G-overhang is proposed to strand-invade into a region of the telomeric dsDNA to form a displacement-loop (D-loop). Telomere proteins are thought to aid in t-loop formation. T-loops have been observed both on isolated telomeric DNA and on telomeric chromatin by electron microscopy [20, 21], but it is currently unclear whether they are present at all telomeres.
3. Shelterin
Telomeres are protected by specialized multi-protein complexes that include proteins tailored to recognize the dsDNA and the G-overhang. The complex from mammalian cells is called shelterin because it shelters the telomere from a series of unwanted activities [17]. Shelterin consists of six proteins, TRF1, TRF2, RAP1, TIN2, TPP1 and POT1 [17, 22](Fig. 1B). TRF1 and TRF2 bind the telomeric dsDNA while POT1 binds the overhang. TIN2 and TPP1 are linker proteins that hold the complex together. TIN2 is the lynchpin that stabilizes the complex by interacting with TRF1, TRF2 and TPP1. TPP1 interacts with POT1 in addition to TIN2, thus forming a link between the G-overhang and duplex binding proteins. RAP1 interacts with TRF2. At first sight, the six protein shelterin complex appears to work as a functional unit because disruption of one component can affect the activities of the remaining components and lead to loss of telomere protection. However, as discussed below, the individual shelterin components have evolved specific and not necessarily overlapping functions in telomere replication and end-protection [23]. Sub-complexes consisting of only 3-5 shelterin components have also been identified but their function remains poorly understood [24, 25].
TRF1 and TRF2 are structurally similar in that they both bind telomeric dsDNA through a SANT/Myb domain found at the C-terminus and they share an internal TRF homology domain (TRFH) [26]. They each form homodimers and oligomers through the TRFH domains but they do not directly interact to form heterodimers or oligomers. Both proteins are essential in mice. Conditional deletion of either protein leads to destabilization of shelterin and the resulting telomere deprotection leads to a telomeric DNA damage response. However, genetic studies have revealed that TRF1 and TRF2 perform very different roles in telomere replication and end-protection. TRF1 helps promote replication through the telomere duplex, possibly by recruiting helicases such as BLM and RTEL [27, 28]. Removal of TRF1 leads to replication fork stalling and ultimately to defects in packaging of the telomeric tract (visible as multi-telomeric signals by fluorescence in situ hybridization (FISH) with telomere probes) and fusions of sister telomeres. In contrast, TRF2 functions to ensure maintenance of the G-overhang, and is needed to prevent G-overhang degradation [29]. TRF2 also plays a key role in overhang generation following DNA replication by recruiting the Apollo nuclease (see below) [29-31]. Deletion of TRF2 leads to overhang loss and rampant telomere fusions via NHEJ [32, 33].
POT1 and TPP1 function together by forming a heterodimer that regulates telomerase activity and general access to the G-overhang [34]. POT1 binds to telomeric G-strand DNA via two oligonucleotide/oligosaccharide binding (OB) folds. Although TPP1 does not bind DNA directly, it increases the binding affinity of POT1 for telomeric DNA by 5-10-fold [35]. TPP1 is also required to localize POT1 to the telomere as POT1 lacks a nuclear localization sequence [36]. A key function of POT1-TPP1 is to prevent the abundant single strand binding protein RPA from binding the G-overhang and eliciting an ATR-mediated DNA damage response [37-40]. If POT1 is removed from the telomere, the overhang becomes coated with RPA which then recruits ATRIP-ATR and other proteins necessary for ATR activation, resulting in cell cycle arrest and cell death. POT1 and TPP1 also function in telomere length regulation [34]. Both proteins regulate telomerase action on the chromosome terminus but they appear to have opposing activities. POT1 can sequester the DNA 3′ end to make it inaccessible to telomerase [39, 41], while TPP1 participates in telomerase recruitment and stimulates telomerase activity by increasing enzyme processivity [42-44]. It is not yet understood how these two activities are coordinated.
TIN2 plays a key role in stabilizing shelterin and maintaining association of the other shelterin components with the telomere. Although TRF1 and TRF2 bind telomeric DNA independently and with high affinity, depletion of TIN2 results in substantial loss of both proteins from the telomere [45-47]. Moreover, the TIN2-TPP1 interaction is responsible for the association of POT1-TPP1 with the duplex region of the telomeric DNA and the resulting high local concentration, which may help POT1 compete with RPA for binding to the G-overhang or base of the t-loop [48]. TIN2 also plays an important role in maintaining chromosome integrity through interactions with non-shelterin proteins. Both TIN2 and TRF1 interact with the cohesin subunit SA1 and this interaction is required to establish and maintain sister chromatid cohesion at telomeres and along the chromosome arms [49]. TIN2 may also anchor telomeres into the nuclear matrix through an extra C-terminal domain present in some splice variants [50]. It is unclear whether TIN2 interactions with either cohesin or the nuclear matrix occur in the context of the canonical shelterin complex or through various TIN2-containing subcomplexes [25]. Interestingly, TIN2 is the only sheterin component thus far to be linked to human disease [51, 52]. As discussed in the accompanying article by A. Bertuch [13], certain TIN2 mutations are found in patients with short telomeres and the pathology associated with dyskeratosis congenita.
The remaining shelterin protein, RAP1, differs from the other five shelterin subunits in that it is the only subunit that is not essential in mice [53, 54]. Deletion of mouse RAP1 leads to increased telomere recombination but does not cause NHEJ or chromosome fusions. The lack of effect on NHEJ suggests that RAP1 functions independently of its partner, TRF2. However, the situation in human cells is somewhat different as here RAP1 seems to function in tandem with TRF2 to prevent NHEJ and chromosome fusions [55]. Like its counterpart in budding yeast, mammalian RAP1 also appears to have various non telomere-related activities. It can form a complex with IKK (IkapaB Kinase) and participate in NF-kapaB activation [56]. Moreover, RAP1 has been found in association with TTAGGG repeats at extra telomeric sites, probably tethered there via TRF2, where it seems to play a role in regulating gene expression [54, 57].
4. The CST complex
Although shelterin is the primary mammalian telomere protein complex, an additional telomere-associated complex has recently been identified in mammalian cells. This complex, known as CST, consists of Conserved Telomere Component 1 (CTC1), STN1 (also known as OBFC1) and TEN1 [58, 59]. Mammalian STN1 and TEN1 are orthologous to Stn1 and Ten1 from budding yeast. In yeast, these proteins interact with Cdc13 to form a complex, also named CST, which is essential for both telomere protection and telomere replication [60, 61]. Budding yeast telomeres are protected by two different protein complexes; the Cdc13/Stn1/Ten1 (CST) complex which binds to the 3′ overhang and the Rap1/Rif1/Rif2 complex that bids to the telomere duplex [62, 63]. Mammalian cells were initially thought to lack a CST complex because orthologs of Cdc13 and Ten1 were difficult to discern in the human genome, and as shelterin also binds the 3′ overhang and plays a role in telomere protection, it was thought to have replaced CST. However, the discovery of a mammalian CST-like complex now suggests that mammals have evolved multiple complexes to coordinate different aspects of telomere biology [64].
The budding yeast CST complex protects the telomere from degradation during G2/M of the cell cycle [65] with deletion of any subunit of the complex leading to C-strand degradation, a DNA damage response and cell cycle arrest [66-68]. Yeast CST also plays a key role in telomere replication by coordinating the recruitment of telomerase and DNA polymerase α (polα) to extend the telomere (see below) [60, 61]. Like POT1, Cdc13 binds telomeric G-strand DNA through two OB folds [69]. However, the overall structure of CST appears to resemble that of RPA rather than shelterin [60, 70]. RPA is also a hetrotrimeric complex that binds ssDNA through a series of OB-folds [71] and the crystal structures of yeast Stn1-Ten1 and RPA30-RPA14 are strikingly similar [72, 73].
Mammalian CST, like yeast CST, binds ssDNA, contains multiple predicted OB-folds, and an NMR structure of the mouse STN1 (OBFC1, PDB1wj5) shows significant structural homology to RPA2 (RPA30) [58, 59, 64]. Knockdown of CTC1 or STN1 leads to telomere defects, such as increased G-overhang length, telomere loss and/or problems with telomere replication. STN1 has been shown to interact with TPP1, suggesting possible coordination between shelterin and the CST complex [74]. In addition to telomere defects, depletion of CST subunits leads to general genomic instability, i.e. chromatin bridges, increased γH2AX staining and defects in replication reinitiation [58, 59, 75]. Interestingly, CTC1 and STN1 were originally identified as pola accessory factors (AAF), AAF132 and AAF42, and were shown to stimulate both the processivity of DNA polα-primase and its affinity for ssDNA templates [76, 77]. Although mammalian CST binds to ssDNA, it does not show sequence specificity for telomeric DNA and not all of the CST in a cell localizes to telomeres [59]. These findings suggest that mammalian CST has both telomeric and non-telomeric functions within the cell.
5. Consequences of telomere deprotection
Telomeres can become deprotected or “uncapped” as a result of shortening or deficiency in one of the shelterin components [5, 78]. In either case, the result is a DNA damage response that can lead to senescence or apoptosis and genome instability. The exact outcome depends on the extent and cause of the deprotection and the p53/Rb status of the cell. Partial telomere deprotection is commonly observed in late passage human primary cells and in some telomerase negative cell lines [5, 79]. In this situation, the de-protected telomeres activate a DNA damage response but do not trigger end-to-end fusion of chromosomes. In p53 and Rb proficient cells, the DNA damage response can lead to cell cycle arrest and senescence or apoptosis. Although partial telomere deprotection frequently correlates with telomere shortening, the physical nature of the defect is unclear and may reflect an overall change in the protective structure rather than actual telomere length [79, 80].
More severe loss of protection, or telomere uncapping, occurs after removal of a telomere protein or after extreme telomere shortening in p53 and/or Rb deficient cells [81, 82]. In either case, cells attempt to repair their chromosome ends via NHEJ, which leads to telomere fusions. The resulting dicentric chromosomes are unstable and break during cell division [83, 84]. This initiates a breakage bridge fusion cycle causing ongoing genome instability (discussed in accompanying article by J. Murnane [85]). Loss of capping has been studied extensively in mouse embryonic fibroblasts (MEFS) after conditional deletion of individual shelterin components. The deletion is usually performed in a p53-/-background to prevent rapid senescence in response to the resulting DNA damage signal [33, 40].
As discussed above, elimination of TRF2 leads to telomere fusions via NHEJ. Following TRF2 removal, the DNA damage response proteins MRN, ATM and 53BP1 are recruited to the exposed telomeres and any pre-existing overhangs are degraded by ERCC1 [29, 86]. The telomeres are then ligated together by the classical NHEJ (C-NHEJ) pathway that involves Ku70/86 and the DNA ligase IV/XRCC4 complex [33, 87]. Under these conditions the level of chromosome fusions can be very high, affecting most telomeres in the cell. When POT1 is removed cells initially exhibit an ATR-mediated DNA damage checkpoint. Cells that escape the checkpoint are then exposed to processes that cause genome instability. First, some chromosomes are subject to end-to-end fusion by the alternate NHEJ (A-NHEJ) pathway that requires MRN and CtIP but not Ku70/86 or DNA ligase IV [87, 88]. Second, cells that escape the checkpoint tend to re-enter S-phase without first undergoing mitosis and so end up with an 8N, 16N or 32N DNA content [88, 89]. Cells depleted of TPP1 have a similar phenotype to a POT1 knockout [40, 48]. This is to be expected as TPP1 depletion results in concomitant loss of POT1 from the telomere.
In human cells, telomere uncapping commonly occurs as a result of extreme telomere shortening due to continued cell division in the absence of telomerase [5]. In cells that lack DNA damage checkpoints, the telomeres eventually become short enough to trigger repair by NHEJ [84]. The resulting telomere fusions and ensuing breakage fusion bridge cycle leads to further genome rearrangements [83]. During tumor development the genomic instability is thought to cause telomerase upregulation in some cells [6]. The critically short telomeres are then elongated and a subset of cells survives the telomere-induced crisis. Genetic studies of mouse cells have demonstrated that fusion of chromosomes with critically short telomeres occurs via the C-NHEJ pathway [87]. This fits with the known role of TRF2 in protecting telomeres from C-NHEJ as short telomeres may be unable to bind sufficient TRF2 to ensure full protection.
A somewhat different form of telomere deprotection is observed in human cells that lack the NHEJ factor Ku [90]. Cells that contain a conditional Ku86 allele exhibit sudden and complete telomere signal loss from most chromosome ends upon Ku86 deletion, although end-to-end fusions are not observed. As the telomere loss is accompanied by the appearance of small circles of telomeric DNA (t-circles) which are a hallmark of telomere recombination [91], Ku may protect human telomeres by suppressing telomere recombination.
6. Telomere replication
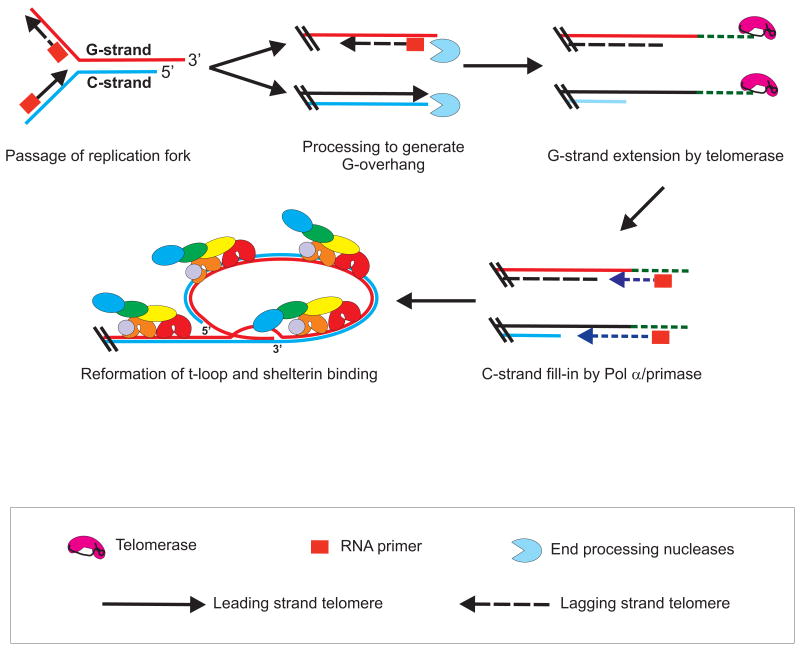
Telomere replication is a critical part of the cell division cycle because failure to regenerate the nucleoprotein architecture required for telomere protection leads to the catastrophic DNA damage response and genomic instability associated with telomere uncapping. Replication of telomeric DNA is a multi-step process that involves passage of a replication fork along the DNA duplex followed by processing of the DNA terminus to generate the 3′ overhang structure needed for shelterin binding, t-loop formation, telomerase action and maintenance of overall telomere length [3]. Formation of the overhang involves nuclease digestion to generate overhangs on both chromosome ends, extension of the G-strand by telomerase and fill-in of the complementary C-strand by DNA polα primase (Fig. 2).
Fig. 2. Telomere Replication.
Telomere replication is a multi-step process which necessitates dynamic opening of the telomeric DNA. First, replication of the telomere duplex occurs via the conventional replication form machinery. Next nucleases cleave the C-strand to generate a G-overhang, which allows telomerase access to the G-strand terminus. The G-strands are then elongated by telomerase followed by C-strand fill-in synthesis, creating a short G-overhang. Finally the telomeres are re-bound by shelterin and the t-loop reforms. See text for further details.
Replication of the duplex region of the telomeric tract appears to pose a problem for the replication machinery, most likely because of the repetitive nature of the DNA sequence and its potential to form secondary structures [3]. Consequently, a number of extra proteins are required in addition to the standard replication machinery to ensure efficient replication of this region of the chromosome. These proteins include TRF1, the RecQ helicases BLM and WRN, the FANCJ family helicase RTEL, the nuclease FEN1 and subunits of the CST complex [28, 64, 92-94]. Depletion of any one of these proteins leads to telomere loss and/or alteration in packaging of the telomeric tract (multi-telomere signals by FISH). As BLM, WRN and FEN1 are known to aid in the recovery of stalled replication forks, these telomere defects are thought to result from fork stalling during replication of the telomeric tract. Support for this hypothesis comes from analysis of mouse cells depleted of TRF1. These cells show both multi-telomere signals on FISH analysis and stalled replication forks by DNA fiber analysis [28].
When the replication fork reaches the end of a chromosome, the telomere generated by lagging strand synthesis will automatically gain a 3′ overhang due to removal of the RNA primer on the terminal Okazaki fragment and failure to position this Okazaki fragment at the chromosome terminus (Fig. 2). However, no such overhang will be formed on the telomere replicated by leading strand synthesis so this must be generated by a series of DNA processing reactions. Studies with budding yeast indicate that the processing steps are similar to those used to resect double-strand breaks during DNA repair [95]. Initiation of resection requires recognition of the DNA terminus by the Mre11-Rad50-Xrs2 (MRX) complex (MRN in humans) and subsequent recruitment of the nucleases Exo1 and/or Dna2. These act in concert with the helicase Sgs1 (BLM/WRN homolog) to cleave the DNA 5′ strand thus creating the 3′ overhang. Currently it is unclear whether EXO1 and DNA2 play a similar role at human telomeres. However, genetic analysis in mice has implicated another repair nuclease, Apollo/SNM1B, in generating overhangs on leading strand telomeres [30, 31]. While Apollo is known for its role in inter-strand crosslink repair, it can associate specifically with telomeres through an interaction with TRF2 [96].
DNA-processing to generate G-overhangs occurs regardless of whether a cell expresses telomerase [97]. In telomerase positive cells, the overhang is then elongated by the addition of new repeats on to the DNA terminus. Telomerase recruitment to human telomere is not fully understood but seems to involve trafficking of telomerase to the telomere in association with Cajal bodies and interaction between telomerase and TPP1 [43, 98-100] (see accompanying articles by J. Chen and D. Shippen [15, 16]). In human cancer cells, telomerase appears to elongate most, if not all, telomeres each cell cycle [18]. Each telomere is extended by 50-60 nt which is similar to the length of DNA added to a primer molecule in vitro by a single telomerase RNP when enzyme processivity is enhanced by POT1/TPP1 [42].
Following telomerase action, the complementary C-strand is filled-in to leave an overhang that ranges in length from (∼)40-400 nt [18, 101]. Studies in yeast indicate that DNA polα is responsible for fill-in synthesis with CST playing a key role in coordinating G-strand extension by telomerase with C-strand fill-in by DNA polα [61, 102]. Telomerase is recruited to the DNA terminus and enzyme activity is enhanced through an interaction between Cdc13 and the telomerase subunit Est1 [103-106]. Cdc13 and Stn1 both interact with DNA polα and these interactions are thought to then recruit the polymerase for fill-in synthesis [102, 107, 108]. Recent data from our lab (F. Wang, unpublished results) suggest that human CST may function in a similar manner. Given the original identification of mammalian CST as a DNA polα affinity factor, we propose that after telomerase action, CST binds the newly added telomeric G-strand and recruits polα to initiate C-strand fill-in [64]. Following C-strand fill-in, additional processing must occur to remove the RNA primer and to generate the CCAATC-5′ sequence found at the 5′ end of most human telomeres [109].
Although the general steps in telomere replication appear well conserved, it is becoming apparent that timing of the actual events differs between species. In human cells, telomeres are replicated throughout S-phase and telomerase-mediated extension of the G-overhang also occurs at this time [18, 110]. However, C-strand fill-in appears to be uncoupled from telomere extension and is delayed until late S/G2. This situation contrasts with what occurs in budding yeast where telomeres are replicated in late S/G2 and there is tight coupling between G-strand extension and C-strand fill in [61]. Telomerase extension and C-strand fill-in are also uncoupled in S. pombe [111]. It is currently unclear what occurs in human cells in the interval between telomerase extension and C-strand fill-in but these findings present an interesting question concerning the status of the telomeric end and how it is protected during this delay.
7. Telomere dynamics and telomere length regulation
In some ways telomeres have opposing functions in the cell as they need to sequester and protect the chromosome terminus from unwanted nuclease and DNA repair activities but during DNA replication they have to make the terminus available to telomerase and the other factors needed for telomere replication. These contradictory requirements have led to the concept of telomeres being dynamic structures that can switch between a closed protected state and a more open extendable state in which the DNA terminus becomes available to replication factors [112]. Evidence for this switch, between extendable and non-extendable states, has comes from studies with budding yeast where telomerase has been shown to preferentially extend short telomeres [113-116]. Changes in telomere conformation are also likely to be cell cycle regulated, which may partially explain why in budding yeast G-overhang processing and telomerase action occur in late S/G2 and require a high level of CDK1 activity [65, 117, 118]. Likewise, increased residence time in an extendable state would explain the enhanced telomere elongation observed in chicken cells when passage through G2 is delayed after Pot1 inactivation [39]. While relatively little is known about cell cycle regulation of telomere structure in vertebrate cells, it is intuitively obvious that the overall chromatin structure must become sufficiently open to allow passage of the replication fork and the t-loop must be disassembled to allow access to telomerase, Apollo and any other DNA processing activities.
After replication factors gain access to the telomeric DNA, final telomere length is determined by the degree of DNA elongation by telomerase versus erosion by incomplete DNA replication, nuclease action and double-strand breaks caused by DNA damage. In human somatic cells that lack telomerase, the rate of telomere shortening reflects a composite of all activities that deplete the telomeric tract. The range is usually 50-200 bp per population doubling but this varies with cell type [101]. In cells that express telomerase, final telomere length depends on a number of additional factors including the level of telomerase in the cell, trafficking of telomerase to the telomere by Cajal bodies, how readily telomerase gains access to the DNA terminus and the presence of molecules that directly stimulate or inhibit telomerase enzyme activity [15]. In human cancer cells, the telomere lengthening and shortening activities are generally balanced so telomere length is fairly constant. However, this is not the case for all telomerase expressing cells.
The importance of the absolute telomerase level in determining telomere length has been demonstrated by artificially raising or lowering the amount of active enzyme in human cancer cells [119, 120]. Overexpression of TERT can lead to rapid telomere elongation while TERT knockdown or inhibition results in telomere shortening. Interestingly the mode of repeat addition is related to the enzyme level [120]. In cells with naturally occurring levels of telomerase, most telomeres are extended each cell cycle by a single telomerase molecule. This molecule synthesizes on average 60 nt of TTAGGG sequence in one processive reaction. However, in cells with very high levels of enzyme, or telomeres that have been artificially shortened, multiple telomerase molecules act on each telomere during one cell cycle. The difference in telomerase action is thought to reflect the likelihood of telomerase molecules gaining access to a telomere while it remains in an accessible state. If telomerase levels are low, the telomere may reassume the closed, non-extendable state before a second molecule can gain access to the DNA terminus. Short telomeres may cycle into the non-extendable state less rapidly while high enzyme levels would increase the likelihood of a second molecule gaining access before the conformation switch.
Highly proliferative cell types such as hematopoetic precursor cells, activated lymphocytes and keratinocytes are thought to reflect a natural situation where telomere length is directly determined by telomerase level. These cell types all express low levels of telomerase but it appears insufficient to compensate for the replication problem [8, 121, 122]. Consequently, they undergo telomere shortening but at a slower rate than would be expected of a telomerase negative cell.
Simply having a high level of telomerase in a cell is not sufficient to ensure that telomeres are extended [120, 123]. In human cells, the enzyme has to be delivered to the telomere via Cajal bodies in a process that remains poorly understood [98, 99]. Once at the telomere, access to the DNA terminus is modulated by shelterin. Regulation of telomerase action by shelterin seems to occur both via non-specific steric effects and through specific interactions between telomerase and shelterin subunits. Overexpression of TRF1 and TRF2 leads to telomere shortening, suggesting that increased packaging of the telomere duplex by shelterin makes the DNA terminus less accessible, perhaps by promoting formation of t-loops or other higher order chromatin structures [124, 125]. In a similar vein, removal of POT1 can lead to telomere elongation, and in some cases this appears to be due to decreased sequestration of the DNA terminus [39, 41, 126, 127].
In contrast, to the apparently non-specific inhibitory activities of TRF1, TRF2 and POT1, TPP1 promotes telomerase action on the chromosome end by binding to the enzyme through a specific OB-fold motif [43]. This interaction can recruit telomerase to the telomere and increases enzyme processivity [42, 44, 128]. While at first sight it appears inefficient for shelterin to have both inhibitory and stimulatory activities within the same complex, it is likely that these activities are regulated in tandem with the structural changes that must take place during telomere replication. Thus, sequestration of DNA terminus by TRF1, TRF2 and POT1 may be relaxed at the time that TPP1 recruits telomerase and enhances enzyme processivity.
8. Conclusions: The interplay between telomere proteins and telomerase in human health
Studies of telomere-related health problems have mostly focused on telomerase because these diseases revolve around improper regulation of telomere length; for example, inappropriate telomere lengthening promotes tumor formation while overly short telomeres give rise to diseases such as dyskeratosis congenita and idiopathic pulmonary fibrosis. In many patients, short telomeres are caused by telomerase subunit mutations that result in decreased enzyme activity in proliferative tissues such as bone marrow, lung and skin. However, while telomerase is essential for telomere maintenance, it is not sufficient to ensure telomeres are preserved in a fully functional state. Shelterin also plays a key role in telomere maintenance as it regulates telomerase access and enzyme activity. Likewise, both shelterin and the complex process of telomere replication are essential to generate and protect the terminal structure that functions as the substrate for telomerase action; i.e. the G-strand overhang.
Out of the six shelterin subunits, thus far only TIN2 mutations have been linked to short telomeres and human disease. Given the key role played by shelterin, this is somewhat surprising. However, the reason may lie in the critical nature of shelterin function. Perhaps, mutations that cause defects in the protective cap structure are lethal because of the resulting DNA damage signaling and genome instability. It is still unclear how the TIN2 mutations found in dyskeratosis congenita patients affect telomerase activity and/or access to cause telomere shortening. However, the mutations do not affect shelterin assembly or telomere capping. Thus, it may be that the only shelterin mutations that can be tolerated will turn out to be in subunits and protein domains that modulate telomerase activity. The OB-fold domain of TPP1 would be a prime candidate and it will be interesting to see if SNP analysis on DNA from patients with short telomeres reveals mutations in this region.
Acknowledgments
The work in our group is supported by NIH grants GM088728 and GM041803 to CMP and Ruth L. Kirchstein National Research Service Awards F32 CA117846 and F32 GM097833 to JAS.
Footnotes
Conflict of Interest: The authors declare they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Lange T, Lundblad V, Blackburn EH. Telomeres. Cold Spring Harbor Laboratory Press; New York: 2006. [Google Scholar]
- 2.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 4.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 5.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 6.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiyama E, Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96:1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osterhage JL, Friedman KL. Chromosome end maintenance by telomerase. J Biol Chem. 2009;284:16061–16065. doi: 10.1074/jbc.R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 11.Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armanios M. Beyond Dyskeratosis Congenita: Pulmonary phenotypes as a manifestation of telomerase insufficiency. Mutation Research. 2011 In Press. [Google Scholar]
- 13.Bertuch A. Dyskeratosis congenita as a disease of telomere mainenance. Mutation Research. 2011 doi: 10.1016/j.mrfmmm.2011.06.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shay JW. Is telomerase a viable target in cancer. Mutation Research. 2011 doi: 10.1016/j.mrfmmm.2011.07.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shippen DE. Telomerase Regulation. Mutation Research. 2011 doi: 10.1016/j.mrfmmm.2011.10.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J. Telomerase Structure and Biogenesis. Mutation Research. 2011 doi: 10.1016/j.mrfmmm.2011.11.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 21.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, Davalos AR, Heo SJ, Rodier F, Zou Y, Beausejour C, Kaminker P, Yannone SM, Campisi J. Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes. J Cell Biol. 2008;181:447–460. doi: 10.1083/jcb.200710028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 30.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell. 2010;39:606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29:2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 34.Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 36.Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 37.Gong Y, de Lange T. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell. 2010;40:377–387. doi: 10.1016/j.molcel.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 39.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 42.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 46.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 47.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 48.Kibe T, Osawa GA, Keegan CE, de Lange T. Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol. 2010;30:1059–1066. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith S. The long and short of it: a new isoform of TIN2 in the nuclear matrix. Cell Cycle. 2009;8:797–798. doi: 10.4161/cc.8.6.8337. [DOI] [PubMed] [Google Scholar]
- 51.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, Tran NL, Speiser LJ, Wong M, Saez E, Schultz P, Chanda SK, Verma IM, Tergaonkar V. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758–767. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- 57.Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011 doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 61.Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 62.Anbalagan S, Bonetti D, Lucchini G, Longhese MP. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet. 2011;7:e1002024. doi: 10.1371/journal.pgen.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vodenicharov MD, Laterreur N, Wellinger RJ. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 2010;29:3007–3019. doi: 10.1038/emboj.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 66.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint [published erratum appears in Mol Cell Biol 1996 Jan;16(1):457] Mol Cell Biol. 1995;15:6128–6238. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. Embo J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 69.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 70.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 71.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 72.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci U S A. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.J.A. Stewart, Wang, F., Chaiken, M., Price, C., Manuscript in preparation.
- 76.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goulian M, Heard CJ. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem. 1990;265:13231–13239. [PubMed] [Google Scholar]
- 78.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 79.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 80.Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol Biol Cell. 2004;15:3709–3718. doi: 10.1091/mbc.E04-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 82.Thanasoula M, Escandell JM, Martinez P, Badie S, Munoz P, Blasco MA, Tarsounas M. p53 prevents entry into mitosis with uncapped telomeres. Curr Biol. 2010;20:521–526. doi: 10.1016/j.cub.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 84.Zou Y, Misri S, Shay JW, Pandita TK, Wright WE. Altered states of telomere deprotection and the two-stage mechanism of replicative aging. Mol Cell Biol. 2009;29:2390–2397. doi: 10.1128/MCB.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murnane JP. Telomere dysfunction and chromosome instability. Mutation Research. 2011 doi: 10.1016/j.mrfmmm.2011.04.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dimitrova N, de Lange T. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009;29:5552–5563. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 89.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem. 2011;149:5–14. doi: 10.1093/jb/mvq119. [DOI] [PubMed] [Google Scholar]
- 92.Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J Biol Chem. 2010;285:27057–27066. doi: 10.1074/jbc.M110.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 94.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tam PP, Nagy A, Lansdorp PM. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 95.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J. 2010;29:2864–2874. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 97.Hemann MT, Greider CW. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 100.Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 102.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 104.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 105.Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DeZwaan DC, Freeman BC. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci U S A. 2009;106:17337–17342. doi: 10.1073/pnas.0905703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 108.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 110.Wright WE, Tesmer VM, Liao ML, Shay JW. Normal human telomeres are not late replicating. Exp Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- 111.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 113.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 114.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 115.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vodenicharov MD, Wellinger RJ. The cell division cycle puts up with unprotected telomeres: cell cycle regulated telomere uncapping as a means to achieve telomere homeostasis. Cell Cycle. 2007;6:1161–1167. doi: 10.4161/cc.6.10.4224. [DOI] [PubMed] [Google Scholar]
- 118.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 119.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao Y, Abreu E, Kim J, Stadler G, Terns MP, Terns RM, Shay JW, Wright WE. Processive and distrubutive extension of human telomeres by telomerase under homeostatic and non-equilibrium conditions. Molecular Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weng NP, Palmer LD, Levine BL, Lane HC, June CH, Hodes RJ. Tales of tails: regulation of telomere length and telomerase activity during lymphocyte development, differentiation, activation, and aging. Immunol Rev. 1997;160:43–54. doi: 10.1111/j.1600-065x.1997.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 122.Buckingham EM, Klingelhutz AJ. The role of telomeres in the ageing of human skin. Experimental Dermatology. 2011;20:297–302. doi: 10.1111/j.1600-0625.2010.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 125.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 127.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 128.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]