Abstract
Enriched environmental conditions induce neuroanatomical plasticity in a variety of vertebrate and invertebrate species. We explored the molecular processes associated with experience-induced plasticity, using naturally occurring foraging behavior in adult worker honey bees (Apis mellifera). In honey bees, the mushroom bodies exhibit neuroanatomical plasticity that is dependent on accumulated foraging experience. To investigate molecular processes associated with foraging experience, we performed a time-course microarray study to examine gene expression changes in the mushroom bodies as a function of days foraged. We found almost 500 genes that were regulated by duration of foraging experience. Bioinformatic analyses of these genes suggest that foraging experience is associated with multiple molecular processes in the mushroom bodies, including some that may contribute directly to neuropil growth, and others that could potentially protect the brain from the effects of aging and physiological stress.
INTRODUCTION
Neuroplasticity, the brain’s ability to respond to experience and modulate behavior, is an important trait in many species. A question of interest is what molecular processes support this capacity. Studies examining transcriptional responses to enriched environments in the laboratory (Rampon et al., 2000; Ronnback et al., 2005; Li et al., 2007; Thiriet et al., 2008) have been valuable in the discovery of molecular processes associated with neuroanatomical plasticity. However, few studies have examined the brain’s response to the extended performance of a specific, naturally occurring, ecologically relevant behavior. This approach allows for the possibility that mechanisms of neuroanatomical plasticity can be understood in the context of changes in neural functioning and behavior that occur throughout the lifespan, much the way increases and decreases in different cognitive abilities occur as a function of aging (McArdle et al., 2002). We used the honey bee to begin to understand how naturally occurring cognitive challenges and sensory stimulation affect molecular processes associated with neuroanatomical plasticity.
Neural development in adult worker honey bees (Apis mellifera) provides an opportunity to study experience-induced neuroanatomical plasticity in a naturalistic setting. The first 1–3 weeks of honey bee worker adult life are spent performing tasks inside the hive, after which bees typically forage for resources (Winston, 1987). This dramatic behavioral shift means that honey bees can be used to examine the response of a mature brain to a sudden and sustained increase in cognitive demand that is highly relevant to their natural lifestyle. Foraging exposes bees to novel sensory stimuli and involves memory-intensive tasks, such as spatial navigation and recognition of floral sources (Menzel, 1990).
The mushroom bodies, a region of the insect protocerebrum that performs multimodal integration of input from primary sensory neuropils and is involved in learning and memory (Fahrbach, 2006), is likely to participate in these cognitive tasks. Adult honey bee mushroom bodies exhibit neuropil growth in response to accumulated foraging experience; expansion is detectable between the first and second week of foraging (Farris et al., 2001; Ismail et al., 2006). This expansion is not caused by adult neurogenesis in the mushroom bodies (Fahrbach et al.,1995). Instead, experience-dependent expansion is accompanied by increased dendritic branching and branch length in the visual input region of the calyx (Farris et al., 2001), and changes in microglomerular structure and density (Krofczik et al., 2008). Behavioral studies show that foraging efficiency also improves with experience (Dukas and Visscher, 1994) and that learning may contribute to this improvement (Dukas, 2008), suggesting a link between anatomical and behavioral change.
Foraging is a physiologically demanding activity, and the average time to death once a bee has commenced foraging is one week (Visscher and Dukas, 1997); foraging-induced neuropil growth therefore represents an extreme example of plasticity in aging individuals. Neuroanatomical plasticity in other species typically decreases with age, but this decrease and the corresponding loss of cognitive ability can sometimes be prevented by experience (van Praag et al., 2000; Kolb, 1998). It is of interest to discover molecular processes that enable persistent neuroanatomical plasticity in the aging brain.
The mushroom body neuropil of adult honey bees also undergoes developmentally related growth prior to the onset of foraging behavior. The mushroom bodies of 1-day-old bees exhibit active neuropil outgrowth and dendritic branching (Farris et al, 2001), and the neuropil shows further expansion in anticipation of foraging onset (Withers et al 1993, 1995; Durst et al, 1994). These phases of growth are apparently distinct from the plasticity observed in older foragers and independent of experience; neuropil expansion in young bees is robust even to extreme sensory deprivation (Fahrbach et al., 1998), whereas neuropil expansion in foragers requires foraging activity (Ismail et al., 2006). Are the same genes responsible for directing initial growth in development and for re-initiating neuroanatomical plasticity in response to experience?
Because knowledge of molecular processes directing adult neuropil growth is so limited, we chose to conduct a genome-wide survey for changes in gene expression that might be related to this type of growth in honey bees. We performed a detailed investigation of the long-term dynamics of gene expression associated with foraging experience, with the idea that some of these genes are potentially related to neuroanatomical plasticity. We used whole-genome microarrays to measure gene expression in the mushroom bodies of honey bees throughout their foraging lifespan.
METHODS
Animals
Bees were observed and collected from apiaries maintained by the University of Illinois Bee Research Facility, Urbana, IL in June and July of 2008. Additional bees were collected for qRT-PCR analysis in August 2009. Experimental honey bee colonies were designed and maintained as described in Ismail et al (2006). Colonies began as one-story Langstroth hives with several empty combs and combs containing larval brood. Frames of honeycomb containing pupae were collected from 25–30 other colonies and stored in a dark, humid incubator at 34°. Adults were removed from the comb within 24 h of emergence, marked on the thorax with paint (Testor’s) and introduced to each experimental colony. In total, 10,000–13,000 marked bees were added over a two-day period to each host colony. Each colony also contained a naturally-mated queen and at least 30,000 unmarked bees.
The majority of honey bee workers die before reaching 12–16 days of active foraging behavior (Visscher and Dukas, 1997). In order to collect adequate numbers of older foragers for analysis, colony demography was manipulated to ensure a large starting population of simultaneously foraging focal bees. This was done by removing non-focal foraging bees from the colony (Huang and Robinson, 1996). Collections for microarray analysis were made from two colonies (two trials). Collections for qRT-PCR came from the same trials as those used for microarray analysis, but from groups of bees that initiated foraging at a later (for trial one) or earlier (for trial two) period. Samples from a third colony, collected a year later, were also used to confirm microarray results.
Behavioral observations
Behavioral observations were performed as in Ismail et al (2006). Briefly, beginning when focal bees were 6–7 days old, the colony entrance was observed daily for 8 h; this allowed focal bees to be identified on the first day they foraged. Focal bees observed returning with a nectar or pollen load were given a second paint mark on the abdomen to indicate their status as foragers. Based on this second paint mark, the number of days spent foraging could be determined for any double-marked bee captured. After a sufficient number of first-day foragers had been marked, brief observations of the colony every 2–3 days ensured that overall foraging activity remained high. Twelve days after this initial marking period, as many as possible of the individuals that had subsequently commenced foraging were also given an abdominal mark. This allowed the identification of age-matched, less-experienced foragers at the end of the experiment.
Collections and dissection
Individuals for microarray and qRT-PCR analyses were flash-frozen in liquid nitrogen. 1-day-old bees were collected after removal from the comb containing brood. Foragers with 4, 8, 12, and 16 days of flight experience were collected upon their return to the hive entrance with a pollen or nectar load. Samples were stored at −80° C between all subsequent processing steps.
Mushroom bodies were isolated as previously described (Sen Sarma et al., 2009). Frozen brains were treated overnight with RNAlater-ice (Ambion) and then dissected over ice. Only the calyces of the mushroom bodies were retained because of the difficulty of separating the peduncles and lobes of the mushroom bodies from the central body. The calyces contain the somata of all intrinsic mushroom body neurons (Kenyon cells) and should contain the majority of mushroom body transcripts.
Microarrays
Microarray sample preparation and hybridization procedures were similar to those described in Alaux et al (2009a). RNA was extracted from tissue using an RNeasy kit (Qiagen) and amplified (from 200 ng) with an Amino Allyl MessageAmp™ II aRNA Amplification kit (Ambion). Amplified RNA (2 µg) was labelled with a ULS aRNA labelling kit (Kreatech), and 60 ng labelled material from each sample was hybridized to honey bee whole genome oligonucleotide arrays using a Maui Hybridization System (Kreatech).
Mushroom body gene expression in five groups (1-day old bees, and bees collected 4, 8, 12, or 16 days after foraging initiation) was compared using a loop design (Supplemental figure). For each of two trials, 12 bees per group were included, and analyzed on an individual basis. Slides were scanned using an Axon 4000B scanner, and images analyzed with GENEPIX software (Agilent Technologies). Spots were removed from analysis if flagged by the GENEPIX software or if the fluorescence intensity was less than the median intensity of the negative control spots. A Loess transformation was performed using Beehive (http://stagbeetle.animal.uiuc.edu/Beehive) to normalize expression intensities.
Real-time RT-PCR
RNA extraction was performed as described for microarray samples. An on-column genomic DNA digest was performed with DNase 1 (Qiagen). cDNA synthesis was performed using 150 ng RNA. Quantification was performed (on samples independent from those used on microarrays) with an ABI Prism 7900 sequence detector, SYBR green detection method (Applied Biosystems), and relative quantification to a genomic DNA standard curve. Expression was normalized to the geometric mean of two constitutively expressed genes used in past studies, eIF3-s8 (Alaux et al., 2009a) and rp49 (Ament et al., 2008).
Statistics
A linear model including both fixed and random effects implemented using restricted maximum likelihood was used to analyze the normalized log2-transformed fluorescence intensities for each gene, accounting for the effects of dye, bee, colony and microarray. An FDR (false discovery rate) correction was used to correct for multiple testing. Both microarray and qRT-PCR analyses used PROC MIXED in SAS. Genes highly expressed in the hypopharyngeal glands, a nearby and potentially contaminating tissue, were filtered from the microarray results, as in previous studies (Alaux et al., 2009a). Clustering analyses were performed using MultiExperiment Viewer, part of the TM4 Microarray Software Suite (Saeed et al., 2006). The R package CMA (Classification for Microarrays), a program designed for class prediction in microarray data sets (Slawski et al., 2008), was used for this purpose; data were pre-filtered for genes with missing values. For Gene Ontology and other informatic functional analyses, known Drosophila melanogaster orthologs of bee genes were used as previously described (Alaux et al., 2009b), and enrichment was identified using the functional annotation clustering tool in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Dennis et al., 2003; Huang et al., 2009). Correlation network analysis was performed using the R package Weighted Gene Co-expression Network Analysis (WGCNA). The forager bee network was constructed with values from all forager time points; the young bee network was constructed using values from day-old individuals. STEM (Short Time-series Expression Miner), a program designed for time-series microarray studies with relatively few time points, was used to identify gene expression profiles (patterns of up- or down-regulation between adjacent time points) that arose more frequently than expected by chance (Ernst and Bar-Joseph, 2006). Clustering and network analyses used log2-transformed estimated values for individual samples, as described previously (Whitfield et al., 2006).
RESULTS
Foraging induces a restricted and temporally focused transcriptional response
We compared gene expression in the mushroom bodies of five groups: 1-day old bees, and foragers with 4, 8, 12, or 16 days of flight experience. A total of 6,219 genes of the 11,761 included in the analysis (53%) were significant in the overall ANOVA (FDR<0.05). Of these, 5,839 genes (50%) were differentially regulated in the mushroom bodies of 1-day-olds and foragers. This large number of regulated genes is consistent with previous comparisons of whole-brain gene expression in foragers and hive bees (Alaux et al., 2009a; Whitfield et al., 2003) or foragers and day-olds (Whitfield et al., 2006; Sen Sarma et al., 2007).
By contrast, the number of genes in the mushroom bodies differentially regulated by increasing amount of foraging experience was much smaller: only 498 genes (4%) were differentially regulated between any two foraging time points, and 239 genes were differentially regulated between adjacent time points (pair-wise comparisons). A total of 200 of these 239 showed significant regulation between 8 and 12 days of foraging. A disproportionate amount of experience-related expression differences thus occurred within the previously identified time frame for experience-dependent neuropil expansion (Ismail et al., 2006).
Analysis of microarray gene expression data through ANOVA relies on expression differences in individual genes, identified by a somewhat arbitrary significance threshold. A relatively new alternative method for understanding expression data, weighted gene co-expression network analysis (WGCNA), can supplement other methods and provide a more complete view of molecular processes associated with a factor of interest. Networks of genes are identified by their coexpression (positive or negative correlation) in multiple individuals across different experimental conditions. Within-group variation, otherwise detrimental to detection of expression differences for a single gene across groups of individuals, can in WGCNA be used to reveal potential interactions between groups of genes; correlation of gene expression is likely to reflect coregulation (Zhang and Horvath, 2005).
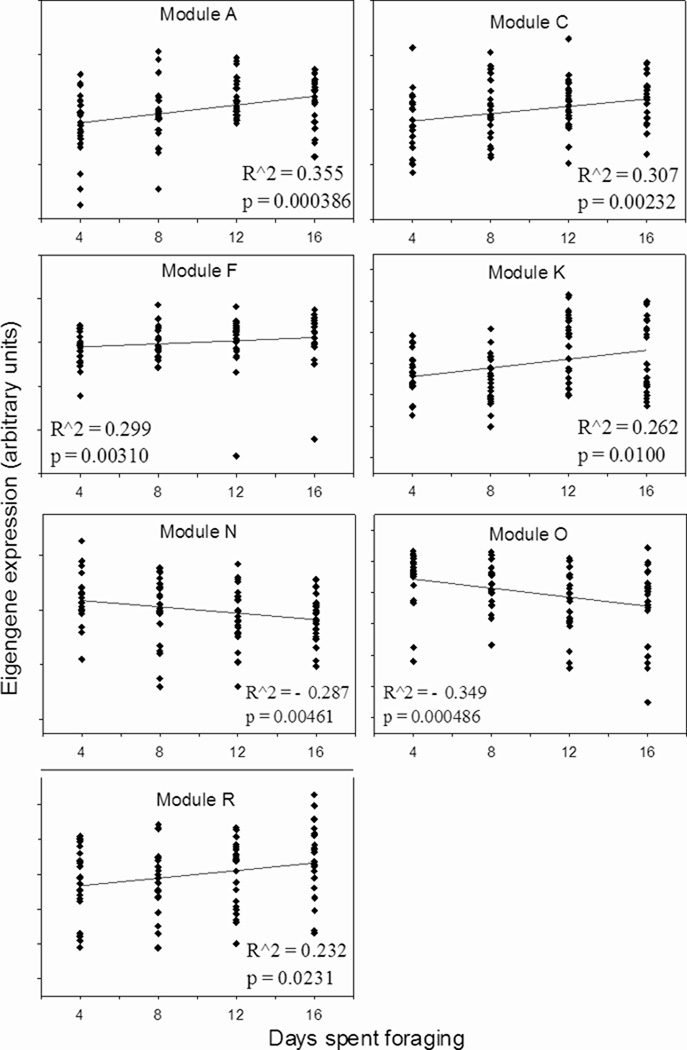
We applied WGCNA to the expression of 9754 genes in individual foragers: the resulting network contained 17 groups, or modules, of genes that exhibit high expression overlap (Figure 1A). The stereotypical expression of genes in a module can be described by the eigengene, which represents the first principal component of variation in expression for those genes. Modules with expression influenced by amount of foraging experience were identified by calculating the correlation between eigengene expression and number of days spent foraging. Expression in five modules showed a positive correlation with amount of foraging experience, while expression in two modules showed a negative correlation (Figure 1A, B). Genes in these modules displayed too much individual variability to be detected by ANOVA, but their membership in foraging-related modules provides support for their regulation by foraging experience.
Figure 1.
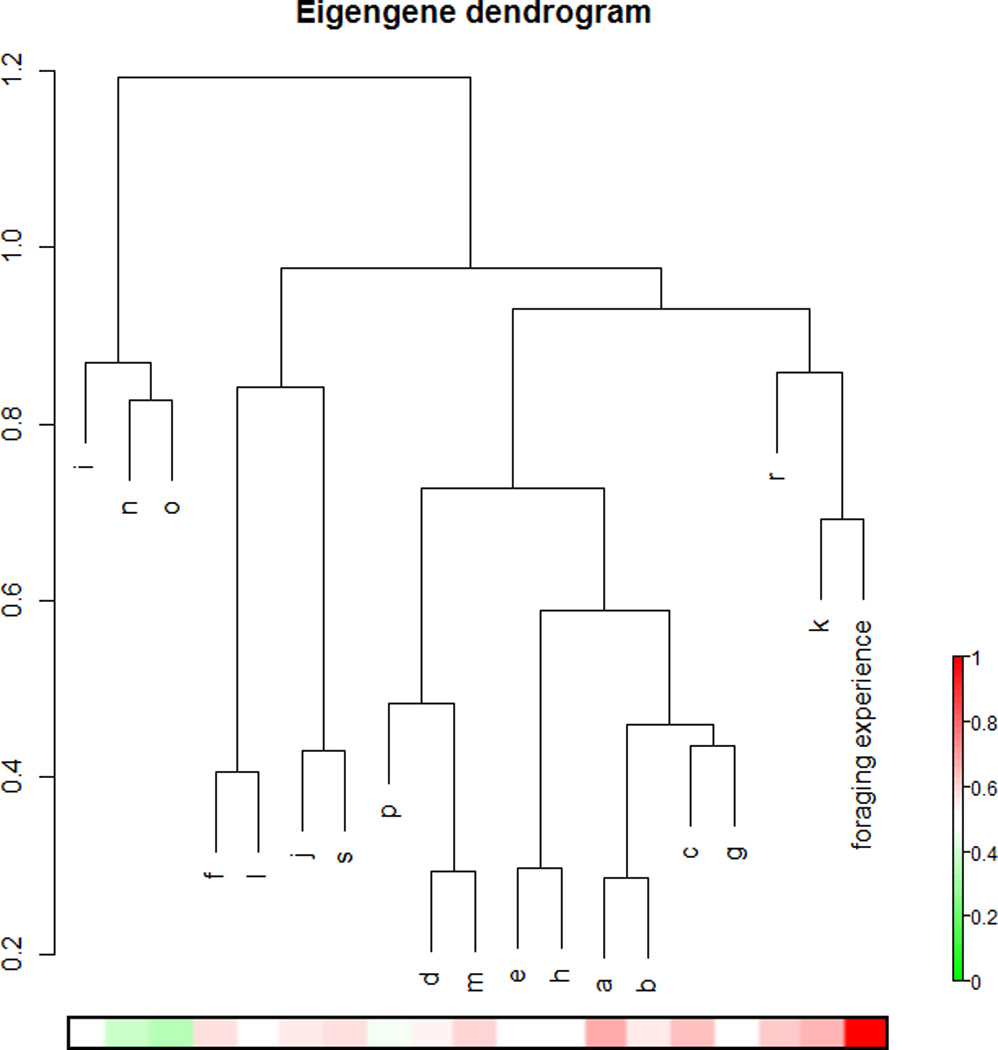
A. Hierarchical clustering of coexpression modules in the forager network. The number of days each individual had foraged was included in the analysis and is shown here as the arm labeled “foraging experience.” The heatmap below the dendrogram shows the R2 value for the correlation between expression in each module with days of foraging experience, according to the legend on the right.
B. Days spent foraging vs. module eigengene expression for each module showing a significant correlation with foraging experience (Spearman’s rho<0.05). Each point represents an individual forager.
We used gene expression data from the mushroom bodies of 1-day-old individuals to construct a separate co-expression network that describes transcriptional regulation at this earlier (“young bee”) developmental stage. Comparison with the forager network revealed that all but one forager module had significant overlap with at least one of the modules in the young bee network. Among foraging-related modules, only one, module K, showed no significant overlap with any of the young bee modules. This result suggests that the genes in module K are involved in disparate functions in 1-day-olds, but act together to perform some activity- or plasticity-related function in forager mushroom bodies. All other foraging-related modules showed significant, >10% overlap with at least one of the young bee modules (Fisher’s exact test, p<0.0005). Modules C, F and R, overlapped with the same large module in 1-day-olds. This widespread overlap indicates that coregulation of expression is similar in the two distinct phases of mushroom body development exhibited by 1-day-old and forager bees.
Individual experience level is reflected in distinct gene expression profiles
Given that the complexity and vagaries of natural foraging behavior lead to considerable individual variability in experience, can foraging-related genes serve as reliable measures of foraging experience? The coincident timing of expression differences in this study and previously observed neuropil expansion suggests a natural division of foragers into phenotypically distinguishable groups: “less experienced” (4- and 8-day foragers) and “more experienced” foragers (12- and 16-day foragers.) Two statistical methods were used to evaluate the strength of this distinction.
An unsupervised clustering method, principal component analysis, was performed on estimated values of foraging-regulated genes for individual foragers. A 2D plot of the two largest components of variation identified by this analysis revealed approximate separation of less and more experienced foragers into two clusters (Figure 2). This separation can be attributed to the largest principal component of variation in foraging-related gene expression (PC1), which describes 21% of total variation, indicating that duration of foraging experience is a moderate but important factor contributing to individual differences in gene expression. Other potential sources of variation in mushroom body gene expression likely include genetic background and uncontrolled aspects of foraging experience.
Figure 2.
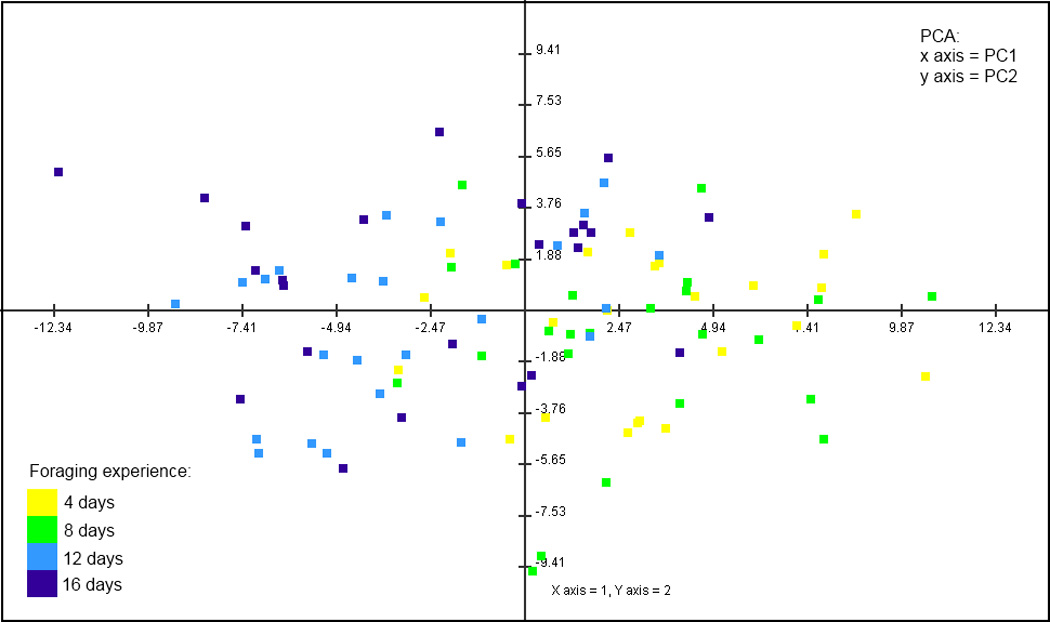
Principal component analysis of foraging individuals shows a division between less (4- and 8-day) and more experienced (12- and 16-day) foragers.
Additional results supporting the division of foragers into these two categories were obtained by class prediction analysis (Leung and Cavalieri, 2003). We used class prediction analysis to select a list of 100 genes best able to classify individual foragers as less or more experienced. An algorithm trained with expression values from our study performed leave-one-out cross-validation with 78.1% accuracy, and the number of incorrect assignments was roughly equal for the two groups. This rate is significantly better than what would be expected by chance (Chi-square test, p<0.0001) and demonstrates the ability of gene expression to accurately predict foraging experience, despite substantial within-group variation. All but four of these 100 most predictive genes were upregulated in more experienced foragers (Figure 3).
Figure 3.
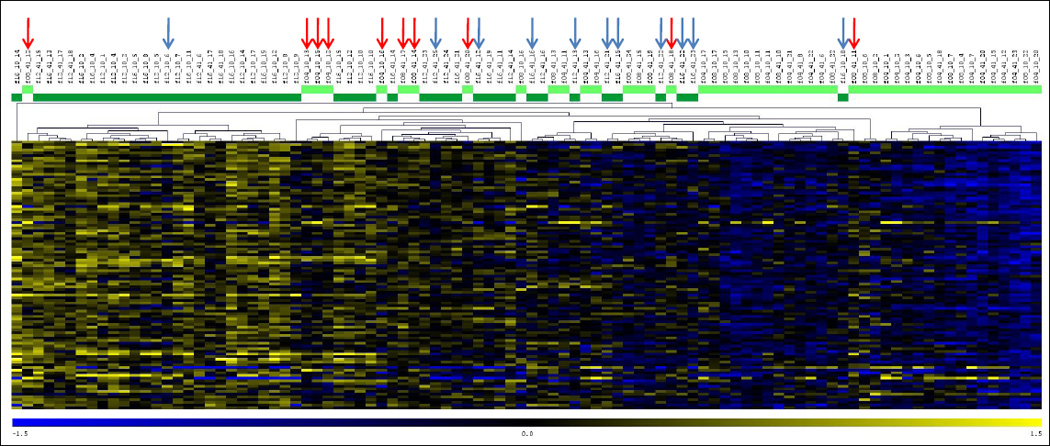
Heat map depicting expression differences for 100 most predictive genes identified by class prediction analysis. The heat map displays expression profiles of all foraging individuals. At the top, light green indicates novice foragers and dark green indicates experienced foragers. Misclassified individuals are marked with red or blue arrows, respectively. Yellow represents upregulation, blue represents downregulation.
Validation of microarray results by qPCR
We selected 10 genes to measure with qRT-PCR to evaluate the microarray results (Table 1). Independently collected samples were used, both from the same colonies as the microarray study and from an additional colony. We also used an additional control group of bees that were the same age as 16-day foragers, but had only 4 days of foraging experience. This group was used to help distinguish between effects of experience and age on gene expression. Four out of 10 genes showed significant differences in the direction predicted by the microarray results (inos, jip, GB13145, and GB30322, Figure 4.) As in the microarray study, expression was increased in bees with greater foraging experience, and greatest in 16-day foragers. Expression in the mushroom bodies of bees of the same age as 16-day foragers, but with minimal flight experience, was comparable to 4-day foragers for each of these four genes. This demonstrates that the increase in expression was likely related to experience itself, not increasing age. Expression of a fifth gene examined, GB12552, showed a weak but significant positive correlation with number of days spent foraging (Spearman’s rho=0.173, p=0.039). Additionally, for two heat shock protein genes, hsp90 and GB14435, expression was increased in 16-day foragers in two out of three trials. In the third trial, an approximately 4-fold upregulation of these genes in 12-day foragers, a much larger change than any observed in previous trials, suggested that these genes were responding to some acute stimulus in these individuals, rather than reflecting the accumulated effect of foraging experience. In summary, 7 out of 10 genes showed expression changes in some way consistent with the microarray results, with inos, jip, GB13145, and GB30322 being the most consistent. Both genes with Drosophila annotations have potential functional connections to neural plasticity. jip encodes JNK interacting protein, a scaffolding protein that modulates MAPK signaling pathways (Dickens et al, 1997). Inositol-3-phosphate is synthesized de novo by the enzyme encoded by inos (Park et al, 2000); inositol signaling is important to neuronal development and plasticity, and alterations in inositol phosphate synthesis affect neural development in mice (Alebous et al, 2009). Inositol signaling has been previously implicated in honey bee foraging (Kucharski and Maleszka, 2002; Whitfield et al, 2003).
Table 1.
Genes selected for validation and further analysis using qRT-PCR. Gene names and putative function based on orthology to Drosophila melanogaster.
| Gene | Name | Putative Function | Drosophila best hit |
|---|---|---|---|
| GB12552 | unknown | FBgn0052512 | |
| GB30322 | similar to insulin-like receptor | FBgn0013984 | |
| GB14758 | hsp90 | response to heat | FBgn0001233 |
| GB13145 | unknown | ||
| GB14435 | response to heat | FBgn0011296 | |
| GB11572 | inos | inositol synthesis | FBgn0025885 |
| GB19694 | samdc | S-adenosylmethionine decarboxylase | FBgn0019932 |
| GB14151 | jip | JNK-interacting protein | FBgn0040281 |
| GB10565 | trbl | kinase-like protein | FBgn0028978 |
| GB18093 | transcription factor | FBgn003 9169 |
Figure 4.
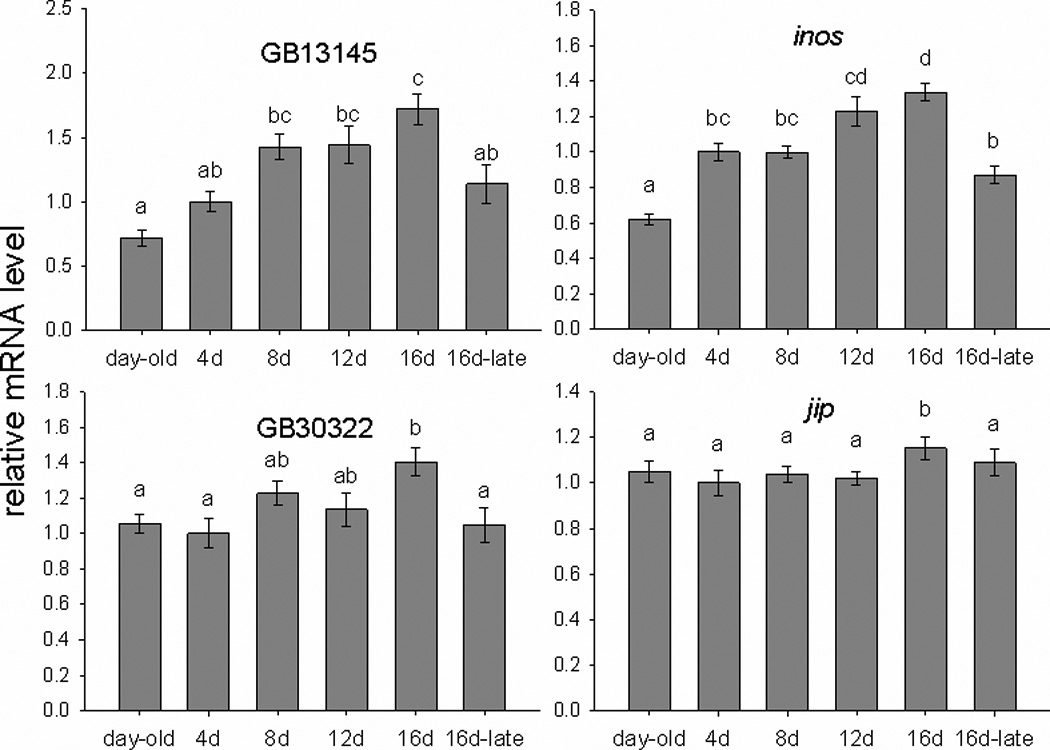
Expression of four genes measured with qRT-PCR. Letters indicate significant differences (p<0.05, mixed model ANOVA). Expression is shown relative to average expression in 4-day foragers, which was normalized to 1. The group name 16d-late indicates bees of the same age as 16-day foragers, but with only 4 days of foraging experience.
Functional processes regulated by foraging experience
To discover functional processes regulated by foraging experience, we first grouped foraging-regulated genes according to their temporal expression patterns; such groups can be valuable for identifying functionally related genes (Wang et al., 2008). We used STEM (Short Time-series Expression Miner) to classify foraging-related genes by their pattern of expression over the 16-day period of foraging experience. Of the 498 foraging-related genes, 382 were clustered into 8 significantly over-represented expression profiles (FDR<0.01, Figure 5); the remaining genes exhibited less common patterns of expression. STEM additionally grouped profiles according to pattern similarity. Significantly overrepresented categories could be described as linearly increasing (profiles 31 and 43), increased in a step-wise fashion in experienced foragers (profiles 23, 17 and 32), linearly decreasing (profiles 12 and 20), or increasing in experienced foragers (profile 16). Expression in the largest and most significant profile, profile 23, closely resembles the pattern of expression seen in the highly predictive genes identified with class prediction analysis (Figure 3).
Figure 5.
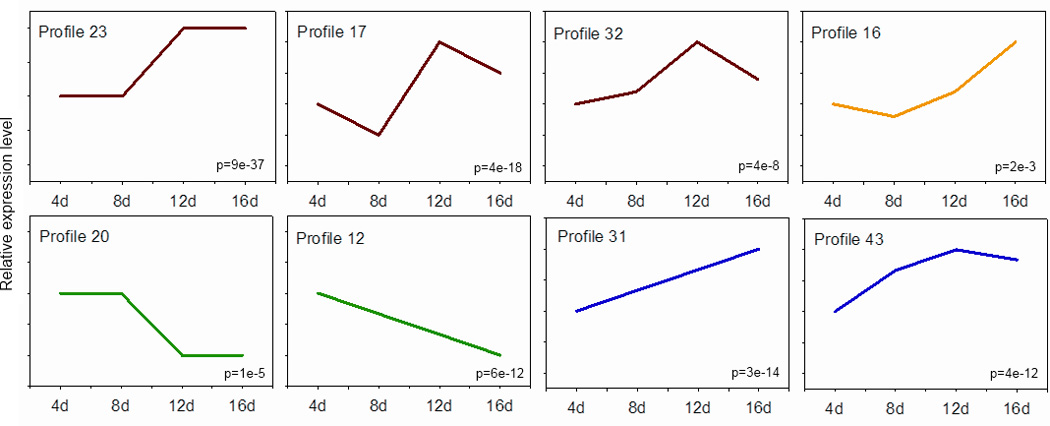
Overrepresented gene expression profiles identified by STEM. Lines represent pattern of expression across the four foraging time points. Profiles with similar expression patterns were assigned the same color. Numbers in the bottom left are p-values representing significance of overrepresentation.
Gene Ontology (GO) enrichment analysis of genes from expression profiles identified by STEM is shown in Table 2. Genes upregulated in experienced foragers (profile 23) were enriched for genes related to protein kinase activity and G-protein coupled receptor signaling. Another group of genes upregulated in experienced foragers, but showing a more cubic trend (profile 17), was enriched for genes involved in protein folding.
Table 2.
Gene Ontology functional processes enriched in temporal expression profiles identified by STEM. Clusters of enriched categories (EASE<0.05) were identified in DAVID, with a representative category from each cluster shown here.
| Profile | Functional Category | Number of genes |
|---|---|---|
| 23 | protein kinase activity | 7 |
| regulation of JNK cascade | 3 | |
| G-protein coupled receptor protein signaling pathway | 6 | |
| nuclear export | 4 | |
| 17 | protein folding | 7 |
| 31 and 43 | nuclear lumen | 6 |
| regulation of transcription, DNA-dependent | 6 | |
| RNA processing | 4 | |
| 12 and 20 | oxidation reduction | 7 |
It was not possible to identify significantly enriched categories in profiles 31 and 43, because each contained a relatively small number of genes. When the two clusters were pooled, however, enriched terms included DNA-dependent regulation of transcription and RNA processing. Several genes in this category (achaete, diminutive and extradenticle) are involved in cell fate determination and patterning in Drosophila melanogaster larval development (Tweedie et al., 2009). Because neurogenesis does not occur in the adult honey bee mushroom bodies and is therefore not the basis for neuropil expansion (Fahrbach et al., 1995), it is not expected that these genes are performing a similar function here. Rather, if functional, they may play an unknown organizational role in adult dendritic morphogenesis.
Another way to strengthen functional interpretation of genes showing a pattern of linear increase is to include those identified through co-expression network analysis. Eigengenes of 4 of the 5 modules positively correlated with foraging experience were clustered into profile 31 by STEM, confirming the association of this broader set of genes with a linearly increasing expression pattern. In order to understand the biological processes represented by these modules, we again used GO enrichment analysis to discover overrepresented functional categories. Representative examples of the most enriched categories are shown in Table 3. Categories in modules positively correlated with foraging included small GTPase mediated signal transduction, nuclear export, protein kinase activity, regulation of RNA splicing, plasma membrane part, actin cytoskeleton organization, and cell morphogenesis. Genes involved in small GTPase mediated signal transduction regulate membrane trafficking, and several, including Rab11 and Cdc42, are involved in neurite outgrowth (Ng and Tang, 2008; Rodal et al., 2008). Another enriched category was plasma membrane part, which consists of genes encoding proteins present in the cellular membrane. Genes in this category included several related to synaptic signaling, including dopamine receptor 2, serotonin receptor 1a, and shaker. Genes in the category nuclear export are involved in transporting mRNA from the nucleus, reinforcing the suggestion of increased transcription associated with foraging experience. Together, these results suggest that foraging-related gene expression in the mushroom bodies is responsible for both directing and providing materials needed for neuropil growth.
Table 3.
Gene Ontology functional processes enriched in modules showing significant correlations with amount of foraging experience. Module F was not significantly enriched for any functional category. Clusters of enriched categories (EASE<0.05) were identified in DAVID, with a representative category from each cluster shown here.
| Module | Functional Category | Number of genes |
|---|---|---|
| A | small GTPase mediated signal transduction | 11 |
| nuclear export | 6 | |
| translation initiation factor activity, nucleic acid binding | 10 | |
| ncRNA metabolic process | 15 | |
| nucleotide binding | 64 | |
| establishment of protein localization | 21 | |
| C | protein kinase activity | 35 |
| regulation of RNA splicing | 13 | |
| ATP-dependent helicase activity | 14 | |
| chromatin modification | 17 | |
| guanylate kinase activity | 5 | |
| synaptic vesicle transport | 15 | |
| regulation of MAPKKK cascade | 10 | |
| learning | 10 | |
| F | chromatin organization | 5 |
| R | plasma membrane part | 93 |
| multicellular organism reproduction | 118 | |
| cellular component morphogenesis | 108 | |
| regulation of nervous system development | 32 | |
| cell motion | 65 | |
| cell adhesion | 38 | |
| learning or memory | 24 | |
| sensory organ development | 69 | |
| tissue morphogenesis | 53 | |
| neuron projection | 17 | |
| growth | 28 | |
| locomotor behavior | 34 | |
| negative regulation of gene expression | 41 | |
| asymmetric protein localization | 14 | |
| actin filament-based process | 33 | |
| molecular adaptor activity | 9 | |
| cation channel activity | 25 | |
| regulation of Ras protein signal transduction | 26 | |
| metal ion binding | 200 | |
| courtship behavior | 12 | |
| gliogenesis | 16 | |
| Notch signaling pathway | 12 | |
| K | protein folding | 7 |
| ATP binding | 10 | |
| epigenetic regulation of gene expression | 5 | |
| N | structural constituent of cuticle | 4 |
| sensory perception of chemical stimulus | 5 | |
| microsome | 3 | |
| extracellular space | 3 | |
| O | mitotic spindle elongation | 6 |
| ATP biosynthetic process | 4 | |
| hexose catabolic process | 3 |
To discover functional processes downregulated by foraging experience, genes in profiles 12 and 20 were also analyzed together. The only overrepresented category was oxidation reduction. This is in agreement with previous findings that metabolic processes are downregulated in forager brains compared with hive bees (Ament et al., 2008; Alaux et al., 2009c). Our result indicates that the decrease in metabolic activity in the mushroom bodies is not only seen during the transition to foraging activity, but is ongoing throughout foraging experience.
Eigengenes of negatively correlated modules N and O were associated with profiles 12 and 20, respectively. These modules were enriched for genes related to mitotic spindle elongation and sensory perception of chemical stimuli. Several genes in the latter category are orthologs of odorant-binding proteins; it is surprising to see expression of these genes in the mushroom bodies. However, our previous studies have also detected expression of these genes in whole brain as well as mushroom bodies (Sen Sarma et al., 2010; Alaux et al., 2009b; Alaux et al., 2009c). In addition, a study examining regional expression of odorant-binding proteins found several expressed in whole bee brain, as well as other parts of the body, suggesting currently unknown functions for these genes (Foret and Maleszka, 2006). The number of genes in enriched categories for both negatively correlated modules was relatively small, hindering biological interpretation of their importance.
DISCUSSION
We investigated the transcriptional response to naturally occurring foraging experience in the mushroom bodies of honey bees. Because neuropil outgrowth is taking place in the mushroom bodies during this time, it is likely that part of this transcriptional response represents molecular processes involved in neuroanatomical plasticity, and our functional analyses of genes identified here are consistent with that interpretation. Future studies could extend and further support these findings. For example, it might be possible to identify a pharmacological inhibitor of one or more of the genes discussed here that could be administered in the field; the ability of such a drug to prevent neuropil expansion in foraging bees would provide evidence for a causal role of the target in MB neuropil outgrowth. Another type of experiment that would provide more associational data, but would be more practical within the constraints of a field experiment, could examine the specificity of potential growth-related genes by examining their expression in brain regions in which no large-scale neuropil growth is occurring. However, neuroanatomical plasticity is present in other regions of the bee brain (Winnington et al., 1996; Sigg et al., 1997; Brown et al., 2002), and it might be difficult to find such regions.
Investigation of multiple time points allowed us to discover a linear increase in gene expression with increasing experience, and stepwise upregulation between 8 and 12 days of foraging. These distinct temporal expression patterns may reflect several distinct molecular processes associated with neuropil outgrowth. Comparison of gene expression networks in the mushroom bodies of 1-day old bees and foragers also suggested that similar molecular processes support outgrowth at different stages of development. Parallels to findings from analyses of enriched environments in the laboratory demonstrate the robustness of the brain’s molecular responses to experience.
Our study identified genes whose expression continually increased in response to foraging experience. The temporal expression pattern of these genes suggests that they may be necessary for dendritic growth, with continued upregulation reflecting accelerated growth in more experienced foragers. Investigation of enriched environment-induced gene expression in mouse cortex identified a set of genes similarly regulated at 2 and 14 days after the onset of experience (Rampon et al., 2000), and genes with functions related to functional neuroplasticity are upregulated throughout the critical period in the visual cortex of mice (Lyckman et al., 2008).
The most common pattern of gene expression we observed in forager mushroom bodies was higher expression in more experienced foragers. Genes with this profile were part of the gene expression signature that could distinguish foragers on the basis of their experience. Functional analysis suggests that these genes could be involved in processes supporting neuropil growth. Included in this group were several genes involved in signal transduction or protein kinase activity, both of which are functional groups related to plasticity in other organisms. These functional categories were also implicated in a laboratory study of genes differentially regulated by enriched environment in the striatum of adolescent mice (Thiriet et al., 2008). Protein kinase activity is well known to be involved in molecular studies of memory and synaptic plasticity, and post-translational modification has been proposed as a mechanism underlying long-term memory consolidation (Routtenberg and Rekart, 2005).
Another possible role for genes exhibiting higher expression in more experienced foragers is the stabilization of newly formed dendrites. Long-term maintenance of dendritic arbors is thought to require constitutive molecular signaling distinct from that responsible for outgrowth (Benard and Hobert, 2009). A few specific genes involved in stabilization have been identified so far, including several protein kinases (Moresco et al., 2005; Emoto et al., 2006). The coincidence of upregulation and neuropil expansion and the overrepresentation of protein kinases among the genes with this expression profile make them candidates for involvement in a mechanism of stabilization in forager mushroom bodies that have experienced recent dendritic growth.
We also found that many foraging-related modules overlapped significantly with modules in the gene expression network for 1-day-old bees, with shared enrichment of neuroanatomical plasticity-related functional categories. These results suggest that similar sets of genes could mediate some aspects of two phases of mushroom body neuropil growth in honey bees that have been documented, the early developmental phase in 1-day-old bees and the later experience-dependent phase (Farris et al., 2001). If this is the case, it might be expected that the same pathways are also involved in the experience-expectant phase of mushroom body neuropil growth that occurs before the onset of foraging in hive bees.
One foraging-related gene module was notable because it did not overlap with any module in the young bee network. Module K in foragers was enriched for protein folding genes, several of which are heat shock protein genes. Expression in this module was significantly correlated with amount of foraging experience. Experienced foragers are rapidly approaching the end of their lifespan (Visscher and Dukas, 1997) and the act of foraging is associated with a measure of accumulation of oxidative damage in the brain (Seehuus et al., 2006). Heat shock proteins upregulated in the mushroom bodies may be necessary to prevent or delay neurological aging. Moderate downregulation of protein-folding genes in 16-day foragers could represent the onset of neurodegenerative processes. The specificity of this module in the forager network, in comparison with the young bee network, reinforces the idea that protein-folding genes are acting together to perform some function unique to forager mushroom bodies.
Given the ability of muscarinic agonists to induce neuropil growth (Ismail et al., 2006) and dendritic arborization (Dobrin et al., 2011) in foragers, we expected that genes directly related to cholinergic signaling would also be among those regulated by foraging experience. We found no significant expression differences among foragers in genes directly related to cholinergic signaling, including AChE, ChaT, VAChAT, and mAchR. However, among the larger set of foraging-regulated genes identified by network analysis were several involved in the regulation of the MAPKKK cascade, the downstream target of which is the MAPK signaling pathway. Upregulation of these genes represents a possible convergence of mechanisms with muscarinic receptor stimulation. In vertebrates, muscarinic signaling modulates synaptic plasticity by increasing intracellular calcium, thereby activating MAPK and other protein kinases (Hamilton and Nathanson, 2001; Gu, 2003). Our results suggest that transcriptional responses to experience may enhance the effects of muscarinic signaling on neuropil growth by enhancing these downstream processes.
The relatively small number of foraging-related genes identified in the mushroom bodies in this study also provides a marked contrast to the much broader transcriptional changes in the honey bee brain associated with changes in behavioral state seen in comparisons of 1-day old and forager gene expression (this study and Alaux et al., 2009b; Whitfield et al., 2003). Several factors could contribute to the relatively small number of significantly regulated foraging-related genes. The study of neuroanatomical plasticity in a naturalistic setting necessitates considerable loss of control over variation in individual experience; this variation may be caused by effects of genetic background, individual differences in foraging activity, as well as the complexity of foraging behavior, which might result in qualitatively different types of experience among individuals visiting different floral sources. It is likely that some real effects of foraging on gene expression may be hidden by within-group variation. However, comparison of our results to those from laboratory studies of other species reveal a general trend for relatively small gene lists associated with experience-dependent neuroanatomical plasticity (Rampon et al., 2000; Thiriet et al., 2008).
We found that foraging experience drives transcriptional changes in genes in the mushroom bodies. Because foraging is a natural behavior, it is expected that the brain’s responses to foraging experience should occur on a time scale consistent with the life history of the bee. This perspective, together with a consideration of the functions of the foraging-related genes we identified in this study, suggests that experience-dependent neuroanatomical changes in the mushroom bodies represent a balance between plasticity in response to novel sensory input, and stability and protection in the face of aging and physiological stress. The balance appears to shift toward the latter in experienced foragers, which is consistent with findings that suggest a qualitative difference in cognitive ability in more experienced foragers (Behrends et al., 2007). Manipulation of genes associated with these processes could lead to a more complete understanding of how neuroplasticity is maintained in the adult and aging brain.
Supplementary Material
Acknowledgements
This work was supported by NIH grant GM073644 to GER and SEF, a University of Illinois Fellowship to CCL, as well as NSF grant EF-25852 (BR Schatz, PI) for a large-scale project on the influences of heredity and the environment on brain gene expression and behavior, to which this work is related. The authors thank the following individuals for their assistance: Karen Pruiett contributed beekeeping expertise; Tom Newman and Trang Nguyen provided assistance with molecular techniques; Jessica Ray and Linda Qi provided invaluable assistance with fieldwork; and Alissa Eisenstein with microarray image processing. Members of the Robinson laboratory and two anonymous readers reviewed the article and made valuable suggestions for its improvement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alaux C, Le Conte Y, Adams HA, Rodriguez-Zas S, Grozinger CM, Sinha S, Robinson GE. Regulation of brain gene expression in honey bees by brood pheromone. Genes Brain Behav. 2009a;8:309–319. doi: 10.1111/j.1601-183X.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- Alaux C, Duong N, Schneider SS, Southey BR, Rodriguez-Zas S, Robinson GE. Modulatory communication signal performance is associated with a distinct neurogenomic state in honey bees. PLoS One. 2009b;4:e6694. doi: 10.1371/journal.pone.0006694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C, Sinha S, Hasadsri L, Hunt GJ, Guzman-Novoa E, DeGrandi-Hoffman G, Uribe-Rubio JL, Southey BR, Rodriguez-Zas S, Robinson GE. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc Natl Acad Sci U S A. 2009c;106:15400–15405. doi: 10.1073/pnas.0907043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament SA, Corona M, Pollock HS, Robinson GE. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci U S A. 2008;105:4226–4231. doi: 10.1073/pnas.0800630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends A, Scheiner R, Baker N, Amdam GV. Cognitive aging is linked to social role in honey bees (Apis mellifera) Exp Gerontol. 2007;42:1146–1153. doi: 10.1016/j.exger.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard C, Hobert O. Looking beyond development: Maintaining nervous system architecture. Curr Top Dev Biol. 2009;87:175–194. doi: 10.1016/S0070-2153(09)01206-X. [DOI] [PubMed] [Google Scholar]
- Brown SM, Napper RM, Thompson CM, Mercer AR. Stereological analysis reveals striking differences in the structural plasticity of two readily identifiable glomeruli in the antennal lobes of the adult worker honeybee. J Neurosci. 2002;22:8514–8522. doi: 10.1523/JNEUROSCI.22-19-08514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Dobrin SE, Herlihy JD, Robinson GE, Fahrbach SE. Muscarinic regulation of kenyon cell dendritic arborizations in adult worker honey bees. Arthropod Struct Dev. 2011 doi: 10.1016/j.asd.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukas R. Life history of learning: Performance curves of honeybees in the wild. Ethology. 2008;114:1195–1200. [Google Scholar]
- Dukas R, Visscher PK. Lifetime learning by foraging honey bees. Anim Behav. 1994;48:1007–1012. [Google Scholar]
- Durst C, Eichmueller S, Menzel R. Development and experience lead to increased volume of subcompartments of the honeybee mushroom body. Behav Neural Biol. 1994;62:259–263. doi: 10.1016/s0163-1047(05)80025-1. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE. Structure of the mushroom bodies of the insect brain. Annu Rev Entomol. 2006;51:209–232. doi: 10.1146/annurev.ento.51.110104.150954. [DOI] [PubMed] [Google Scholar]
- Fahrbach SE, Moore D, Capaldi EA, Farris SM, Robinson GE. Experience-expectant plasticity in the mushroom bodies of the honeybee. Learn Mem. 1998;5:115–123. [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Strande JL, Robinson GE. Neurogenesis is absent in the brains of adult honey bees and does not explain behavioral neuroplasticity. Neurosci Lett. 1995;197:145–148. doi: 10.1016/0304-3940(95)11913-h. [DOI] [PubMed] [Google Scholar]
- Farris SM, Robinson GE, Fahrbach SE. Experience- and age-related outgrowth of intrinsic neurons in the mushroom bodies of the adult worker honeybee. J Neurosci. 2001;21:6395–6404. doi: 10.1523/JNEUROSCI.21-16-06395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera) Genome Res. 2006;16:1404–1413. doi: 10.1101/gr.5075706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q. Contribution of acetylcholine to visual cortex plasticity. Neurobiol Learn Mem. 2003;80:291–301. doi: 10.1016/s1074-7427(03)00073-x. [DOI] [PubMed] [Google Scholar]
- Hamilton SE, Nathanson NM. The M1 receptor is required for muscarinic activation of mitogen-activated protein (MAP) kinase in murine cerebral cortical neurons. J Biol Chem. 2001;276:15850–15853. doi: 10.1074/jbc.M011563200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (England) 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Z, Robinson GE. Regulation of honey bee division of labor by colony age demography. Behavioral Ecology and Sociobiology. 1996;39:147–158. [Google Scholar]
- Ismail N, Robinson GE, Fahrbach SE. Stimulation of muscarinic receptors mimics experience-dependent plasticity in the honey bee brain. Proc Natl Acad Sci U S A. 2006;103:207–211. doi: 10.1073/pnas.0508318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb Brain plasticity and behavior. Annu Rev Psychol. 1998;49:43–64. doi: 10.1146/annurev.psych.49.1.43. [DOI] [PubMed] [Google Scholar]
- Krofczik S, Khojasteh U, de Ibarra NH, Menzel R. Adaptation of microglomerular complexes in the honeybee mushroom body lip to manipulations of behavioral maturation and sensory experience. Dev Neurobiol. 2008;68:1007–1017. doi: 10.1002/dneu.20640. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Molecular profiling of behavioural development: differential expression of mRNAs for inositol 1,4,5-trisphosphate 3-kinase isoforms in naive and experienced honeybees (Apis mellifera) Brain Res Mol Brain Res. 2002;99:92–101. doi: 10.1016/s0169-328x(01)00325-4. [DOI] [PubMed] [Google Scholar]
- Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–659. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Li C, Niu W, Jiang CH, Hu Y. Effects of enriched environment on gene expression and signal pathways in cortex of hippocampal CA1 specific NMDAR1 knockout mice. Brain Res Bull. 2007;71:568–577. doi: 10.1016/j.brainresbull.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Lyckman AW, Horng S, Leamey CA, Tropea D, Watakabe A, Van Wart A, McCurry C, Yamamori T, Sur M. Gene expression patterns in visual cortex during the critical period: Synaptic stabilization and reversal by visual deprivation. Proc Natl Acad Sci U S A. 2008;105:9409–9414. doi: 10.1073/pnas.0710172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle JJ, Ferrer-Caja E, Hamagami F, Woodcock RW. Comparative longitudinal structural analyses of the growth and decline of multiple intellectual abilities over the life span. Dev Psychol (United States) 2002;38:115–142. [PubMed] [Google Scholar]
- Menzel R. Learning, memory, and "cognition" in honey bees. In: Kesner RP, Olton DS, editors. Neurobiology of comparative cognition. Hillsdale, NJ: Erlbaum Associates, Inc.; 1990. pp. 237–292. [Google Scholar]
- Moresco EM, Donaldson S, Williamson A, Koleske AJ. Integrin-mediated dendrite branch maintenance requires abelson (abl) family kinases. J Neurosci. 2005;25:6105–6118. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EL, Tang BL. Rab GTPases and their roles in brain neurons and glia. Brain Res Rev. 2008;58:236–246. doi: 10.1016/j.brainresrev.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Rampon C, Jiang CH, Dong H, Tang YP, Lockhart DJ, Schultz PG, Tsien JZ, Hu Y. Effects of environmental enrichment on gene expression in the brain. Proc Natl Acad Sci U S A. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnback A, Dahlqvist P, Svensson PA, Jernas M, Carlsson B, Carlsson LM, Olsson T. Gene expression profiling of the rat hippocampus one month after focal cerebral ischemia followed by enriched environment. Neurosci Lett. 2005;385:173–178. doi: 10.1016/j.neulet.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Rosenzweig MR. Modification of brain circuits through experience. In: Bermudez-Rattoni F, editor. Neural plasticity and memory: From genes to brain imaging. Boca Raton (FL): Taylor & Francis Group, LLC.; 2007. [Google Scholar]
- Routtenberg A, Rekart JL. Post-translational protein modification as the substrate for long-lasting memory. Trends Neurosci. 2005;28:12–19. doi: 10.1016/j.tins.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush Jz. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Seehuus SC, Krekling T, Amdam GV. Cellular senescence in honey bee brain is largely independent of chronological age. Exp Gerontol. 2006;41:1117–1125. doi: 10.1016/j.exger.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Sarma M, Whitfield CW, Robinson GE. Species differences in brain gene expression profiles associated with adult behavioral maturation in honey bees. BMC Genomics. 2007;8:202. doi: 10.1186/1471-2164-8-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Sarma M, Rodriguez-Zas SL, Hong F, Zhong S, Robinson GE. Transcriptomic profiling of central nervous system regions in three species of honey bee during dance communication behavior. PLoS One. 2009;4:e6408. doi: 10.1371/journal.pone.0006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Sarma M, Rodriguez-Zas SL, Gernat T, Nguyen T, Newman T, Robinson GE. Distance-responsive genes found in dancing honey bees. Genes Brain Behav. 2010;9:825–830. doi: 10.1111/j.1601-183X.2010.00622.x. [DOI] [PubMed] [Google Scholar]
- Sigg Activity-dependent changes to the brain and behavior of the honey bee, apis mellifera (L.) The Journal of Neuroscience. 1997;17:7148. doi: 10.1523/JNEUROSCI.17-18-07148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawski M, Daumer M, Boulesteix AL. CMA: A comprehensive bioconductor package for supervised classification with high dimensional data. BMC Bioinformatics. 2008;9:439. doi: 10.1186/1471-2105-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet N, Amar L, Toussay X, Lardeux V, Ladenheim B, Becker KG, Cadet JL, Solinas M, Jaber M. Environmental enrichment during adolescence regulates gene expression in the striatum of mice. Brain Res. 2008;1222:31–41. doi: 10.1016/j.brainres.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H FlyBase Consortium. FlyBase: Enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH. Neural consequences of environmental enrichment. Nat Rev Neurosci. 2000;1:191–198. doi: 10.1038/35044558. [DOI] [PubMed] [Google Scholar]
- Visscher PK, Dukas R. Survivorship of foraging honey bees. Insectes Soc. 1997;44:1–5. [Google Scholar]
- Wang X, Wu M, Li Z, Chan C. Short time-series microarray analysis: Methods and challenges. BMC Syst Biol. 2008;2:58. doi: 10.1186/1752-0509-2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, Leconte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci U S A. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnington Structural plasticity of identified glomeruli in the antennal lobes of the adult worker honey bee. Journal of Comparative Neurology. 1996;365:479. doi: 10.1002/(SICI)1096-9861(19960212)365:3<479::AID-CNE10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Winston ML. The biology of the honey bee. Cambridge, MA: Harvard University Press; 1987. [Google Scholar]
- Withers GS, Fahrbach SE, Robinson GE. Selective neuroanatomical plasticity and division of labour in the honeybee. Nature. 1993;364:238–240. doi: 10.1038/364238a0. [DOI] [PubMed] [Google Scholar]
- Withers GS, Fahrbach SE, Robinson GE. Effects of experience and juvenile hormone on the organization of the mushroom bodies of honeybees. J Neurobiol. 1995;26:130–144. doi: 10.1002/neu.480260111. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


