Abstract
Multi-drug resistance protein (MRP) 4, an ATP-binding cassette (ABC) transporter, has broad substrate specificity. It facilitates the transport of bile salt conjugates, conjugated steroids, nucleoside analogs, eicosanoids, and cardiovascular drugs. Recent studies in liver carcinoma cells and hepatocytes showed that MRP4 expression is regulated by the aryl hydrocarbon receptor (AhR) and nuclear factor E2-related factor 2 (Nrf2). The AhR has particular importance in the lung and is most commonly associated with the up-regulation of cytochrome P-450 (CYP)-mediated metabolism of benzo[a]pyrene (B[a]P) to reactive intermediates. Treatment of H358, human bronchoalveolar, cells with 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) or (−)-benzo[a]pyrene-7,8-dihydro-7,8-diol (B[a]P-7,8-dihydrodiol), the proximate carcinogen of B[a]P, revealed that MRP4 expression was increased compared to control. This suggested that MRP4 expression might contribute to the paradoxical decrease in (+)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene-2′-deoxyguanosine ((+)-anti-trans-B[a]PDE-dGuo) DNA-adducts observed in TCDD-treated H358 cells. We have now found that decreased MRP4 expression induced by a short hairpin RNA (shRNA), or chemical inhibition with probenecid, increased (+)-anti-trans-B[a]PDE-dGuo formation in cells treated with (−)-B[a]P-7,8-dihydrodiol, but not the ultimate carcinogen (+)-anti-trans-B[a]PDE. Thus, up-regulation of MRP4 increased cellular efflux of (−)-B[a]P-7,8-dihydrodiol, which attenuated DNA-adduct formation. This is the first report identifying a specific MRP efflux transporter that decreases DNA damage arising from an environmental carcinogen.
Keywords: benzopyrene, environmental carcinogen, LC-MS, aryl hydrocarbon receptor
1. Introduction
Multi-drug resistance protein 4 (MRP4) is an ABC class C (ABCC4) transporter, which acts as a transmembrane efflux transporter (Ritter et al., 2005; Russel et al., 2008). In humans, there are some 48 ABC transporters that have been assigned to 7 sub-families (ABCA to ABCG), based on their sequence homology (Gradhand and Kim, 2008). MRP4 is typically known for the transport of nucleoside-analogues, particularly anti-virals, cyclic nucleotides such as cAMP, prostaglandins (PGs), bile salts, and conjugated steroids, such as estradiol 17-β-D-glucuronide (Zelcer et al., 2003; van Aubel et al., 2002; Reid et al., 2003). The MRP family is comprised of nine ABC transporters (ABCC1-9) of which MRP4 has the broadest substrate specificity (Kruh et al., 2007). However, a specific function for MRP4 has yet to be established (Russel et al., 2008). MRP4, MRP5, and MRP9 differ from the other MRPs as they possess only 12 transmembrane α-helices and lack the extra membrane-spanning domain at the N-terminus (Kruh et al., 2007; Gradhand and Kim, 2008). Many studies of MRP4 expression and substrate specificity have focused on its role in the liver and kidney. However, MRP4 is unique in its dual membrane localization and diverse tissue expression (Russel et al., 2008; Torky et al., 2005).
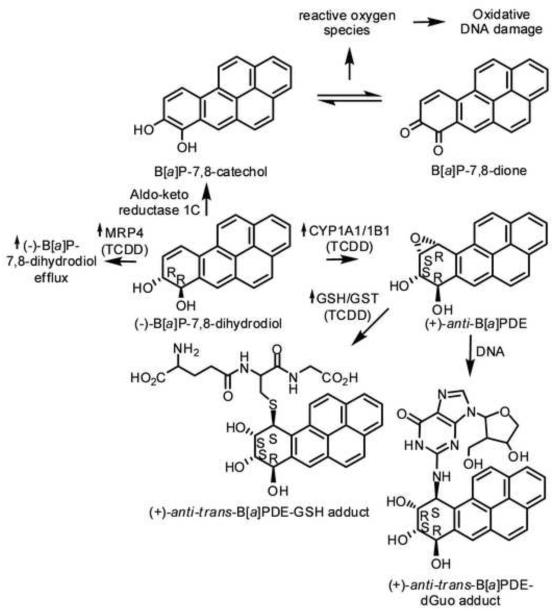
Recent studies have shown that the AhR and Nfr2 are key regulators of human MRP4 expression in human liver, HepG2 cells, and hepatocytes (Xu et al., 2010). Thus, MRP4 was induced by TCDD-treated HepG2 cells and primary hepatocytes obtained from two different individuals. TCDD is best known for its ability to bind the AhR, which translocates to the nucleus and heterodimerizes with the AhR nuclear transporter, and binds to an enhancer element that up-regulates the transcription of CYPs 1A/1B1 (Nebert and Karp, 2008). AhR-mediated activation of CYP1A/1B1 is critical for the activation of many xenobiotics during phase 1 metabolism in the liver and the lung. Numerous previous studies have shown that AhR is expressed at high levels in the lung (Chiba et al., 2011). We have also shown in H358, human lung bronchoalveolar cells that CYP1A1/1B1 are responsible for the metabolism of benzo[a]pyrene (B[a]P), to the ultimate carcinogen (+)-(B[a]PDE (Jiang et al., 2007). Detoxification of B[a]PDE occurs through glutathione (GSH) S-transferase (GST)-mediated adduct formation or further hydrolysis by epoxide hydrolase to the corresponding tetraols (Uno et al., 2004; Hukkanen et al., 2002; Zhang et al., 2006). B[a]PDE that escapes detoxification is able to enter to the nucleus and react with DNA to form adducts, particularly at the exocyclic amines of 2′-deoxyguanosine (dGuo) and 2′-deoxyadenosine (dAdo) (Fig. 1) (Ruan et al., 2007).
Fig. 1. Pathway of B[a]P Adduct Formation.
Metabolism of the proximate carcinogen (−)-B[a]P-7,8-dihydrodiol to the ultimate carcinogen (+)-anti-B[a]PDE by CYP1A1/1B1 and to B[a]P-7,8-catechol by aldo-keto reductases of the 1C family. (+)-anti-trans-B[a]PDE is able to enter the nucleus and form the DNA-adduct (+)-anti-trans-B[a]PDE-dGuo. Alternatively, (+)-anti-trans-B[a]PDE can be detoxified through GST-mediated GSH adduct formation. B[a]P-7,8-catechol can redox cycle to B[a]P-7,8-dione, which generates reactive oxygen species that can cause oxidative DNA damage.
Reports in the literature have linked exposure of B[a]P, a ubiquitous environmental polycyclic aromatic hydrocarbon (PAH), to DNA-adduct formation and to subsequent genetic alterations, which can promote the progression of multistage carcinogenesis. Much of this research was performed in liver cells or primary hepatocytes. Paradoxically, CYP1A1 knockout mice produce more B[a]P-derived DNA adducts when treated with B[a]P than wild-type mice. These data were interpreted to support a role for CYP1A1 in B[a]P detoxication rather than its activation (Uno et al., 2004). Interestingly we have shown that TCDD-pretreatment of human H358 lung cells also resulted in decreased B[a]P-derived DNA-adduct formation. However, this was due in part to TCDD-mediated increase in GST activity in the lung cells, which led to decreased DNA-adduct formation (Fig. 1) (Gelhaus et al., 2011). Despite intense research over the last two decades, a clear consensus of inter-individual differences in the risk for lung cancer through the CYP activation pathway of B[a]P has not emerged. Another possible mechanism for reduced DNA-adduct formation in TCDD pretreated cells might involve an increased export of B[a]P metabolites or B[a]PDE-GSH conjugates due to transporter induction by TCDD (Baird et al., 2005; Grimmer and Bohnke, 1975). Nevertheless, the exact mechanism, by which this could occur remains to be characterized (Peluso et al., 1998; Mastrangelo et al., 1996; Zmirou et al., 2000; Georgiadis and Kyrtopoulos, 1999; Bostrom et al., 2002; Pozzoli et al., 2004; Castano-Vinyals et al., 2004). Previous studies have shown that inhibition of ABCB1 (P-glycoprotein) and MRP1 (ABCC1) with verapamil, probenecid, or PSC833 increased B[a]PDE-DNA-adduct formation in MCF-7 cells (Myllynen et al., 2007). Unfortunately, the changes in MRP4 expression at the mRNA or protein level were not addressed in this study so it is not clear whether flavanoids only inhibited the transporter or whether they affected expression as well. There is also evidence from mouse models that there is cross-talk between the AhR and Nrf2 pathways (Yeager et al., 2009). These findings lead us to investigate whether MRP4 expression could be induced by TCDD (an AhR ligand) in lung cells and whether its inhibition would attenuate (+)-anti-trans-B[a]PDE-dGuo adduct formation.
2. Materials and Methods
2.1. Reagents
Caution: All PAHs are potentially hazardous and should be handled in accordance with NIH guidelines for the Use of Chemical Carcinogens. Cell culture medium and reagents were all obtained from Invitrogen Co. (Carlsbad, CA) except fetal bovine serum from (FBS) Hyclone (Logan, Utah). The (−)-B[a]P-7,8-dihydrodiol, (±)-anti-B[a]PDE, and TCDD were obtained from the NCI Chemical Carcinogen Standard Reference Repository (Midwest Research Institute, Kansas City, Missouri) and dGuo, dAdo, probenecid, and dimethylsulfoxide (DMSO) were purchased from Sigma Aldrich (St. Louis, MO). [15N5]-dGuo and [15N5]-dAdo were purchased from Cambridge Isotope Laboratories (Andover, MA). [15N5]-B[a]PDE-dGuo and [15N5]-B[a]PDE-dAdo were synthesized as previously described (Ruan et al., 2007). [2H9]-PGE2 and PGE2 were purchased from the Cayman Biochemical Company (Ann Arbor, MI). The purity and identity of all B[a]P metabolites was established by liquid chromatography-mass spectrometry (LC-MS) as previously described (Ruan et al., 2007; Jiang et al., 2007). Ammonium acetate and formic acid were HPLC grade and the methanol, hexanes, and isopropanol were Optima grade (Fisher Scientific, Waltham, MA). All water was purified through a MilliQ reverse osmosis system (Millipore, Billerica, MA).
2.2. Cell culture
Human bronchoalveolar, H358, cells were obtained from the American Type Culture Collection (ATCC #CRL-5807) and maintained in RPMI 1640 nutrient mixture with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and were passaged every 3 days at a 1:3 dilution. Cultured cells with passage number of 20 or less were used in the experiments to reduce variability during cell culture.
2.3. MRP4 shRNA knockdown
Five shRNA pLKO lentiviral vectors were purchased from Open Biosystems (Huntsville, AL) and will herein be referred to as 52164, 52165, 52166, 52167, and 52168. These five vectors along with pLKO expressing non-specific sequence vector were grown in 500 mL of LB with 3 mL of E. coli shaking overnight at 37°C in preparation for a DNA Maxi-prep. The DNA maxi-prep was performed using a Qiagen Plasmid Maxi kit (Valencia, CA) following the manufacturer’s instructions. Lentiviral vectors were transfected into 293T cells along with Rev, PMDL, and VSVG plasmids for viral production. After 8 h the media was changed and the virus was allowed to grow for 2 days. Media containing the virus was filtered through a 0.45 μm filter and aliquoted into 50 mL conical tubes. The virus was spun at 25,000 × g for 1.5 h, the media was decanted, and the virus was reconstituted in 200 mL of cold PBS with 0.05% BSA. It was aliquoted into tubes and stored at −80 °C until used. Initially, 52164 was chosen to transfect H358 cells under experimental conditions due to reduced MRP4 expression by Western analysis (data not shown). H358 cells were plated, 300,000 cells per well, into 4-12 well plates along with 10 μL of 52164 MRP4 shRNA virus in one plate and Scramble empty virus in another. On the second day, 3 wells were combined and transferred to one 10 cm plate to give 4 sets of 10 cm plates. Transfection of 52164 and knockdown of MRP4 in H358 shMRP4 cells was confirmed by Western blots as described in 2.4.
2.4. Immunohistochemistry
Briefly, protein was extracted from H358 or H358 shMRP4 cells using RIPA buffer and protease inhibitors. Protein was quantified using a Pierce BCA analysis kit (Rockford, IL). A gradient denaturing gel was run at 60 V for 2.5 hours (4-12% Bis-Tris) and protein was transferred for 2 h on ice in the cold room at constant amperage (200 mA) onto a PVDF membrane. Chemiluminescent detection was conducted using an primary MRP4 mAb (Abcam, Cambridge, MA) and ECL Amersham rabbit anti-mouse HRP-linked secondary (GE Healthcare Life Sciences, Piscataway, NJ). MCM3, a mini-chromosome maintenance protein, with a molecular weight of 100 kDA, was used as the loading control. The primary antibody for MCM3 was a polyclonal goat (Bethyl Laboratories, Montgomery, TX) and the secondary used was a donkey anti-goat HRP-linked (Santa Cruz Biotechnology, Santa Cruz, CA). Western blots were performed in triplicate, but only one representative experiment is shown in the figures.
2.5. Reverse-transcriptase polymerase chain reaction (RT-PCR)
Cell plates were treated with DMSO (vehicle control), 10 nM TCDD (48 h) or 2 μM (−)-B[a]P-7,8-dihydrodiol (24 h). RNA was extracted using the Ambion RNA Aqueous kit (Foster City, CA) following the manufacturer’s protocol and stored at −80°C until cDNA preparation. RNA was quantified by UV and cDNA was isolated from the four most abundant samples from each category using the Applied Biosystems High Capacity cDNA Reverse Transcription kit (Carlsbad, CA). For RT-PCR, the Applied Biosystems Step One instrument was used with the Taqman Master Mix and Applied Biosystems Gene Expression Assays for MRP4 and GAPDH (endogenous control) following the manufacturer’s instructions. In addition to the samples (n = 3 for each treatment), three no template controls containing all of the reagents except the cDNA were also run as a negative control. Relative quantitation of the genes of interest was performed by the ΔΔCt method.
2.6. TCDD and B[a]P metabolite treatments
H358, H358 shMRP4, or H358 Scramble (empty vector) cells were treated with 0.3% DMSO (control vehicle), 10 nM TCDD (48 h), 2 μM (−)-B[a]P-7,8-diol (24 h), or pretreated with 10 nM TCDD (24 h) before 2 μM (−)-B[a]P-7,8-dihydrodiol (24 h) in 10 mL of the culture medium. Additionally, H358, H358 shMRP4, or H358 Scramble were treated with 2 μM (±)-B[a]PDE for 24 h with or without 10 nM TCDD pretreatment (24 h). Cells were washed with PBS and scraped for immunohistochemistry or DNA-adduct analysis. Experiments were conducted in triplicate.
2.7. Probenecid inhibition
H358 cells (in triplicate) were pretreated with 1 mM probenecid for 24 h before treatment with 2 μM (−)-B[a]P-7,8-diol. After 24 h, H358 cells were washed with PBS and then scraped for DNA extraction and hydrolysis as described below. (+)-anti-trans-B[a]PDE-dGuo was quantified by stable isotope dilution LC-selected reaction monitoring (SRM)/MS and normalized to total DNA bases as described in 2.10.
2.8. PGE2 analysis
H358 cells and H358 shMRP4 cells were treated in triplicate with 10 μM arachidonic acid (AA) for 4 h. Additionally, a second set of H358 cells were pretreated with 1 mM probenecid for 24 h before 10 μM AA treatment for 4 h. PGE2 was quantified using a stable isotope dilution targeted lipidomics method previously developed in our lab (Lee and Blair, 2007). Briefly, lipids were extracted from 3 mL of media using 5 mL of diethyl ether. The ether extraction was dried under nitrogen and the electron capture reagent, pentafluorobenzyl bromide, was used to derivatize the lipids. After derivatization, samples were dried under nitrogen and reconstituted in 100 μL of hexanes/ethanol (95:5; v/v) and 10 μL was injected onto a Waters 2690 Separation module (Milford, MA) coupled to a Thermo Fisher TSQ Quantum Ultra (West Palm Beach, FL). Analysis of PGE2 was performed in negative ion mode using electron capture atmospheric pressure chemical ionization (ECAPCI) following a chiral normal phase separation (Lee and Blair, 2007).
2.9. DNA extraction and hydrolysis
Total DNA from H358 cells was extracted using DNAzol® (Invitrogen, Carlsbad, CA), by following the manufacturers protocol. Extracted DNA was stored at 4°C, overnight in 75% ethanol. Samples were spun at 8000 rpm for 3 min and the 75% ethanol was decanted leaving the DNA pellet. The pellet was then brought up in 500 μL of 10 mM Tris/100 mM MgCl2 pH 7.4 to which 10 μL of DNAse 1 (Calbiochem, Gibbstown, NJ) was added. This was incubated in a water bath at 37 °C for 1.5 h. Following the incubation, the pH was adjusted to pH 9 with 75 μL of 0.2 M glycine buffer, along with 10 μL of phosphodiesterase (Worthington Biochemical Co., Lakewood, NJ) and this was incubated for 2 h at 37 °C. For the last step in DNA hydrolysis, 75 μL of shrimp alkaline phosphatase (SAP) buffer and 75 μL of 50 mM Tris-HCl (pH 7.4) was added with 15 μL of SAP (Hoffman La-Roche, Nutley, NJ) and this was incubated for another 2 h at 37 °C. Before evaporation, 12.5 ng of [15N5]-B[a]PDE-dGuo internal standard was added to each sample. Samples were evaporated by Speedvac and reconstituted in 200 μL methanol/H2O (1:1 v/v) and filtered through Costar Spin-X nylon filters (Corning, Corning, NY) for analysis by LC-MS and LC-UV.
2.10. LC-electrospray ionization (ESI)/SRM/MS
LC-MS analyses of (+)-anti-trans-B[a]PDE-dGuo adducts were conducted using an Agilent 1100 HPLC system (Agilent Technology, Palo Alto, CA) equipped with a CTC autosampler (Leap Technology, Carrboro, NC) and an MDS-Sciex API-4000 triple-quadrupole mass spectrometer (Applied Biosystems, Foster City, CA). The HPLC method employed a YMC J’sphere M80 column (2.0 mm I.D. × 150 mm, 4 μm) using 5 mM NH4OAc buffer containing 0.02% formic acid as mobile phase A and methanol as mobile phase B at a flow rate of 200 μL/min. The gradient started from 49% B to 51% B in 20 min, followed by 51% B to 65% B in 10 min, and continued to 100%B in 5 min. The following mass spectrometer parameters were set: collision gas 10 units, curtain gas 30 units, ion source gas #1 30 units, ion source gas #2 30 units, ion spray voltage 5.0 kV, ionization temperature 500 °C, decluster potential 50 V, entrance potential 8 V, collision energy 40 eV, collision cell exit potential 22 V. MRM analyses were conducted in positive ESI mode. The following transitions were monitored for (+)-anti-trans-B[a]PDE-dGuo: m/z 570.2 (MH+, (+)-anti-trans-B[a]PDE-dGuo) → m/z 257.1 (MH+-dGuo-H2O-CO) and m/z 575.2 ([15 N5]-B[a]PDE-dGuo) → m/z 257.1 (MH+-[15N5]-dGuo-H2O-CO).
2.11. Quantification of DNA-adducts
Calibration curves of the DNA-adducts were prepared for the four stereoisomers of authentic B[a]PDE-N2-dGuo to make standard mixtures of different concentrations (0.1, 0.2, 0.5, 1.0, 2.5, 5.0, and 10.0 ng of each). The corresponding B[a]PDE-[15N5]-N2-dGuo stereoisomers (10 ng of each) were then added to the standards and to the experimental samples. Standard solutions underwent the same sample preparation and analytical procedures as the experimental samples as described above. Calibration curves were calculated with a linear regression analysis of the peak area ratios of standard versus the internal standard. Adduct levels were calculated by interpolation from the calibration curve and normalized with the total base content in each sample. Levels are reported as number of (+)-anti-trans-B[a]PDE-dGuo adducts per 106 normal bases.
2.12. DNA base analysis
LC-UV chromatography for DNA base analysis was conducted on a Hitachi Elite Lachrom HPLC system equipped with a UV detector. The separation employed a Phenomenex Luna C18(2) column (4.6 mm I.D. × 150 mm, 5 μm). Solvent A was aqueous 5mM NH4OAc with 0.02% formic acid and solvent B was methanol. The flow rate was 0.9 mL/min and DNA bases were separated under isocratic conditions, 15% B for 20 min. The UV detector monitored the absorbance at 260 nm. For calibration curves, solutions containing certain concentrations of authentic DNA bases (0.0, 2.0, 5.0, 10.0, 20.0, and 50.0 μg/mL) were subjected to HPLC analysis. The injection volume was 20 μL. Calibration curves were calculated with a linear regression analysis of the peak areas of authentic standards. A portion of the digested sample was subjected to the same HPLC procedure for base analysis. DNA base levels were calculated by interpolation from the calibration curve.
2.13. Statistics
Changes in mRNA expression and PGE2 secretion were subjected to a one way analysis of variance (ANOVA) with Tukey’s post hoc test. For the studies on the effects of chemicals and shMRP4 on (+)-anti-trans-B[a]PDE-dGuo formation in H358 cells, a two-way ANOVA model was used to analyze the data. The interaction of two factors was first examined and then the main effects. No interaction effects were found. The sizes of the effects were estimated using linear contrasts after fitting multiple linear regression models and tested using Wald tests. A two tailed t-test was employed to assess the effect of scrambled shMRP4 on DNA-adduct formation. p-values of <0.05 were considered to be statistically significant. Adduct levels and PGE2 concentrations are given as values ± standard deviation.
3. Results
3.1. MRP4 expression in H358 cells
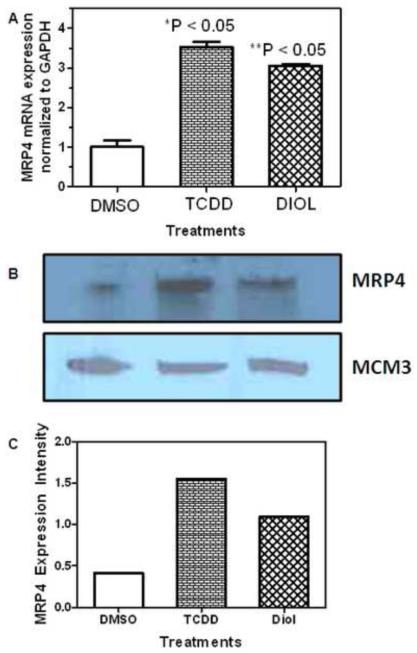
H358 cells were treated with DMSO vehicle, 10 nM TCDD for 48 h or 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h. RT-PCR was performed to determine the relative expression of MRP4 after treatment using the ΔΔCt relative quantitation method. The relative mRNA expression for H358 cells with the 10 nM TCDD treatment resulted in the greatest fold change (3.53) when compared with the DMSO (0.3%) treated cells and the 2 μM (−)-B[a]P-7,8-dihydrodiol treated cells (Fig. 2A). MRP4 expression was also monitored at the protein level. Western blot analysis revealed the same pattern as was observed with mRNA (Fig. 2B). Thus, TCDD treatment (Lane 2) resulted in the highest protein expression when compared with DMSO control (Lane 1) and (−)-B[a]P-7,8-dihydrodiol (Lane 3) treatments (Fig. 2B). The relative protein intensity was normalized to MCM3 (loading control) and plotted as protein intensity compared with treatment in Fig. 2C.
Fig. 2. MRP4 Expression in H358 Cells.
(A) MRP4 mRNA expression. GAPDH was used as a control for the RT-PCR and relative mRNA expression was determined using the ΔΔCt method. The error bars show standard deviations. (*) indicates p <0.05 compared to control and (**) indicates p < 0.05 compared to control; n=3 for each treatment. (B) MRP4 protein expression determined by Western blot for one representative analysis. MCM3 was used as a loading control. (C) Quantitative analysis of image intensity of Western blots in Figure 2B. Analyses were conducted in triplicate using H358 WT cells, H358 cells treated with 10 nM TCDD for 48 h, or H358 cells treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h.
3.2 Inhibition of B[a]P-7,8-dihydrodiol-derived DNA-adducts in H358 cells
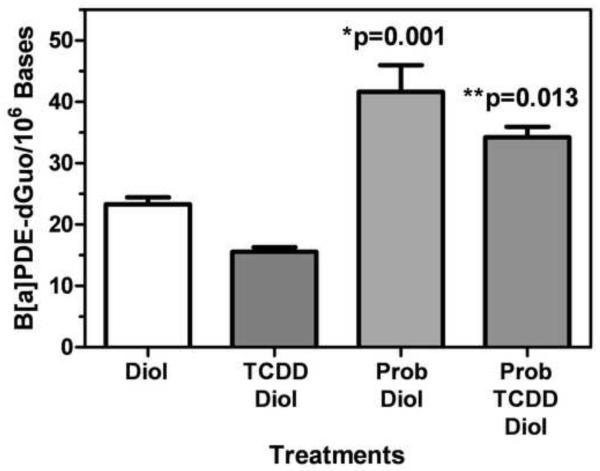
The cells were pre-treated with either 1 mM probenecid and/or 10 nM TCDD for 24 h before treatment with 2 μM (−)-B[a]P-7,8-dihydrodiol (24 h) or with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h without pretreatment. DNA was extracted and hydrolyzed for DNA-adduct analysis by LC-SRM/MS and base analysis by LC-UV. The (+)-anti-trans-B[a]PDE-dGuo adducts were normalized to total DNA and are reported as DNA-adducts per 106 bases. TCDD treatment was associated with a decrease in (+)-anti-trans-B[a]PDE-dGuo by 7.6 adducts/106 bases (95% CI: −12.779, −2.401) that did not reach statistical significance (p=0.055) when compared to control B[a]P-7,8-dihydrodiol treatment alone (Fig. 3). Probenecid treatment resulted a significant increase in (+)-anti-trans-B[a]PDE-dGuo to 41.65 ± 7.52 adducts/106 bases (95% CI: 10.43, 26.29, p=0.001) compared to control B[a]P-7,8-dihydrodiol treatment alone (23.30 ± 2.00 adducts/106 bases) (Fig. 3). TCDD attenuated the effect of probenecid to 34.22 ± 2.96 adducts/106 bases but the combined effects of TCDD and probenecid were still significantly higher than treatment with B[a]P-7,8-dihydrodiol treatment alone (95 % 2.99, 18.85, p=0.013).
Fig. 3. Measurement of (+)-anti-trans-B[a]PDE-dGuo in H358 cells after chemical inhibition.
H358 cells were treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h with or without 1 mM probenecid for 24 h and/or 10 nM TCDD for 24 h. (+)-anti-trans-B[a]PDE-dGuo adducts were measured by LC-SRM/MS and normalized by total DNA. Plots show means and standard deviation, (*) indicates p=0.001 compared to control and (**) indicates p=0.013 compared to control; n = 3 for each treatment.
3.3. Effect of MRP4 inhibition on B[a]P-7,8-dihydrodiol-derived DNA-adducts
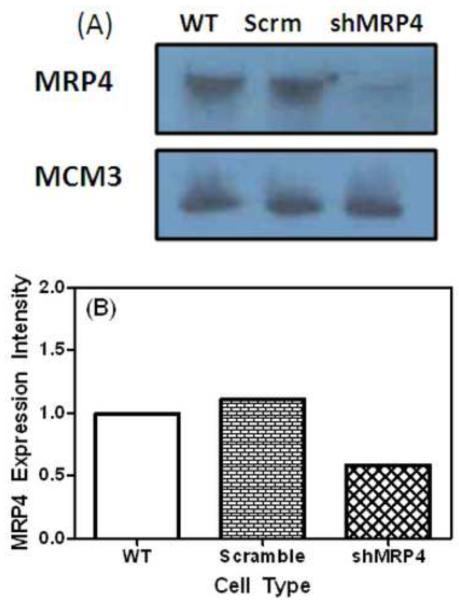
In order to show that the increase in (+)-anti-trans-B[a]PDE-dGuo adducts was a result of the specific inhibition of MRP4, a lentiviral introduction of shRNAs targeting MRP4 (52164) approach was used. shMRP4 (pLKO vector) and other shRNA-expressing lentiviruses were produced and titered as described (Gilad et al., 2010). For shRNA lentivirus infections, 5X105 cells were plated on 6 well plates and infected with concentrated virus at a multiplicity of infection of 10 to 20, typically yielding 95% to 98% transduction. A Western blot analysis for the knockdown of MRP4 using the lentiviral vector was conducted (Fig. 4A) and the amount of MRP4 knockdown calculated (Fig. 4B). Protein expression was monitored for wild type H358 (Lane 1), H358 containing the scramble vector (Lane 2) and H358 cells containing the shMRP4 vector (Lane 3). MCM3 was used as the loading control (Fig. 4A).
Fig. 4. MRP4 KD Western blot.
(A) Expression of MRP4 in H358 WT cells (Lane 1), H358 cells transduced with scramble vector (Lane 2) and H358 cells transduced with shMRP4 vector (Lane 3). MCM3 was used as a loading control. (B) The relative quantitation of protein expression.
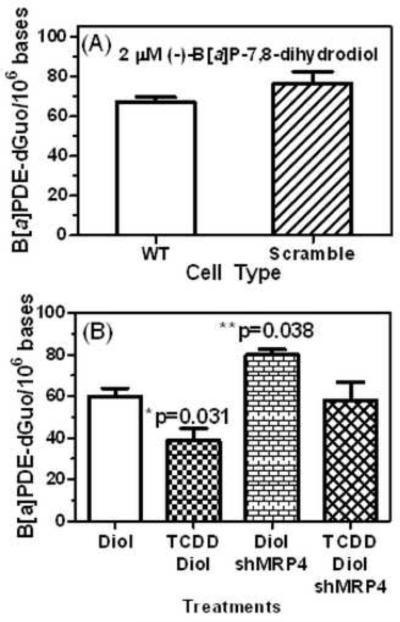
H358 wild type, scramble, and cells containing shMRP4 were treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h with or without 10 nM TCDD pretreatment for 24 h. There was no difference in the level of (+)-anti-trans-B[a]PDE-dGuo adducts formed in H358 WT cells (67.1 ± 4.0 adducts/106 cells) or H358 scramble (empty vector) cells (76.4 ± 10.4 adducts/106 cells, p = 0.2245) treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h (Fig. 5A). The increase in basal production of DNA-adducts in the control wild type cells (Fig. 5A) compared with the control cells used in the probenecid study (Fig. 3) was ascribed to cells having undergone different numbers of passages. In keeping with our previous data (Gelhaus et al., 2011), B[a]P-7,8-dihydroidiol and TCDD treatment was associated with a significant decrease in (+)-anti-trans-B[a]PDE-dGuo by 21.17 adducts/106 bases (95% CI: −39.82, −2.52, p=0.031) compared to control B[a]P-7,8-dihydrodiol treatment alone (Fig. 5B). Conversely, B[a]P-7,8-dihydrodiol and shMRP4 treatment was associated with a significant increase in (+)-anti-trans- of 20.11 adducts/106 bases (95% CI: .46, 38.76, p=0.038) compared to control B[a]P-7,8-dihydrodiol treatment alone (Fig. 5B). This is similar to the effect observed with chemical inhibition of MRP4 by probenecid (Fig. 3). Pretreatment of the H358 shMRP4 cells with TCDD attenuated the formation of (+)-anti-trans-B[a]PDE-dGuo to 58.26 ± 14.80 adducts/106 cells (Fig. 5B) so that the combined effects of shMRP4 cells with TCDD were no longer significantly different from control B[a]P-7,8-dihydrodiol treatment alone (p=0.841).
Fig. 5. Measurement of (+)-anti-trans-B[a]PDE-dGuo in H358 cells after shMRP4.
(A) There was no difference in the level of DNA-adduct formation in H358 WT cells and H358 cells transduced with the scramble vector. (B) The levels of DNA-adducts formed after H358 cells were treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h with or without 10 nM TCDD for 24 h. (+)-anti-trans-B[a]PDE-dGuo adducts were quantified by LC-SRM/MS and normalized by total DNA. Plots show means and standard deviations, (*) indicates p=0.031 and (**) indicates p=0.38 compared with control; n = 3 for each treatment.
3.4 Effect of MRP4 inhibition on B[a]PDE-derived DNA-adducts
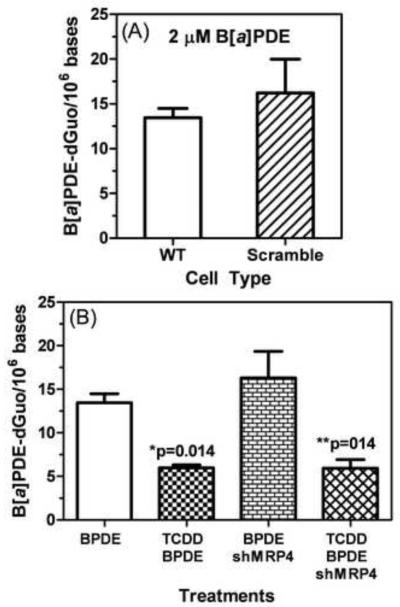
H358, H358 scramble, and H358 shMRP4 were treated with 2 μM (±)-B[a]PDE for 24 h with or without 10 nM TCDD for 24 h. The levels of (+)-anti-trans-B[a]PDE-dGuo adducts in H358 wild type (13.46 ± 1.80 adducts/106 cells) and H358 scramble cells (16.21 ± 6.53 adducts/106 cells, p=0.596) after treatment with 2 μM (±)-anti-trans-B[a]PDE for 24 h were similar (Fig. 6A). Knock down of MRP4 with the shRNA resulted in no significant increase in (+)-anti-trans-B[a]PDE-dGuo formation when cells were treated with 2 μM (±)-anti-trans B[a]PDE for 24 h (Fig. 6B, p=0.270). This contrasts with the significant increase in DNA-adducts that were observed when cells were treated with the B[a]PDE metabolic pre-cursor, (−)-B[a]P-7,8-dihydrodiol (Fig. 5B). However, anti-trans-B[a]PDE and TCDD treatment was associated with a significant decrease in (+)-anti-trans-B[a]PDE-dGuo by 7.5 adducts/106 bases (95% CI: −13.04, −1.97, p=0.014) compared to control anti-trans-B[a]PDE treatment alone (Fig. 6B). Similarly, treatment with B[a]PDE, TCDD, and shMRP4 was also associated with a significant decrease in (+)-anti-trans-B[a]PDE-dGuo of 7.5 adducts/106 bases (95% CI: −13.05, −1.97, p=014) compared to control B[a]PDE treatment alone (Fig. 6B).
Fig. 6. Measurement of (+)-anti-trans-B[a]PDE-dGuo in H358 cells after shMRP4 with (±)-B[a]PDE treatment.
(A) H358 WT and H358 cells transduced with scramble vector were treated with 2 μM (±)-anti-trans-B[a]PDE. There was no difference in DNA-adduct formation. (B) H358 WT and H358 cells transduced with shMRP4 were also treated with 2 μM (±)-B[a]PDE with and without 10 nM TCDD pretreatment for 24 h. (+)-anti-trans-B[a]PDE-dGuo adducts were quantified by LC-SRM/MS and normalized by total DNA. Plots show means and standard deviations, (*) indicates p=0.014 compared to control and (**) indicates p=0.14 compared to control; n = 3 for each treatment.
3.5. Effect of MRP4 inhibition on PGE2 efflux
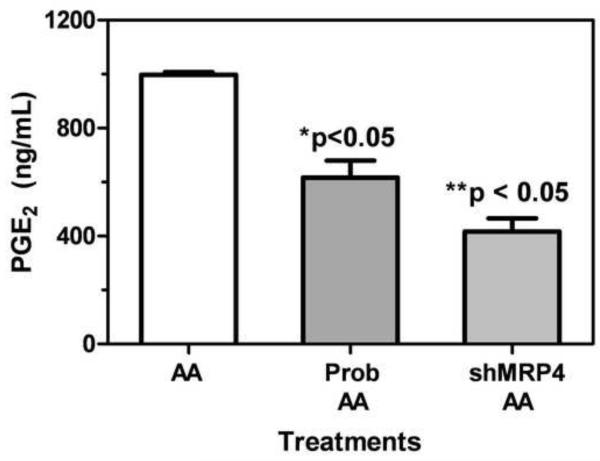
Previous studies have demonstrated that MRP4 is able to control the cellular efflux of PGE2 (Reid et al., 2003; Lin et al., 2008). In order to further confirm that MRP4 activity had been modulated in our model system, H358 wild type and H358 shMRP4 cells were treated with 10 μM AA for 4 h. Additionally, H358 wild type cells were pretreated with 1 mM probenecid for 24 h before 10 μM AA treatment for 4 h. PGE2 generated by cyclooxygenase mediated AA metabolism was then quantified using a stable isotope dilution ECAPCI-SRM/MS (Fig. 7). Results show that levels of PGE2 (ng/mL) found in the media were significantly less in H358 cells pretreated with 1 mM probenecid for 24 h (616.9 ± 107.7 ng/mL, p=9.3 × 10−7) compared to H358 cells only treated with AA (997.3 ± 17.1 ng/mL). Treatment of H358 shMRP4 cells with 10 μM AA for 4 h also resulted in a decrease in PGE2 found in the media (416.5 ± 84.3 ng/mL, p=2.4 × 10−6) compared to H358 wild type cells that had undergone the same treatment.
Fig. 7. PGE2 transport.
H358 WT cells treated with 1 mM probenecid for 24 h, and H358 cells transduced with shMRP4 were treated with 10 μM AA for 4 h. After AA treatment for 4 h, 3 mL of media was taken and lipids were extracted for PGE2 analysis by chiral normal phase LC-ECAPCI/SRM/MS as described previously (Lee and Blair, 2007). (*) indicates p<0.05 compared to control and (**) indicates p<0.05 compared to control; n=3.
4. Discussion
We showed previously that levels of (+)-anti-trans-B[a]PDE-dGuo in wild type H358 cells decreased with TCDD pretreatment (Ruan et al., 2007). These paradoxical findings are in agreement with the observation that treatment of CYP1A1 knockout mice with B[a]P results in increased B[a]P-adduct formation. One explanation is that CYP1A1 plays a more important role in B[a]P detoxication rather than its activation. These previous experiments were conducted either in liver cells or liver tissue and it was assumed that the same mechanism for decreased adduct formation would apply to all cell-types and organs. In our microarray studies with lung cells, we have shown that pre-treatment with TCDD results in up-regulation of CYP1A1 and 1B1 mRNA by 80- and 12-fold, respectively, when compared with (−)-B[a]P-7,8-dihydrodiol treatment alone (Gelhaus et al., 2011). These results were confirmed with RT-PCR using the same treatment conditions of H358 cells as described in this paper (data not shown). Surprisingly, it was shown that the decrease in (+)-anti-trans-B[a]PDE-dGuo levels in H358 lung cells pretreated with TCDD resulted in part from TCDD induction of phase 2 metabolism through activation of a highly stereoselective GST in the lung cells (Fig. 1). This in turn led to higher B[a]PDE-GSH adducts.
It has been demonstrated that in murine models, as well as in human cell lines, there is crosstalk between AhR and Nrf2 pathways (Yeager et al., 2009; Miao et al., 2005; Niestroy et al., 2011). In Caco-2, human colon carcinoma, cells, it was suggested that the AhR might be upstream in the signaling cascade and that its activation was necessary for Nrf2 activation. A similar situation could occur in the TCDD-treated H358 cells in which there is an increase in (±)-B[a]PDE-GSH adducts compared to H358 cells treated with (−)-B[a]P-7,8-dihydrodiol alone (Niestroy et al., 2011; Gelhaus et al., 2011). MRP4 was identified as part of the AhR-Nrf2 gene battery in addition to many of the typical genes involved in drug metabolism. Thus, TCDD treatment of human hepatocarcinoma, HepG2, cells and primary hepatocytes resulted in induced MRP4 expression at mRNA and protein levels (Xu et al., 2010). Therefore, it seemed feasible that the TCDD-mediated decrease of (+)-anti-trans-B[a]PDE-dGuo adducts could have arisen from a combination of increased CYP1A1/1B1 metabolism to B[a]PDE coupled with increased B[a]PDE detoxification (Gelhaus et al., 2011) and increased cellular efflux of B[a]PDE or its metabolic pre-cursor through up-regulation of MRP4 expression.
RT-PCR and Western blot analyses of H358 cells with DMSO (vehicle), 10 nM TCDD for 48 h, or 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h revealed an increase in MRP4 at the mRNA and protein levels for both TCDD and (−)-B[a]P-7,8-dihydrodiol treated cells (Fig. 2). TCDD treatment resulted in the largest increase in MRP4 expression (Fig. 2). When H358 cells were pretreated with 1 mM probenecid for 24 h to inhibit MRP4, (+)-anti-trans-B[a]PDE-dGuo DNA-adduct levels significantly increased compared to H358 cells treated with 2 μM (−)-B[a]P-7,8-dihydrodiol alone (Fig. 3). Furthermore, pretreatment with 10 nM TCDD, simultaneously with 1 mM probenecid attenuated this affect and DNA-adduct levels decreased; however, they were still significantly higher than H358 cells treated for 24 h with 2 μM (−)-B[a]P-7,8-dihydrodiol alone or a combination of TCDD and 2 μM (−)-B[a]P-7,8-dihydrodiol (Fig. 3), adding further credence to the suggestion that MRP4 plays an important role in controlling B[a]PDE-mediated DNA-adduct formation. These findings are also consistent with the concept that TCDD can inhibit B[a]PDE-mediated DNA-adduct formation by two separate pathways in human lung cells and that they counteract the increased adduct formation that would arise through increased CYP1A1/1B1-mediated B[a]PDE formation. First, the previously characterized increase in B[a]PDE GSH-adduct formation would detoxify B[a]PDE and prevent DNA-adduct formation (Fig. 2). Second, TCDD-induced MRP4 expression would cause increased efflux of (−)-B[a]P-7,8-dihydrodiol, decrease its conversion to B[a]PDE and so further decrease DNA-adduct formation. Interestingly, treatment of the H358 cells with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h induced an increase in MRP4 expression (Fig. 2) without a concomitant increase in the formation of B[a]PDE-GSH adducts (data not shown). Inhibition of the MRP4 with probenecid resulted in a significant increase in B[a]PDE-dependent DNA-adduct formation (Fig. 3). Pre-treatment with TCDD prior to probenecid, slightly attenuated the increase in DNA-adduct formation, but it did not reach statistical significance. Therefore, MRP4 rather than GSH-adduct formation appeared to be primarily responsible for controlling DNA-adduct formation in the (−)-B[a]P-7,8-dihydrodiol-treated lung cells.
MRP4 mRNA is expressed most abundantly in humans in prostate glandular epithelial cells with a more moderate abundance in other tissues including lung (Lee et al., 1998; Lee et al., 2000). MRP protein is expressed to some extent in normal lung tissue, but its expression is markedly up-regulated in certain diseases such as pulmonary hypertension (Hara et al., 2011) and cancer (Norris et al., 2005; Ho et al., 2008). Previous studies have established that there are high levels of expression in human lung adenocarcinoma A549 cells (Torky et al., 2005). We have now shown that MRP4 also expressed in significant amounts in human bronchoalveolar carcinoma H358 cells (Fig. 2). Therefore, the H358 cells provided a useful model system to examine the effects of MRP4 in modulating B[a]P-dependent DNA-adduct formation in human lung cells. However, the potential role of this pathway to modulate DNA-adduct formation will be limited to situations where MRP4 protein expression is up-regulated.
In order to specifically examine the role of MRP4 in B[a]PDE-mediated DNA-adduct formation, a lentiviral knockdown was constructed. A Western blot analysis confirmed that H358 cells transduced with scramble vector did not alter the expression of MRP4; whereas the knockdown using shMRP4 resulted in a 41% decrease in MRP4 expression (Fig. 4). To confirm that H358 cells transduced with the scramble vector were metabolically the same as H358 control cells, both were treated with 2 μM (−)-B[a]P-7,8-dihydrodiol for 24 h and (+)-anti-trans-B[a]PDE-dGuo adducts were quantified. There was no statistically significant difference in DNA-adduct formation in control H358 cells when compared to the cells transduced with scramble vector (Fig. 5A). In contrast, knockdown of MRP4 using shRNA technology resulted in a significant increase in (+)-anti-trans-B[a]PDE-dGuo adducts (Fig. 5B) much like the chemical inhibition of MRP4 with probenecid (Fig. 3). Pretreatment of shMRP4 cells with 10 nM TCDD for 24 h decreased the level of (+)-anti-trans-B[a]PDE-dGuo adducts to levels observed with B[a]P-7,8-dihydrodiol alone (Fig. 5B). This provides additional evidence to suggest that both MRP4-dependent efflux and GST-mediated detoxication are important for preventing the DNA-adduct formation, which arises from further metabolism of the proximate carcinogen, (−)-B[a]P-7,8-dihydiol (Fig. 1).
In order to identify the substrate for MRP4 efflux, H358 cells were also treated with (±)-anti-trans-B[a]PDE for 24 h with or without 10 nM TCDD pretreatment in wild type and shMRP4 cells (Fig. 6). There was no difference in B[a]PDE-dependent DNA-adduct formation in wild type cells compared to those transduced with the scramble lentiviral vector (Fig. 6A). As with (−)-B[a]P-7,8-dihydrodiol treatment of H358 cells pretreated with 10 nM TCDD for 24 h, there was a significant decrease in (+)-anti-trans-B[a]PDE-dGuo adducts compared with control untreated cells (Fig. 6B). However, unlike (−)-B[a]P-7,8-dihydrodiol-treated shMRP4 cells (Fig. 5B) there was no increase in the (+)-anti-trans-B[a]PDE-dGuo adducts in shMRP4 cells treated with (±)-anti-trans-B[a]PDE (Fig. 6B). Furthermore, pretreatment of shMRP4 cells with 10 nM TCDD resulted in attenuation of the DNA-adducts to levels that were significantly lower than those observed with (±)-anti-trans-B[a]PDE alone (Fig. 6B). This contrasts markedly with TCDD pretreatment prior to shMRP4/B[a]P-7,8-dihydrodiol where there was no significant difference in DNA-adduct levels compared to treatment with B[a]P-7,8-dihydrodiol alone (Fig. 5B). Therefore, inhibition of MRP4 had little effect on DNA-adduct formation when cells were treated with (±)-anti-trans-B[a]PDE. These results revealed that (+)-anti-trans-B[a]PDE is a poor MRP4 substrate and that TCDD-mediated inhibition of DNA-adduct formation occurs primarily through activation of GST-mediated detoxication of (+)-anti-trans-B[a]PDE (Fig. 1).
MRP4 is widely known for its ability to act as an efflux pump and transport nucleoside analogues, including drugs used against HIV as well as cAMP and cGMP (van Aubel et al., 2002). Torky et al. detected MRP4 as a membrane protein in A549 cells, another lung carcinoma cell line (Torky et al., 2005). Other groups have shown that MRP4 is able to control the transport of PGE1 and PGE2, steroid conjugates, and folate (Reid et al., 2003; Torky et al., 2005; Zelcer et al., 2003; Lemos et al., 2009). To further confirm that H358 lung cells expressed functional MRP4, we examined PGE2 efflux into the media. We found that PGE2 efflux occurred when H358 cells were treated with 10 μM AA (Fig. 7). Inhibition of MRP4 with 1 mM probenecid for 24 h or shMRP4 resulted in a significant decrease in PGE2 efflux (Fig. 7).
While many substrates have been suggested, MRP4 function has not been fully characterized (Russel et al., 2008). The finding that MRP4 can transport (−)-B[a]P-7,8- dihydrodiol is in keeping with previous studies that have identified MRP4 as a transporter for structurally-related flavanoids and steroids (Wu et al., 2005; Zelcer et al., 2003). We have now clearly shown that MRP4 is responsible for the cellular efflux of the proximal B[a]P carcinogen, B[a]P-7,8-dihydrodiol, preventing its CYP-dependent conversion to the ultimate carcinogen anti-B[a]PDE (Fig. 1) and its aldo-keto reductase-mediated conversion to B[a]P-7,8-catechol (Park et al., 2008).
Our study further highlights the balance between carcinogen activation, detoxification, and transport. It is now clear that TCDD-mediated activation of GST in human lung cells attenuates DNA-adduct formation, which results from the ultimate carcinogen, B[a]PDE (Gelhaus et al., 2011). On the other hand, TCDD-mediated up-regulation of MRP4 increases cellular efflux of the proximate carcinogen (−)-B[a]P-7,8-dihydrodiol, which decreases formation of the ultimate carcinogen, and attenuates DNA-adduct formation. MRP4 offers a novel potential therapeutic target because up-regulation of its expression would inhibit B[a]P-mediated DNA-adduct formation as well as the oxidative DNA damage that arises from redox cycling of B[a]P-7,8-catechol to B[a]P-dione (Fig. 1) (Park et al., 2008; Mangal et al., 2009). This is the first report showing that inhibition of a specific MRP efflux transporter can increase the DNA damage in lung cells that arises from CYP-mediated activation of an environmental carcinogen. It was reported previously that probenecid increased B[a]P-mediated DNA damage in breast cancer cells although the precise mechanism was not explored in detail (Myllynen et al., 2007). In addition, probenecid is known to inhibit organic anion transporters of the Scl22a family (Sweet, 2005). Consequently, probenecid was found to cause a modest reduction in 1-hydroxymethylpyrene-mediated DNA-adduct formation in the kidneys of rats with a concomitant increase in DNA-adduct formation in the lungs (Monien et al., 2009).
In earlier studies, both MRP1 and MRP2 were found to be involved in the efflux of B[a]PDE-GSH-adducts (Srivastava et al., 2002). Paradoxically, MRP inhibitors are being developed as therapeutic agents to prevent the efflux of anti-cancer agents and help overcome acquired multi-drug resistance in tumors (Perez-Tomas, 2006; Lee, 2010). Therefore, it will be important to determine whether the new drugs that emerge from these development programs are MRP4 inhibitors so that their potential for causing CYP-mediated increases in DNA damage can be monitored.
Highlights.
H358 expression of MRP4 increased with TCDD and (−)-B[a]P-7,8-dihydrodiol treatment
(−)-B[a]P-7,8-dihydrodiol-derived DNA-adduct levels increase with probenecid inhibition
(−)-B[a]P-7,8-dihydrodiol-derived DNA-adduct levels increase with shMRP4 inhibition
B[a]PDE-derived DNA-adduct levels did not increase in MRP4 inhibited cells
MRP4 decreases DNA damage arising from the environmental carcinogen B[a]P
Acknowledgments
We would like to acknowledge A. Clementina Mesaros and Sankha Basu for their careful reading of this manuscript and Eric Brown for assistance with lentiviral vector production. This project was supported by the National Institutes of Health National Institute of Environmental Health Sciences [5F32ES016683, P30ES013508, R01ES015857] and the National Cancer Institute [R01CA130038, R25CA101871].
Abbreviations
- AhR
aryl hydrocarbon receptor
- ARE
anti-oxidant response element
- B[a]P
benzo[a]pyrene
- (±)-anti-B[a]PDE, (±)-anti-7
8-dihydroxy-9, 10-epoxy, 7, 8, 9, 10-tetrahydrobenzo[a]pyrene
- (+)-anti-trans-B[a]PDE-dGuo
(+)-anti-trans-B[a]P-7,8-diol-9,10-epoxide-2′-deoxyguanosine
- (−)-B[a]P -7,8-diol
(−)-trans-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene
- CYP
cytochrome P450
- dAdo
2′-deoxyadenosine
- dGuo
2′-deoxyguanosine
- ESI
electrospray ionization
- FBS
fetal bovine serum
- LC-MS
liquid chromatography-mass spectrometry
- LC-UV
liquid chromatography-ultraviolet
- MCM3
mini-chromosome maintenance protein 3
- MRP4/ABCC4
multi-drug resistance-related protein 4
- Nrf2
nuclear factor erythroid 2 related factor 2
- PAH
polycyclic aromatic hydrocarbon
- PG
prostaglandin
- RPMI 1640
Roswell Park Memorial Institute 1640
- RT-PCR
reverse-transcriptase polymerase chain reaction
- shRNA
small hairpin RNA
- SRM
selected reaction monitoring
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflict to declare
References
- Baird WM, Hooven LA, Mahadevan B. Carcinogenic Polycyclic Aromatic Hydrocarbon-DNA Adducts and Mechanism of Action. Environ. Mol. Mutagen. 2005;45:106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer Risk Assessment, Indicators, and Guidelines for Polycyclic Aromatic Hydrocarbons in the Ambient Air. Environ. Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Vinyals G, D’Errico A, Malats N, Kogevinas M. Biomarkers of Exposure to Polycyclic Aromatic Hydrocarbons From Environmental Air Pollution. Occup. Environ. Med. 2004;61:e12. doi: 10.1136/oem.2003.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Uchi H, Yasukawa F, Furue M. Role of the Arylhydrocarbon Receptor in Lung Disease. Int. Arch. Allergy Immunol. 2011;155(Suppl 1):129–134. doi: 10.1159/000327499. [DOI] [PubMed] [Google Scholar]
- Gelhaus SL, Harvey RG, Penning TM, Blair IA. Regulation of Benzo[a]Pyrene-Mediated DNA- and Glutathione-Adduct Formation by 2,3,7,8-Tetrachlorodibenzo-p-Dioxin in Human Lung Cells. Chem. Res. Toxicol. 2011;24:89–98. doi: 10.1021/tx100297z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis P, Kyrtopoulos SA. Molecular Epidemiological Approaches to the Study of the Genotoxic Effects of Urban Air Pollution. Mutat. Res. 1999;428:91–98. doi: 10.1016/s1383-5742(99)00035-6. [DOI] [PubMed] [Google Scholar]
- Gradhand U, Kim RB. Pharmacogenomics of MRP Transporters (ABCC1-5) and BCRP (ABCG2) Drug Metab Rev. 2008;40:317–354. doi: 10.1080/03602530801952617. [DOI] [PubMed] [Google Scholar]
- Grimmer G, Bohnke H. Polycyclic Aromatic Hydrocarbon Profile Analysis of High-Protein Foods, Oils, and Fats by Gas Chromatography. J. Assoc. Off Anal. Chem. 1975;58:725–733. [PubMed] [Google Scholar]
- Hara Y, Sassi Y, Guibert C, Gambaryan N, Dorfmuller P, Eddahibi S, Lompre AM, Humbert M, Hulot JS. Inhibition of MRP4 Prevents and Reverses Pulmonary Hypertension in Mice. J. Clin. Invest. 2011;121:2888–2897. doi: 10.1172/JCI45023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LL, Kench JG, Handelsman DJ, Scheffer GL, Stricker PD, Grygiel JG, Sutherland RL, Henshall SM, Allen JD, Horvath LG. Androgen Regulation of Multidrug Resistance-Associated Protein 4 (MRP4/ABCC4) in Prostate Cancer. Prostate. 2008;68:1421–1429. doi: 10.1002/pros.20809. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and Regulation of Xenobiotic-Metabolizing Cytochrome P450 (CYP) Enzymes in Human Lung. Crit Rev. Toxicol. 2002;32:391–411. doi: 10.1080/20024091064273. [DOI] [PubMed] [Google Scholar]
- Jiang H, Gelhaus SL, Mangal D, Harvey RG, Blair IA, Penning TM. Metabolism of Benzo[a]Pyrene in Human Bronchoalveolar H358 Cells Using Liquid Chromatography-Mass Spectrometry. Chem. Res. Toxicol. 2007;20:1331–1341. doi: 10.1021/tx700107z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruh GD, Belinsky MG, Gallo JM, Lee K. Physiological and Pharmacological Functions of Mrp2, Mrp3 and Mrp4 As Determined From Recent Studies on Gene-Disrupted Mice. Cancer Metastasis Rev. 2007;26:5–14. doi: 10.1007/s10555-007-9039-1. [DOI] [PubMed] [Google Scholar]
- Lee CH. Reversing Agents for ATP-Binding Cassette Drug Transporters. Methods Mol. Biol. 2010;596:325–340. doi: 10.1007/978-1-60761-416-6_14. [DOI] [PubMed] [Google Scholar]
- Lee K, Belinsky MG, Bell DW, Testa JR, Kruh GD. Isolation of MOAT-B, a Widely Expressed Multidrug Resistance-Associated Protein/Canalicular Multispecific Organic Anion Transporter-Related Transporter. Cancer Res. 1998;58:2741–2747. [PubMed] [Google Scholar]
- Lee K, Klein-Szanto AJ, Kruh GD. Analysis of the MRP4 Drug Resistance Profile in Transfected NIH3T3 Cells. J. Natl. Cancer Inst. 2000;92:1934–1940. doi: 10.1093/jnci/92.23.1934. [DOI] [PubMed] [Google Scholar]
- Lee SH, Blair IA. Targeted Chiral Lipidomics Analysis by Liquid Chromatography Electron Capture Atmospheric Pressure Chemical Ionization Mass Spectrometry (LC-ECAPCI/MS) Methods Enzymol. 2007;433:159–174. doi: 10.1016/S0076-6879(07)33009-7. [DOI] [PubMed] [Google Scholar]
- Lemos C, Kathmann I, Giovannetti E, Belien JA, Scheffer GL, Calhau C, Jansen G, Peters GJ. Cellular Folate Status Modulates the Expression of BCRP and MRP Multidrug Transporters in Cancer Cell Lines From Different Origins. Mol. Cancer Ther. 2009;8:655–664. doi: 10.1158/1535-7163.MCT-08-0768. [DOI] [PubMed] [Google Scholar]
- Lin ZP, Zhu YL, Johnson DR, Rice KP, Nottoli T, Hains BC, McGrath J, Waxman SG, Sartorelli AC. Disruption of CAMP and Prostaglandin E2 Transport by Multidrug Resistance Protein 4 Deficiency Alters CAMP-Mediated Signaling and Nociceptive Response. Mol. Pharmacol. 2008;73:243–251. doi: 10.1124/mol.107.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangal D, Vudathala DK, Park JH, Lee SH, Penning TM, Blair IA. Analysis of 7,8-Dihydro-8-Oxo-2′-Deoxyguanosine in Cellular DNA During Oxidative Stress. Chem. Res. Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo G, Fadda E, Marzia V. Polycyclic Aromatic Hydrocarbons and Cancer in Man. Environ. Health Perspect. 1996;104:1166–1170. doi: 10.1289/ehp.961041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional Regulation of NF-E2 P45-Related Factor (NRF2) Expression by the Aryl Hydrocarbon Receptor-Xenobiotic Response Element Signaling Pathway: Direct Cross-Talk Between Phase I and II Drug-Metabolizing Enzymes. J. Biol. Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- Monien BH, Muller C, Bakhiya N, Donath C, Frank H, Seidel A, Glatt H. Probenecid, an Inhibitor of Transmembrane Organic Anion Transporters, Alters Tissue Distribution of DNA Adducts in 1-Hydroxymethylpyrene-Treated Rats. Toxicology. 2009;262:80–85. doi: 10.1016/j.tox.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Myllynen P, Kurttila T, Vaskivuo L, Vahakangas K. DNA Damage Caused by Benzo(a)Pyrene in MCF-7 Cells Is Increased by Verapamil, Probenecid and PSC833. Toxicol. Lett. 2007;169:3–12. doi: 10.1016/j.toxlet.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Karp CL. Endogenous Functions of the Aryl Hydrocarbon Receptor: Intersection of CytochromeP450 (CYP1)-Metabolized Eicosanoids and AHR Biology. J. Biol. Chem. 2008;283:36061–36065. doi: 10.1074/jbc.R800053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niestroy J, Barbara A, Herbst K, Rode S, van LM, Roos PH. Single and Concerted Effects of Benzo[a]Pyrene and Flavonoids on the AhR and Nrf2-Pathway in the Human Colon Carcinoma Cell Line Caco-2. Toxicol. In Vitro. 2011;25:671–683. doi: 10.1016/j.tiv.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Norris MD, Smith J, Tanabe K, Tobin P, Flemming C, Scheffer GL, Wielinga P, Cohn SL, London WB, Marshall GM, Allen JD, Haber M. Expression of Multidrug Transporter MRP4/ABCC4 Is a Marker of Poor Prognosis in Neuroblastoma and Confers Resistance to Irinotecan in Vitro. Mol. Cancer Ther. 2005;4:547–553. doi: 10.1158/1535-7163.MCT-04-0161. [DOI] [PubMed] [Google Scholar]
- Park JH, Mangal D, Tacka KA, Quinn AM, Harvey RG, Blair IA, Penning TM. Evidence for the Aldo-Keto Reductase Pathway of Polycyclic Aromatic Trans-Dihydrodiol Activation in Human Lung A549 Cells. Proc. Natl. Acad. Sci. USA. 2008;105:6846–6851. doi: 10.1073/pnas.0802776105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M, Merlo F, Munnia A, Valerio F, Perrotta A, Puntoni R, Parodi S. 32P-Postlabeling Detection of Aromatic Adducts in the White Blood Cell DNA of Nonsmoking Police Officers. Cancer Epidemiol. Biomarkers Prev. 1998;7:3–11. [PubMed] [Google Scholar]
- Perez-Tomas R. Multidrug Resistance: Retrospect and Prospects in Anti-Cancer Drug Treatment. Curr. Med. Chem. 2006;13:1859–1876. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- Pozzoli L, Gilardoni S, Perrone MG, De GG, De RM, Vione D. Polycyclic Aromatic Hydrocarbons in the Atmosphere: Monitoring, Sources, Sinks and Fate. I: Monitoring and Sources. Ann. Chim. 2004;94:17–32. doi: 10.1002/adic.200490002. [DOI] [PubMed] [Google Scholar]
- Reid G, Wielinga P, Zelcer N, van d. H., I, Kuil A, de HM, Wijnholds J, Borst P. The Human Multidrug Resistance Protein MRP4 Functions As a Prostaglandin Efflux Transporter and Is Inhibited by Nonsteroidal Antiinflammatory Drugs. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9244–9249. doi: 10.1073/pnas.1033060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter CA, Jedlitschky G, Meyer Zu SH, Grube M, Kock K, Kroemer HK. Cellular Export of Drugs and Signaling Molecules by the ATP-Binding Cassette Transporters MRP4 (ABCC4) and MRP5 (ABCC5) Drug Metab. Rev. 2005;37:253–278. doi: 10.1081/dmr-200047984. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Gelhaus SL, Penning TM, Harvey RG, Blair IA. Aldo-Keto Reductase- and Cytochrome P450-Dependent Formation of Benzo[a]Pyrene-Derived DNA Adducts in Human Bronchoalveolar Cells. Chem. Res. Toxicol. 2007;20:424–431. doi: 10.1021/tx060180b. [DOI] [PubMed] [Google Scholar]
- Russel FG, Koenderink JB, Masereeuw R. Multidrug Resistance Protein 4 (MRP4/ABCC4): a Versatile Efflux Transporter for Drugs and Signalling Molecules. Trends Pharmacol. Sci. 2008;29:200–207. doi: 10.1016/j.tips.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Srivastava SK, Watkins SC, Schuetz E, Singh SV. Role of Glutathione Conjugate Efflux in Cellular Protection Against Benzo[a]Pyrene-7,8-Diol-9,10-Epoxide-Induced DNA Damage. Mol. Carcinog. 2002;33:156–162. [PubMed] [Google Scholar]
- Sweet DH. Organic Anion Transporter (Slc22a) Family Members As Mediators of Toxicity. Toxicol. Appl. Pharmacol. 2005;204:198–215. doi: 10.1016/j.taap.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Torky AR, Stehfest E, Viehweger K, Taege C, Foth H. Immuno-Histochemical Detection of MRPs in Human Lung Cells in Culture. Toxicology. 2005;207:437–450. doi: 10.1016/j.tox.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW. Oral Exposure to Benzo[a]Pyrene in the Mouse: Detoxication by Inducible Cytochrome P450 Is More Important Than Metabolic Activation. Mol. Pharmacol. 2004;65:1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- van Aubel RA, Smeets PH, Peters JG, Bindels RJ, Russel FG. The MRP4/ABCC4 Gene Encodes a Novel Apical Organic Anion Transporter in Human Kidney Proximal Tubules: Putative Efflux Pump for Urinary CAMP and CGMP. J. Am. Soc. Nephrol. 2002;13:595–603. doi: 10.1681/ASN.V133595. [DOI] [PubMed] [Google Scholar]
- Wu CP, Calcagno AM, Hladky SB, Ambudkar SV, Barrand MA. Modulatory Effects of Plant Phenols on Human Multidrug-Resistance Proteins 1, 4 and 5 (ABCC1, 4 and 5) FEBS J. 2005;272:4725–4740. doi: 10.1111/j.1742-4658.2005.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Aryl Hydrocarbon Receptor and NF-E2-Related Factor 2 Are Key Regulators of Human MRP4 Expression. Am. J. Physiol Gastrointest. Liver Physiol. 2010;299:G126–G135. doi: 10.1152/ajpgi.00522.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the ‘TCDD Inducible AhR-Nrf2 Gene Battery’. Toxicol. Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcer N, Reid G, Wielinga P, Kuil A, van d. H., I, Schuetz JD, Borst P. Steroid and Bile Acid Conjugates Are Substrates of Human Multidrug-Resistance Protein (MRP) 4 (ATP-Binding Cassette C4) Biochem. J. 2003;371:361–367. doi: 10.1042/BJ20021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Pelletier RD, Wong YN, Sugawara M, Zhao N, Littlefield BA. Preferential Inducibility of CYP1A1 and CYP1A2 by TCDD: Differential Regulation in Primary Human Hepatocytes Versus Transformed Human Cells. Biochem. Biophys. Res. Commun. 2006;341:399–407. doi: 10.1016/j.bbrc.2005.12.203. [DOI] [PubMed] [Google Scholar]
- Zmirou D, Masclet P, Boudet C, Dor F, Dechenaux J. Personal Exposure to Atmospheric Polycyclic Aromatic Hydrocarbons in a General Adult Population and Lung Cancer Risk Assessment. J. Occup. Environ. Med. 2000;42:121–126. doi: 10.1097/00043764-200002000-00004. [DOI] [PubMed] [Google Scholar]