Abstract
New template-based self-propelled gold/nickel/polyaniline/platinum (Au/Ni/PANI/Pt) microtubular engines, functionalized with the Concanavalin A (ConA) lectin bioreceptor, are shown to be extremely useful for the rapid, real-time isolation of Escherichia coli (E. coli) bacteria from fuel-enhanced environmental, food and clinical samples. These multifunctional microtube engines combine the selective capture of E. coli with the uptake of polymeric drug-carrier particles to provide an attractive motion-based theranostics strategy. Triggered release of the captured bacteria is demonstrated by movement through a low-pH glycine-based dissociation solution. The smaller size of the new polymer-metal microengines offers convenient, direct and label-free optical visualization of the captured bacteria and discrimination against non-target cells.
Keywords: Nanomachines, microengines, lectin, E. coli isolation, theranostics, drug delivery, biodetection, complex samples
Major threats to human health from E. coli infection have led to urgent demands to develop highly efficient strategies for isolating and detecting this microorganism in connection to food safety, medical diagnostics, water quality, and counter terrorism.1-3E. coli and other pathogenic bacteria are commonly detected using traditional culture techniques, microscopy, luminescence, enzyme-linked immunosorbent assay (ELISA), biochemical tests and/or the polymerase chain reaction (PCR). These techniques, however, are time-consuming, labor-intensive, and inadequate as they lack the ability to detect bacteria in real time.4,5 Thus, there is an urgent need for alternative platforms for the rapid, sensitive, reliable and simple isolation and detection of E. coli.
Here we present a new nanomotor strategy for isolating pathogenic bacteria from peroxide-fuel containing clinical, environmental and food samples, involving the movement of lectinfunctionalized microengines. The motion and power of self-propelled synthetic and natural nano/microscale motors have been exploited recently as an attractive route for transporting target biomaterials.6-9 The limitations of biological motors for engineered microchip environments6,10 have motivated the use of synthetic nanomotors for diverse biomedical and bioanalytical applications.7,11,12 Particularly attractive for such applications are bubble-propelled microtube engines owing to their efficient movement in fuel-enhanced biological fluids and their large towing forces.13-16 Functionalizing photolitographically-prepared rolled-up microtube engines with oligonucleotide or antibody receptors has recently been shown to be extremely effective for capturing and isolating target biomolecules and cells from biological samples.15,17-19 Yet, the preparation of these rolled-up microengines is complex and costly, requiring the use of clean-room facilities. Recently, Gao et al.20 have developed smaller (8 μm) and highly efficient microtube engines, which can be mass-produced through a low-cost membrane template electrodeposition technique, and offer ultrafast speeds (> 300 body lengths/s) along with low fuel requirements (down to 0.2 %) in optimal conditions.20
The efficient bacterial isolation platform, described in this paper, relies on the attractive behavior of these easily prepared microengines along with their functionalization with lectin receptors. Lectins are readily available glycoproteins that offer an attractive route for recognizing carbohydrate constituents of bacterial surface, via selective binding to cell-wall mono- and oligosaccharide components.21 For example, ConA, the lectin extracted from Canavalia ensiformis and used in the present work, is a mannose- and glucose-binding protein that is capable of recognizing specific terminal carbohydrates of Gram-negative bacteria such as the E. coli surface polysaccharides.22,23 Furthermore, lectins have been recently used as the biosensor recognition elements for bacterial detection24-30 but not in connection to nanomachines or nanoscale motion-based isolation.
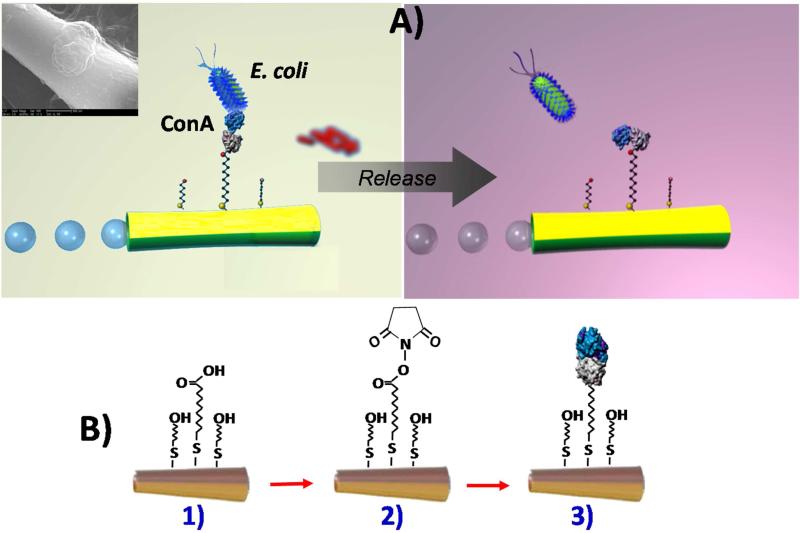
As illustrated in Figure 1A (left), the presented nanoscale bacteria isolation strategy utilizes the movement of ConA-functionalized microengines to scour, interact and isolate pathogenic bacteria from distinct complex samples. A second goal of the present work is to demonstrate a bacterial target unloading scheme through the controlled release of the captured bacteria from a moving synthetic microengine. Such triggered release has been accomplished using a low-pH glycine solution that is able to dissociate the lectin-bacteria complex (Figure 1A, right). Finally, we present the ability of the microengines to capture and transport simultaneously both the target bacteria as well as polymeric drug-carrier spheres (using the lectin and magnetic interactions, respectively). Such dual action, coupling the E. coli isolation with ‘on-the-spot’ therapeutic action adds a completely new and unique theranostics (‘identify and eradicate’) capability to nanomachine platforms. Overall, the template-prepared lectin-modified microengines hold considerable promise for diverse biomedical, food safety, and biodefense applications.
Figure 1.
Lectin-modified microengines for bacteria isolation. Schemes depicting A) the selective pick-up, transport and release of the target bacteria by a ConA-modified microengine, and B) surface chemistry involved on the microengines functionalization with the lectin receptor. Upon encountering the cells, the ConA-functionalized microengines recognize the E. coli cell walls by O-antigen structure binding-allowing for selective pick-up and transport. Inset (in Scheme A, top left side), a SEM image of a portion of a ConA-modified microengine loaded with an E. coli cell. Scheme A, right side: Release of the capture bacteria by navigation in a 10 mM glycine solution, pH 2.5. Scheme B, Steps involved in the microengines gold surface functionalization: 1) self-assembling of MUA/MCH binary monolayer; 2) activation of the carboxylic terminal groups of the MUA to amine-reactive esters by the EDC and NHS coupling agents; 3) reaction of NHS ester groups with the primary amines of the ConA to yield stable amide bonds.
The fabrication of the microengines involves a template-based electrodeposition of a polyaniline (PANI)/Pt bilayer microtube and e-beam vapor deposition of outer Ni/Au layers which are essential for magnetic navigation and surface functionalization. As illustrated in Figure 1B, such functionalization involves the self assembly of alkanethiols and subsequent conjugation of the lectin receptor. The template fabrication process results in 8 μm-long microtube engines that are substantially smaller than common rolled-up tubular microengines. The relative similar dimensions of the microengine and the target bacteria (~2 μm long ×0.5 μm in diameter) permit convenient real-time optical visualization of the isolation process without the need for additional labeling. By varying the membrane pore size and deposition time, the microengine's aspect ratio can be tailored for meeting the needs of specific target bacteria detection paradigms. Similar to rolled-up microengines,15,18,31-33 the template-prepared microengines are propelled efficiently in different media via the expulsion of oxygen bubbles generated from the catalytic oxidation of hydrogen peroxide fuel at the inner Pt layer.20 The high speed of the template-prepared PANI/Pt bilayer microtube engines in the present working medium (300 μm/s) is reduced by ca. 50% after the vapor deposition of the outer Ni/Au layers to around 150 μm/s.
Next, the microengine functionalization protocol was optimized for efficient lectin-bacteria interaction and locomotion. As illustrated in Figure 1B, functionalization was accomplished by conjugating the lectin to the outer gold surface of the microengines via a self-assembled monolayer (SAM). A mixture of 11-mercatoundecanoic acid (MUA) and 6-mercaptohexanol (MCH) was used to create the binary SAM while 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS) chemistry was used to activate the MUA carboxyl terminated groups for conjugation with ConA (see Methods Section in the Supporting Information for additional details). To promote favorable target accessibility while minimizing non-specific adsorption, the binary SAM was prepared using optimal alkanethiol concentrations of 0.25 mM MUA and 0.75 mM MCH.34 The relatively low thiol concentrations ensure minimal poisoning of the inner catalytic platinum surface and hence a high catalytic activity.35 Such surface modification of the Au/Ni/PANI/Pt microtubular engines resulted in an additional speed reduction, down to 80 μm/s. This speed is sufficient to perform cellular towing tasks and is relatively faster than previously reported rolled-up microengines.31,32 After the SAM activation, the lectin receptor was immobilized via NHS/EDC coupling using a binding buffer (BB) solution containing 9 mg/ml of ConA. This step did not affect the microengine speed. Overall, despite of the ~75% total reduction in speed from the entire modification process, the microengine's speed and related force were sufficient to carry multiple bacteria or different cargoes.
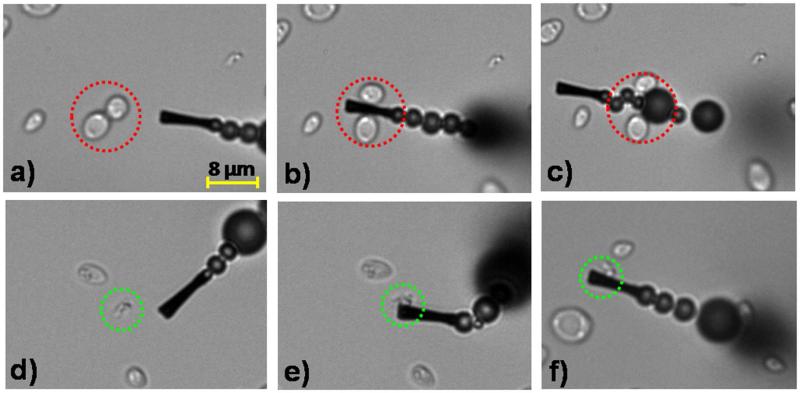
The specific binding of the ConA-modified microengines to E. coli was examined first in inoculated human urine samples. These urine samples were inoculated with E. coli target bacteria along with a 5-fold excess Saccharomyces cerevisiae (S. cerevisiae), a species of yeast frequently responsible for yeast infections and UTIs. Figure 2 and Supporting Video S1 demonstrate the selective binding and transport of the rod-shaped (~2 μm length) gram-negative E. coli bacteria (delineated by green dotted circles). In contrast, the modified microengine does not capture the round-shaped S. cerevisiae cells (~5 μm in diameter) even when multiple contacts occur (delineated by red dotted circles) (Figure 2, a–c and Supporting Video S1A). The distinct size (~2 μm vs 5 μm) and shape (rod vs round) of the target E. coli and control S. cerevisiae, respectively, allow clear optical visualization and discrimination between the target and non-target cells during the motor navigation (see Supporting Video S1A). Note again that the similar size scale of the microengine and bacteria facilitates real-time visualization of the binding process (see Figure 2 and Supporting Video S1A). This selective and rapid capture mechanism is attributed to the nearly instantaneous recognition of the sugar moieties on the bacterial cell wall by the lectin-modified microengine (Figures 2d-f). Indeed, the multivalent binding of ConA to the O-antigen E. coli surface favors strong adhesion of E. coli to the ConA-modified microengine surface23,24 as illustrated clearly in the SEM image depicted in Figure 1A (top left side). To further corroborate the specificity of the ConA-modified microengines, we tested human urine samples inoculated with both E. coli and a 5-fold excess of another urinary pathogen Staphylococcus aureus (S. aureus), (i.e., a small, round, gram-positive UTI-related bacteria)36 (see Supporting Video S1B). Both capture experiments results with S. cerevisiae and S. aureus controls in urine specimens demonstrate the high specificity of the ConA-modified microengines towards E. coli and their ability to selectively capture and transport microorganisms from complex clinical samples.37
Figure 2.
Selective interaction between the ConA-functionalized microengines and the E. coli target bacteria in a fuel-enhanced and E. coli inoculated human urine sample. Time-lapse images - taken from Supporting Video S1A- before, during, and after interaction of the ConA-modified microengines with S. cerevisiae negative control (a–c, respectively) and E. coli target (d-f, respectively) cells. Urine samples are inoculated with E. coli (2.25×107 colony forming units (cfu/ml) or 4.5×104 cfu on the glass slide) and a 5-fold excess of S. cerevisiae and finally diluted 4 times in the glass slide to include the functionalized microengines and the fuel solutions (See Methods Section for additional details). Final fuel conditions: 7.5% (w/v) H2O2, 1.25% (w/v) Triton X-100. The E. coli and S. cerevisiae cells are accented by dashed green and red circles, respectively.
Additional experiments were performed with “control” microengines (i.e., without the immobilized ConA; prepared as described in the SI Methods Section) to demonstrate that the surface-confined lectin is solely responsible for the bacterial isolation. For example, Supporting Video S2 demonstrates that these “control” microengines do not capture E. coli even after multiple contacts with the bacteria while navigating in the BB solution containing a 10-fold excess of E. coli (compared to solutions commonly used with the ConA-modified microengines). These results, along with mixture experiments involving large excess of a gram-positive bacteria or yeast, clearly demonstrate that the capture of the E. coli occurs through the specific lectin-carbohydrate recognition and confirm the high specificity of surface-confined ConA towards lipopolysaccharide O-antigens characteristics of gram-negative bacteria.23,24,26,29,38 The high selectivity, illustrated in Figure 2 and in multiple Supporting Videos (S1A, S1B and S2), is attributed not only to the high affinity of the lectin to the target bacteria but also to the effective minimization of non-specific binding associated with the highly dense hydrophilic SAM surface coating. These data also confirm that the hydrogen peroxide fuel and Triton X-100 surfactant, essential for the microengine movement, do not compromise the specific lectin-bacterial cell wall interaction or the integrity of the assembled binary SAM.
The reproducibility of the new motion-based isolation was investigated by using 5 different batches of ConA-modified microengines following identical processing steps. The results (not shown) demonstrated very small (~5%) differences in the bacteria capture efficiency among different batches of modified microengines, demonstrating the reliability of their fabrication, modification and movement processes. While lectin-bacteria binding often requires long (30-60 min) incubation times,30,39 the microengine-induced localized convection appears to dramatically accelerate the binding process. Short contact times with the target bacteria (on the order of seconds) are thus sufficient for its effective capture. The new microengine platform thus presents a unique approach for meeting the need for rapid, direct and real time isolation of biological agents.
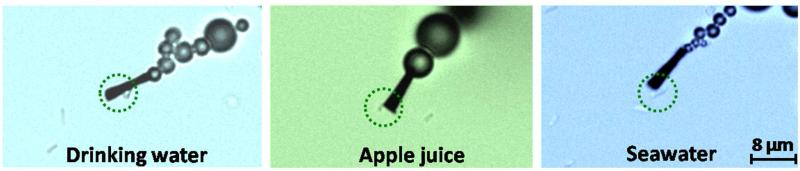
The practical utility of the new microengine approach towards diverse applications was illustrated by the ability of the lectin-modified microengines to recognize the target bacteria in different fuel-enhanced and E-coli-inoculated real samples. These samples included common beverages (ranging from drinking water to apple juice) and environmental matrices (such as seawater). For example, Figure 3 and the corresponding Supporting Video S3, clearly illustrate that the functionalized microengines display an immediate ‘on the fly’ E. coli capture upon contacting the target bacteria in each of these real samples. A successful pick-up rate (during the first engine-cell contact) of nearly 90% (n=50) has been observed in all the tested matrices. In rare occasions (less than 1% of the times) the cells were inhaled into the front opening of the modified microrocket (i.e., captured non-specifically). Overall, the results of Figures 2 and 3 demonstrate the ability of the modified microengines to pick-up bacteria in the presence of diverse conditions, different environments and matrix effects, including low and high sugar concentrations (as in drinking water and apple juice, respectively) as well as high salt (seawater and urine) environments. Note also (from Table 1) that the average speed of the functionalized microengines varies with the specific sample matrix. Despite small decrease in the microengine speed (~10%) after the E. coli capturing, efficient propulsion and transport of the captured bacteria are maintained using the different sample matrices.
Figure 3.
Isolation of the E. coli target bacteria from different real samples using ConA-functionalized microengines. Images -taken from Supporting Video S3- demonstrating the E. coli pick-up and transport in peroxide-fuel containing samples: a) drinking water, b) apple juice and c) seawater samples inoculated with E. coli 1.8×107 cfu/mL (3.6×104 cfu on the glass slide). Other conditions, as in Fig. 2. E. coli cells are accented by dashed green circles.
Table 1.
Estimated speed of the ConA-functionalized microengines in different media inoculated with 1.8×107 cfu/ml of E. coli.
| Media* | Average speed (n = 5), μm/s |
|---|---|
| BB | 50.0 ± 28.7 |
| Drinking water | 27.3 ± 9.3 |
| Apple juice | 30.6 ± 11.0 |
| Seawater | 33.9 ± 13.0 |
Media finally diluted 4 times after mixing with the ConA-microengines and the fuel solutions required by the bioassay. Fuel composition: 7.5% (w/v) H2O2 + 1.25% (w/v) Triton X-100.
The interaction between the bacterial cell and the ConA-modified microengines is not only highly selective but also very robust. For example, Supporting Video S4 illustrates the ability of the microengines to pick up and carry an E. coli cell that had adhered to the glass slide, while Supporting Video S5 displays the simultaneous transport of multiple bacteria by a single ConA-modified microengine. The drag force needed to perform such tasks can be calculated via Stokes’ law.15 For example, a drag force of 1 pN can be estimated for a microengine moving at a 33.9 μm/s in seawater, assuming a cylindrical microengine of 8 μm in length and 2 μm in diameter.22 This force is sufficient to remove a bacterial cell firmly fixated to the glass slide, as clearly demonstrated in Supporting Video S4. Note that in the multiple-bacteria transport process, illustrated in Supporting Video S5, all the captured bacteria are confined only to half of the microengine, reflecting that only ~50% of its outer surface is covered by the gold layer due to the one-sided e-beam deposition process and the corresponding one-sided functionalization with the lectin receptor. Nevertheless, Supporting Videos S4 and S5 demonstrate both the strong lectin/bacteria interaction and the high towing capacity of the lectin-modified microengines.
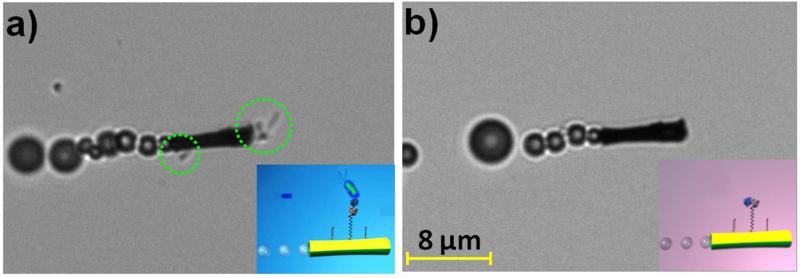
The ability to isolate and unload target bacteria is critical for identifying pathogenic bacteria serotypes. To facilitate the release of the captured bacteria, the loaded ConA-modified microengines were moved through a low-pH glycine-based dissociation solution that disrupts the sugar-lectin complex40-42 (Figure 1A, right). Figure 4 illustrates images, obtained from Supporting Video S6A, of the lectin-modified microengines before (a) and after (b) 20 min navigation in this dissociation solution. Note that such prolonged continuous movement is observed without replenishing the fuel. The multiple bacteria confined to the moving microengine are clearly released from its surface after movement in the low-pH glycine-based dissociation solution. The efficient removal of the captured bacteria is attributed to the unfolding of ConA in this low pH-solution, thereby dissociating the sugar-lectin complex and releasing the captured bacteria for subsequent re-use.40-42 The unfolding of the immobilized bioreceptor in this low-pH solution is supported also by the inability of the ConA-modified microengines to capture target bacteria in this medium while contacting it multiple times, as illustrated in Supporting Video S6B. These mild dissociation conditions were shown not to affect the sugar binding capacity of the immobilized lectin25,43,44 or the integrity of the assembled binary monolayer. Thus, a reversible refolding to the original active ConA conformation is expected after navigating in the BB solution, indicating great promise for reusing the same modified microengines in new isolation experiments.
Figure 4.
Release of the captured bacteria from the ConA-modified microengines. Images taken from Supporting Video S6A showing an E. coli-ConA-modified microengine before a) and after b) incubation (20 min) in a 10 mM glycine solution (pH 2.5). Other experimental conditions, as in Fig. 2. E. coli cells are accented by dashed green circles.
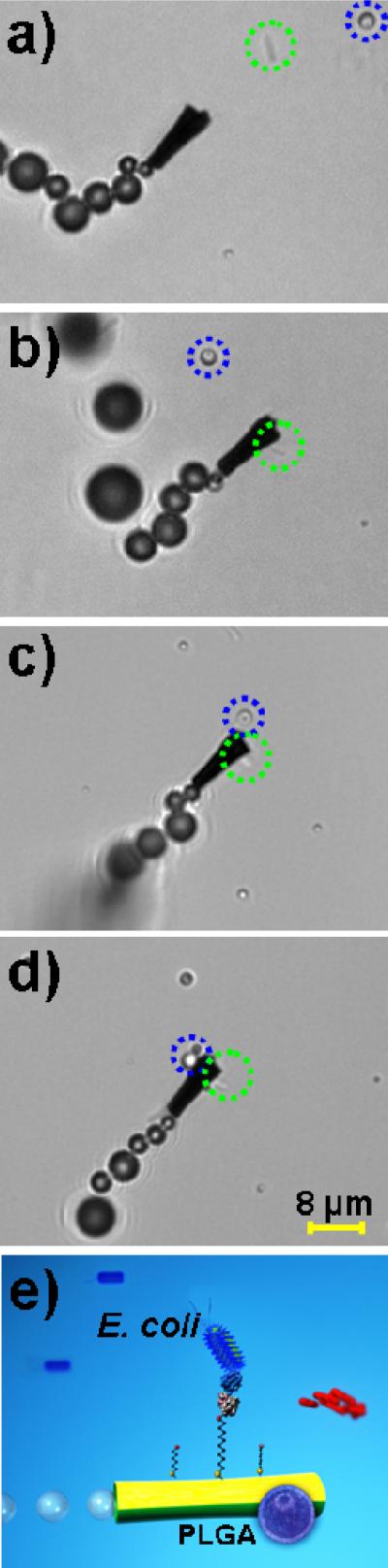
The feasibility of lectin-modified microengines to perform multiple tasks was finally explored. Here, we combine the ability to capture the target bacteria cell with simultaneous transport of common therapeutic polymeric particles towards the creation of the first self-propelling multi-functional (therasonics) device. Common drug carrier microspheres, poly D,L-lactic-co-glycolic acid (PLGA) microparticles, containing magnetic iron-oxide nanoparticles, were employed due to their potential to deliver antimicrobial drugs.45 Figure 5 and Supporting Video S7 demonstrate this dual capture and transport functionality, by displaying a ConA-modified microengine picking-up and transporting both an E. coli cell and a PLGA magnetic particle. The ConA-modified microengine is capable of first identifying and capturing a noxious bacteria (i.e., E. coli) (b), next capturing the magnetic polymeric drug carrier (c), and finally transporting both cargos at the same time (d). Eventually both captured cargoes can be released simultaneously or independently in a specified location for eradication or identification. Alternatively, future therasonics nanomotor schemes could be developed to navigate a PLGA-loaded microengine in the bacteria-containing sample. Overall, the preliminary data of Figure 5 demonstrate the unique multi-functionality potential of the modified-microengines for future applications including UTI diagnosis and treatment or water and food quality control.
Figure 5.
Dual capture and transport of E. coli and polymeric (PLGA) drug-carrier particles. Images taken from Supporting Video S7 with a ConA-modified microengine picking-up first the target bacteria (b), followed by the PLGA magnetic loading (c) and transport of both cargoes (d). Cartoon depicting the dual capture capability of a ConA-modified microengine (e). Other experimental conditions, as in Fig. 2. Attached E. coli cell and PLGA microparticle are accented by dashed green and blue circles, respectively.
While a large fraction of the bacteria are expected to remain viable but not culturable (VBNC) after exposure to the peroxide fuel level,46,47 the new nanomotor ‘swim and catch’ strategy allows for isolation of different bacteria populations (independent of their viability or culturability). Such VBNC E. coli cells, the predominant bacterial population under our working conditions, are not detectable by common culture methods but may retain their capacity to grow and cause infection.47-51 Hence, this nanomotor method can avoid time-consuming PCR protocols and false negatives associated with culture-based methods when detecting VBNC and non-viable bacteria. The ability to detect potentially infectious VBNC E. coli and dead cells is also attractive for tracing the source of an outbreak to identify enterotoxigenic or enterohemorrhagic E. coli serotypes such as Shiga toxin (Stx)-producing E. coli O157:H7.49 Whenever needed, new fuel-free magnetically-driven nanomotors,52 functionalized with the lectin receptor, could also be used to ensure full viability of the isolated bacteria.
In conclusion, we have demonstrated the use of new synthetic microengines, functionalized with lectin receptors, for the efficient isolation of target bacteria from diverse real samples. These modified template-prepared microengines offer very attractive capabilities for autonomous loading, directional transport and bacterial unloading (‘catch and release’) towards subsequent reuse, along with efficient and simultaneous transport of drug nanocarriers. Such ability to perform simultaneously bacteria isolation and directed drug-delivery may help in designing miniaturized theranostics microsystems, integrating dual capture of target species, and transporting and releasing target species in a controlled fashion within spatially separated zones. The incorporation of the new microengine-based bacterial isolation protocol into microchannel networks could lead to microchip operations involving real-time isolation of specific bacteria, and subsequent bacteria lysis and unequivocally identification (by 16S rRNA gene analysis), potentially down to the single-cell level. The diverse capabilities of these lectin-modified hybrid microengines make them extremely attractive for a wide-range of fields, including food and water safety, infectious disease diagnostics, biodefense, and clinical therapy treatments.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Science Foundation (Award Number CBET 0853375), the National Institutes of Health (Award U01 AI075565), and the Laboratory Directed Research and Development program at Sandia National Laboratories. S.C. and J.O. acknowledge the support from Programa Becas Complutense del Amo (2010-2011) and a Beatriu de Pinós postdoctoral fellowship (Government of Catalonia), respectively. M.G. thanks the University Autónoma of Barcelona (UAB) and the Spanish MICINN. The authors would like to acknowledge also B. Chuluun and C. Halford for their technical assistance.
Footnotes
Supporting Information. Supporting Videos description, detailed Methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Laurino P, Kikkeri R. Nano Lett. 2011;11:73–78. doi: 10.1021/nl102821f. [DOI] [PubMed] [Google Scholar]
- 2.Liao JC, Mastali M, Gau V, Suchard MA, Møller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, McCabe ERB, Churchill BM, Haake DA. J. Clin Microbiol. 2006;44:561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guven B, Basaran-Akgul N, Temur E, Tamer U, Boyacı IH. Analyst. 2011;136:740–748. doi: 10.1039/c0an00473a. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Ruan C, Kanayeva D, Lassiter K, Li Y. Nano Lett. 2008;8:2625–2635. doi: 10.1021/nl080366q. [DOI] [PubMed] [Google Scholar]
- 5.Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, Evoy S. Analyst. 2011;136:486–492. doi: 10.1039/c0an00697a. [DOI] [PubMed] [Google Scholar]
- 6.Fischer T, Agarwal A, Hess H. Nat. Nanotechnol. 2009;4:162–166. doi: 10.1038/nnano.2008.393. [DOI] [PubMed] [Google Scholar]
- 7.Wang J. ACS Nano. 2009;3:4–9. doi: 10.1021/nn800829k. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt C, Vogel V. Lab on a Chip. 2010;10:2195–2198. doi: 10.1039/c005241h. [DOI] [PubMed] [Google Scholar]
- 9.Pumera M. Nanoscale. 2010;2:1643–1649. doi: 10.1039/c0nr00287a. [DOI] [PubMed] [Google Scholar]
- 10.Hiyama S, Inoue T, Shima T, Moritani Y, Suda T, Sutoh K. Small. 2008;4:410–415. doi: 10.1002/smll.200700528. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Balasubramanian S, Kagan D, Manesh KM, Campuzano S, Wang J. Nat. Commun. 2010;1:36. doi: 10.1038/ncomms1035. doi: 10.1038/ ncomms1035. [DOI] [PubMed] [Google Scholar]
- 12.Kagan D, Laocharoensuk R, Zimmerman M, Clawson C, Balasubramanian S, Kang D, Bishop D, Sattayasamitsathit S, Zhang L, Wang J. Small. 2010;6:2741–2747. doi: 10.1002/smll.201001257. [DOI] [PubMed] [Google Scholar]
- 13.Manesh KM, Cardona M, Yuan R, Clark M, Kagan D, Balasubramanian S, Wang J. ACS Nano. 2010;4:1799–1804. doi: 10.1021/nn1000468. [DOI] [PubMed] [Google Scholar]
- 14.Solovev AA, Sánchez S, Pumera M, Mei YF, Schmidt OG. Adv. Funct. Mat. 2010;20:2430–2435. [Google Scholar]
- 15.Balasubramanian S, Kagan D, Hu CJ, Campuzano S, Jesus Lobo-Castañon M, Lim N, Kang DY, Zimmerman M, Zhang L, Wang J. Angew. Chem. Int. Ed. 2011;50:4161–4164. doi: 10.1002/anie.201100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei YF, Solovev AA, Sanchez S, Schmidt OG. Chem. Soc. Rev. 2011;40:2109–2119. doi: 10.1039/c0cs00078g. [DOI] [PubMed] [Google Scholar]
- 17.Sánchez S, Solovev AA, Schulze S, Schmidt OG. Chem. Commun. 2011;47:698–700. doi: 10.1039/c0cc04126b. [DOI] [PubMed] [Google Scholar]
- 18.Kagan D, Campuzano S, Balasubramanian S, Kuralay F, Flechsig G-U, Wang J. Nano Letters. 2011;11:2083–2087. doi: 10.1021/nl2005687. [DOI] [PubMed] [Google Scholar]
- 19.Orozco J, Campuzano S, Kagan D, Ming Z, Gao W, Wang J. Anal. Chem. 2011;83:7962–7969. doi: 10.1021/ac202029k. [DOI] [PubMed] [Google Scholar]
- 20.Gao W, Sattayasamitsathit S, Orozco J, Wang J. J. Am. Chem. Soc. 2011;133:11862–11864. doi: 10.1021/ja203773g. [DOI] [PubMed] [Google Scholar]
- 21.Haseley SR. Anal. Chim. Acta. 2002;457:39–45. [Google Scholar]
- 22.Loaiza OA, Lamas-Ardisana PJ, Jubete E, Ochoteco E, Loinaz I, Cabañero G, García I, Penadés S. Anal. Chem. 2011;83:2987–2995. doi: 10.1021/ac103108m. [DOI] [PubMed] [Google Scholar]
- 23.He X, Zhou L, He D, Wang K, Cao J. Analyst. 2011;136:4183–4191. doi: 10.1039/c1an15413c. [DOI] [PubMed] [Google Scholar]
- 24.Shen Z, Huang M, Xiao C, Zhang Y, Zeng X, Wang PG. Anal. Chem. 2007;79:2312–2319. doi: 10.1021/ac061986j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safina G, van Lier M, Danielsson B. Talanta. 2008;77:468–472. [Google Scholar]
- 26.Lu Q, Lin H, Ge S, Luo S, Cai Q, Grimes CA. Anal. Chem. 2009;81:5846–5850. doi: 10.1021/ac9008572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamella M, Campuzano S, Parrado C, Reviejo AJ, Pingarrón JM. Talanta. 2009;78:1303–1309. doi: 10.1016/j.talanta.2009.01.059. [DOI] [PubMed] [Google Scholar]
- 28.Wan Y, Zhang D, Hou B. Talanta. 2009;80:218–223. doi: 10.1016/j.talanta.2009.06.057. [DOI] [PubMed] [Google Scholar]
- 29.Gao J, Liu D, Wang Z. Anal. Chem. 2010;82:9240–9247. doi: 10.1021/ac1022309. [DOI] [PubMed] [Google Scholar]
- 30.Grünstein D, Maglinao M, Kikkeri R, Collot M, Barylyuk K, Lepenies B, Kamena F, Zenobi R, Seeberger PH. J. Am. Chem. Soc. 2011;133:13957–13966. doi: 10.1021/ja2036767. [DOI] [PubMed] [Google Scholar]
- 31.Mei Y, Huang G, Solovev AA, Bermúdez Ureña E, Mönch I, Ding F, Reindl T, Fu RKY, Chu PK, Schmidt OG. Adv. Mater. 2008;20:4085–4090. [Google Scholar]
- 32.Solovev AA, Mei Y, Bermúdez Ureña E, Huang G, Schmidt OG. Small. 2009;5:1688–1692. doi: 10.1002/smll.200900021. [DOI] [PubMed] [Google Scholar]
- 33.Li JX, Huang GS, Ye MM, Li ML, Liu R, Mei YF. Nanoscale. 2011 DOI:10.1039/C1NR10840A. [Google Scholar]
- 34.Briand E, Salmain M, Herry JM, Perrot H, Compère C, Pradier C-M. Biosens. & Bioelectron. 2006;22:440–448. doi: 10.1016/j.bios.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Bartholomew CH, Agrawal PK, Katzer JR. Advances in Catalysis. Vol. 31. Academic Press; 1982. pp. 135–242. [Google Scholar]
- 36.Muder RR, Brennen C, Rihs JD, Wagener MM, Obman A, Stout JE, Yu VL. CID. 2006;42:46–50. doi: 10.1086/498518. [DOI] [PubMed] [Google Scholar]
- 37.Pistole TG. Ann. Rev. Microbiol. 1981;35:85–111. doi: 10.1146/annurev.mi.35.100181.000505. [DOI] [PubMed] [Google Scholar]
- 38.Gao J, Liu C, Liu D, Wang Z, Dong S. Talanta. 2010;81:1816–1820. doi: 10.1016/j.talanta.2010.03.052. [DOI] [PubMed] [Google Scholar]
- 39.Serra B, Gamella M, Reviejo AJ, Pingarrón JM. Anal. Bioanal. Chem. 2008;391:1853–1860. doi: 10.1007/s00216-008-2141-6. [DOI] [PubMed] [Google Scholar]
- 40.Švitel J, Dzgoev A, Ramanathan K, Danielsson B. Biosens. & Bioelectron. 2000;15:411–415. doi: 10.1016/s0956-5663(00)00099-3. [DOI] [PubMed] [Google Scholar]
- 41.Duverger E, Frison N, Roche AC, Monsigny M. Biochimie. 2003;85:167–179. doi: 10.1016/s0300-9084(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 42.Witten KG, Rech C, Eckert T, Charrak S, Richtering W, Elling L, Simon U. Small. 2011;7:1954–1960. doi: 10.1002/smll.201100492. [DOI] [PubMed] [Google Scholar]
- 43.Murthy BN, Sinha S, Surolia A, Indi SS, Jayaraman N. Glycoconj. J. 2008;25:313–321. doi: 10.1007/s10719-007-9076-6. [DOI] [PubMed] [Google Scholar]
- 44.Yakovleva ME, Safina GR, Danielsson B. Anal. Chim. Acta. 2010;668:80–85. doi: 10.1016/j.aca.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Lecaroz MC, Blanco-Prieto MJ, Campanero MA, Salman H, Gamazo C. Antimicrob. Agents Chemother. 2007;51:1185–1190. doi: 10.1128/AAC.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arana I, Orruño M, Pérez-Pascual D, Seco C, Muela A, Barcina I. FEMS Microbiol. Ecol. 2007;62:1–11. doi: 10.1111/j.1574-6941.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- 47.Oliver JD. FEMS Microbiol. Rev. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 48.McDougald D, Rice SA, Weichart D, Kjelleberg S. FEMS Microbiol. Ecol. 1998;25:1–9. [Google Scholar]
- 49.Fode-Vaughan KA, Maki JS, Benson JA, Collins ML. P. Lett. Appl. Microb. 2003;37:239–243. doi: 10.1046/j.1472-765x.2003.01386.x. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Armisen T, Servais P. J. Microbiol. Meth. 2004;58:269–279. doi: 10.1016/j.mimet.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Rowan NJ. Trends Food Sci. Technol. 2004;15:462–467. [Google Scholar]
- 52.Gao W, Sattayasamitsathit S, Manesh KM, Weihs D, Wang J. J. Am. Chem. Soc. 2010;132:14403–14405. doi: 10.1021/ja1072349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.