Abstract
Aberrant activation of the Hedgehog (Hh) signaling pathway is one of the most prevalent abnormalities in human cancer. Tumors with cell autonomous Hh activation (e.g., medulloblastomas) can acquire secondary mutations at the Smoothened (Smo) antagonist binding pocket, which render them refractory to conventional Hh inhibitors. A class of Hh Pathway Inhibitors (HPIs) has been identified that block signaling downstream of Smo; one of these compounds, HPI-1, is a potent antagonist of the Hh transcription factor Gli1, and functions independent of upstream components in the pathway. Systemic administration of HPI-1 is challenging due to its minimal aqueous solubility and poor bioavailability. We engineered a polymeric nanoparticle from poly(lactic-co-glycolic acid) (PLGA) conjugated with polyethylene glycol (PEG), encapsulating HPI-1 (NanoHHI). NanoHHI particles have an average diameter ~60nM, forms uniform aqueous suspension, and improved systemic bioavailability compared to the parent compound. In contrast to the prototype targeted Smo antagonist, HhAntag (Genentech), NanoHHI markedly inhibits the growth of allografts derived from Ptch−/+;Trp53−/− mouse medulloblastomas that harbor a SmoD477G binding site mutation (P<0.001), which is accompanied by significant downregulation of mGli1, as well as bona fide Hh target genes (Akna, Cltb, Olig2). Notably, NanoHHI combined with gemcitabine also significantly impedes the growth of orthotopic Pa03C pancreatic cancer xenografts that have a ligand-dependent, paracrine mechanism of Hh activation, when compared to gemcitabine alone. No demonstrable hematological or biochemical abnormalities were observed with NanoHHI administration. NanoHHI should be amenable to clinical translation in settings where tumors acquire mutational resistance to current Smo antagonists.
Keywords: Hedgehog, smoothened, Gli1, HPI-1, polymeric nanoparticle, NanoHHI, HhAntag
Introduction
The Hh signaling pathway plays a critical role in development as well as in mature tissue homeostasis (1). Aberrant activation of the Hh pathway is commonly observed in many human cancers, and it is implicated in tumor initiation as well as tumor progression (2, 3). Hh activation in human cancers can occur either via a ligand-independent mechanism, as is observed in so-called “Gorlin syndrome” cancers (basal cell carcinoma and medulloblastoma), or by a ligand-dependent mechanism, which has been implicated in many solid tumors (4, 5). Ligand-independent pathway activation is usually a consequence of loss-of-function mutations in the gene encoding the Hh inhibitory receptor Patched (Ptch), or less commonly, a gain-of-function mutation in the gene encoding the essential Hh signal transduction receptor Smoothened (Smo) (6–8). In contrast, mutational activation of the pathway is rare in endodermal tumors such as pancreatic cancer that produce aberrantly high levels of Hh ligand (2). As recent evidence has shown, most of the pathway activity in such cancers appears to be restricted to the juxta-tumoral stromal cells, rather than the neoplastic cells per se (9, 10). Irrespective of the mechanism of pathway activation, de-repression of Smo from Ptch initiates an intracellular cascade that culminates in the nuclear translocation of Gli transcription factors, and the major transcriptional activator in human cancers appears to be Gli1, a product of the GLI1 oncogene (11).
In the area of clinical oncology, small molecule antagonists of Hh signaling have emerged as a more promising targeted approach to cancer therapy. Recently, there has been compelling evidence to suggest that cancer cells can acquire resistance to Smo antagonists through secondary mutations in Smo. Most notably, Yauch and colleagues recently reported on a patient with metastatic medulloblastoma, who initially had a dramatic response to GDC-0449, but subsequently relapsed with refractory disease (12). Sequencing of the relapsed tumor DNA identified a secondary mutation in an extracellular loop of the Smo heptahelical bundle (SmoD473H), which abrogates the binding of GDC-0449. Strikingly, in a Ptch−/+;p53−/− mouse model of medulloblastoma selected in vivo for resistance to GDC-0449, an acquiredSmoD477G alteration was identified, which is orthologous to the aspartic acid residue at position 473 in humans, and similarly disrupts antagonist binding. Of note, neither mutation has an impact on the overall level of Hh activation, suggesting that these are not independently “oncogenic”. Independent experiments in murine medulloblastoma models have also reported comparable acquired mutations in Smo that confer Hh inhibitor resistance (13, 14). Additionally, other mechanisms of resistance to Smo antagonists have also been reported, including amplification of GLI oncogenes that occur downstream of the Smo receptor (13, 14), thus allowing cancer cells to bypass Hh blockade by the current compendium of Smo antagonists.
In light of this emerging evidence on mechanisms of secondary resistance to Smo antagonists, there is a pressing need to identify a new generation of Hh inhibitors that block signaling downstream of Smo. In 2009, Hyman and colleagues identified a series of four Hh Pathway Inhibitors (a.k.a., HPIs 1–4), which block signaling at diverse points downstream of Smo, including Gli processing, stability and trafficking to the primary cilium (15). One of these compounds, HPI-1, is a potent antagonist of both endogenous activator Gli proteins (Gli1/2), and can also abrogate Hh signaling in the setting of exogenous Gli overexpression. Based on its mechanism of action, we can postulate that HPI-1 will circumvent acquired mutational resistance to “conventional” Smo inhibitors.
Despite the promising in vitro findings, however, the in vivo translation of HPI-1 is likely to be hampered by its highly lipophilic nature and poor aqueous solubility, thereby impairing systemic bioavailability. To harness the full therapeutic potential of HPI-1, we have generated a polymer nanoparticle-encapsulated formulation of HPI-1 (“NanoHHI”), which overcomes the barriers to systemic bioavailability. NanoHHI has been engineered using poly(lactic-co-glycolic acid) (PLGA) conjugated with polyethylene glycol (PEG), both of which are considered as generally regarded as safe (GRAS) components by the United States Food and Drug Administration (USFDA) (16). NanoHHI demonstrates strikingly higher systemic bioavailability compared to HPI-1 alone upon parenteral administration, with no apparent histopathological or biochemical evidence of toxicities in mice. Of importance, NanoHHI blocks Hh signaling in cells with ectopic expression of the human SmoD473H allele, and significantly inhibits the in vivo growth of murine medullobastoma allografts harboring the acquired murine SmoD477G mutation, both of which confer resistance to targeted Smo antagonists (12). NanoHHI also inhibits the growth of orthotopic human pancreatic cancer xenografts that harbor a wild type Smo allele, by potentiating the effects of gemcitabine in the orthotopic milieu. Thus, NanoHHI represents a promising new therapeutic formulation for treatment of human cancers with primary or secondary resistance to Smo antagonists.
Materials and Methods
Materials
Poly(lactic-co-glycolic acid) (PLGA) conjugated with polyethylene glycol (PEG), i.e. PLGA-PEG (5050 DLG, mPEG 5000) was purchased from Lakeshore Biomaterials (Birmingham, Alabama). Dichloromethane, acetone, and polyvinyl alcohol PVA18k (87–89% hydrolyzed) were obtained from Sigma Aldrich (St. Louis, Missouri). HPI-1 was synthesized according to the reported procedure (15); the nanoparticulated formulation NanoHHI was stored as a lyophilized powder at 4°C, and dissolved in PBS on the day of use. Gemcitabine (NetQem LLC, Research Triangle Park, North Carolina) was stored at 4°C and dissolved in sterile NaCl (0.9% w/v) on the day of use. HhAntag (Genentech, South San Francisco, California), a parental drug of the lead clinical compound GDC-0449 (17), was freshly formulated as a suspension in 0.5% methylcellulose, 0.2% Tween-80 (MCT).
Cell lines and plasmids
Pa03C (a.k.a. LZ10.7), a low-passage metastatic human pancreatic cancer cell line, was cultured as described (18); the authentication of this cell line was based on representative validation of previously described whole exome mutational profiling data (19). Either wild type Smo or SmoD473H mutant cDNA was cloned into pRK5 mammalian expression vector, and stable clones of HEK293 cells expressing empty pRK5, pRK5-Smowt, or pRK5-SmoD473H were generated for Hh reporter assays. Murine medulloblastoma allografts derived from Ptch−/+;p53−/−mice harboring either Smowt or SmoD477G alleles were generated as described previously, for in vivo studies comparing HhAntag and NanoHHI (12).
Formulation of HPI-1 loaded PLGA-PEG nanoparticles (NanoHHI)
NanoHHI was prepared using a modification of the oil-in-water (o/w) emulsion solvent evaporation method (20). Briefly, 3g of PLGA-PEG and 60mg of HPI-1 was dissolved in 30 mL of dichloromethane and acetone (8:2), and the resulting solution was added to 0.4% polyvinyl alcohol (150mL). The mixture was sonicated for 3 minutes with stirring (20W, 4°C). The suspension was stirred at room temperature for 4 hours to evaporate organic solvents, and complete evaporation of organic solvents was achieved by using the rotary evaporator. The resulting suspension was ultra-centrifuged at 40,000rpm for 45 minutes. The precipitated nanoparticle pellet was washed 3 times with ultra pure water and re-suspended in ultra pure water by mild sonication, and centrifuged at 3,000rpm for 5 minutes to remove large aggregates or any non-encapsulated drug. The supernatant NanoHHI was flash frozen on dry ice, and lyophilized to obtain a dry NanoHHI powder amenable to long term storage. Drug loading and loading efficiency were determined by dissolving dried nanoHHI in 80% methanol in water and quantifying by HPLC. Drug loading percentage (L) was calculated as L = (mdrug/mtotal) × 100, where mdrug and mtotal are the mass of drug and loaded nanoparticles (nanoHHI), respectively. Loading efficiency (E) was calculated as E = (mencap/mdrug) × 100 where mencap and mdrug are the mass of drug encapsulated and the total mass of drug initially loaded, respectively.
Size determination and in vitro release kinetics of NanoHHI
Size analysis of NanoHHI was performed using a Malvern Zetasizer (Malvern, Worcestershire, United Kingdom), and transmission electron microscopy (TEM) images obtained using a Hitachi 7600 (Hitachi Ltd., Tokyo, Japan). Release kinetics studies were performed using the dialysis method, as described (21). Briefly, NanoHHI containing 10mg equivalent of HPI-1 dispersed in 5mL ultra pure water was transferred into a dialysis bag with a molecular cutoff of 12kDa. The bag was suspended in 100mL of release medium (50% v/v of ethanol, water) in a container, with stirring at 50rpm at 37°C. A 300 μL aliquot was withdrawn at predetermined time intervals and replaced with fresh release medium. Finally these samples were analyzed by high-performance liquid chromatography (HPLC) using a Waters HPLC system (Milford, Massachusetts). As HPI-1 is poorly soluble in water, ethanol was used in the release medium to ensure sink conditions.
Pharmacokinetic analyses of parenteral NanoHHI compared with free HPI-1 including brain distribution of NanoHHI
To compare the in vivo pharmacokinetics of NanoHHI versus free HPI-1, we conducted three independent studies, each via a different route of administration: First, for the intraperitoneal (i.p.) route, two cohorts of three mice each were administered a single i.p. injection of either 25 mg/kg free HPI-1 suspended in corn oil or 25 mg/kg NanoHHI equivalent. Second, for the intravenous (i.v.) route, two cohorts of four mice each were administered either 30 mg/kg free HPI-1 in 40% ethanol or 30 mg/kg NanoHHI equivalent. Third, for the oral bioavailability study, two arms of four mice each were administered a single per oral (p.o.) dose of either 30 mg/kg free HPI-1 suspended in corn oil or 30 mg/kg NanoHHI, via oral gavage. Finally, to assess the ability of NanoHHI to cross the blood brain barrier (BBB), two cohorts of three mice each were administered a single intravenous injection of 30 mg/kg NanoHHI (this study was performed in replicate in order to collect terminal brain samples at two different time points following injection). All of the above studies were conducted using non-tumor bearing CD1 mice. Blood samples (50μL) were obtained from the mice at predetermined time intervals post-injection, using EDTA-coated Microvette CB300 capillary tubes (Braintree Scientific). The plasma was separated by centrifuging the blood samples at a speed of 1000×g for 5 minutes. Brain samples were collected immediately following euthanasia at 10 and 30 min post injection.
HPI-1 levels were estimated using liquid chromatography tandem mass spectrometry (LC-MS/MS). Tissue homogenates were prepared at a concentration of 200 mg/mL in plasma prior to extraction. HPI-1 was extracted by acetonitrile-n-butyl chloride (1:4, v/v) and separated on a Waters (Milford, MA) X-Terra MS C18 (50 × 2.1 mm, 3.5μm) column with acetonitrile/water mobile phase (80:20, v/v) containing 0.1% formic acid using isocratic flow at 0.15mL/min for 5 minutes. Plasma calibration curves were prepared over the range of 0.01–2 μg/mL or 0.06 to 12 μg/g, with a 1:1000 dilution being accurately quantitated. Concentration-time data were evaluated using a non-compartmental approach using individual profiles (WinNonlin Professional, version 5.2 software, Pharsight Corporation).
Allograft and Xenograft Studies
All small animal experiments described conformed to the guidelines of the Animal Care and Use Committee of Johns Hopkins University. Mice were maintained in accordance with the guidelines of the American Association of Laboratory Animal Care.
Establishment and treatment of subcutaneous mouse medulloblastoma allografts
Flanks of 5–6 weeks old male athymic nu/nu mice (Harlan Laboratories, Indianapolis, IN) were injected with a single cell suspension (2x106 cells) of either SmoWT; Ptch+/−; Trp53−/− or SmoD477G; Ptch+/−; Trp53−/− in a total volume of 200μL PBS/Matrigel (BD Biosciences, 1:1 (v/v), prechilled to 4°C). One week after the injection of tumor cells, subcutaneous tumor volumes (V) were measured with digital calipers (Fisher Scientific) and calculated using the formula V = 1/2(ab2), where a is the biggest and b is the smallest orthogonal tumor diameter. Mice with successfully engrafted SmoWT or SmoD477G allografts were then randomized into 3 cohorts of 6 animals each and administered one the following regimens: (i) void PLGA-PEG nanoparticles, (ii) NanoHHI at a dose of 30 mg/kg i.p. twice daily, and (iii) HhAntag at a dose of 100 mg/kg p.o., once daily. NanoHHI dosing of 30 mg/kg was chosen after multiple dose tolerance study (data not shown). The allografted mice were monitored daily for any signs of toxicity and behavioral abnormalities during the treatment. Tumor size and body weight were measured every other day. At the end of treatment, allografts were harvested and preserved for mRNA extraction and qRT-PCR analysis of Hh gene targets.
Establishment and treatment of orthotopic Pa03C pancreatic cancer xenografts
The generation of orthotopic Pa03C human pancreatic cancer xenografts by surgical implantation in athymic mice has been described previously by our group (18). Three weeks after surgical orthotopic implantation, the presence of intra-pancreatic “primary” tumors was confirmed by ultrasound scan (Vevo660, VisualSonics). Twenty-eight mice with demonstrable “primary” xenografts were then randomized into four cohorts, with seven mice per arm, as follows: (i) void PLGA-PEG nanoparticles, (ii) NanoHHI at a dose of 30 mg/kg i.p. twice daily, (iii) gemcitabine at a dose of 20 mg/kg i.p. twice weekly, or (iv) the combination of NanoHHI (30 mg/kg i.p. twice daily) and gemcitabine (20 mg/kg i.p. twice weekly). Both agents were administered for a period of four weeks. At the end of therapy, primary tumors were carefully excised from the pancreas and weighed. Portions of treated xenografts were also preserved for mRNA extraction and qRT-PCR for Hh gene targets (primer sequences for human and mouse genes are readily available upon request).
Results
Physicochemical characterization and in vitro release kinetics of NanoHHI
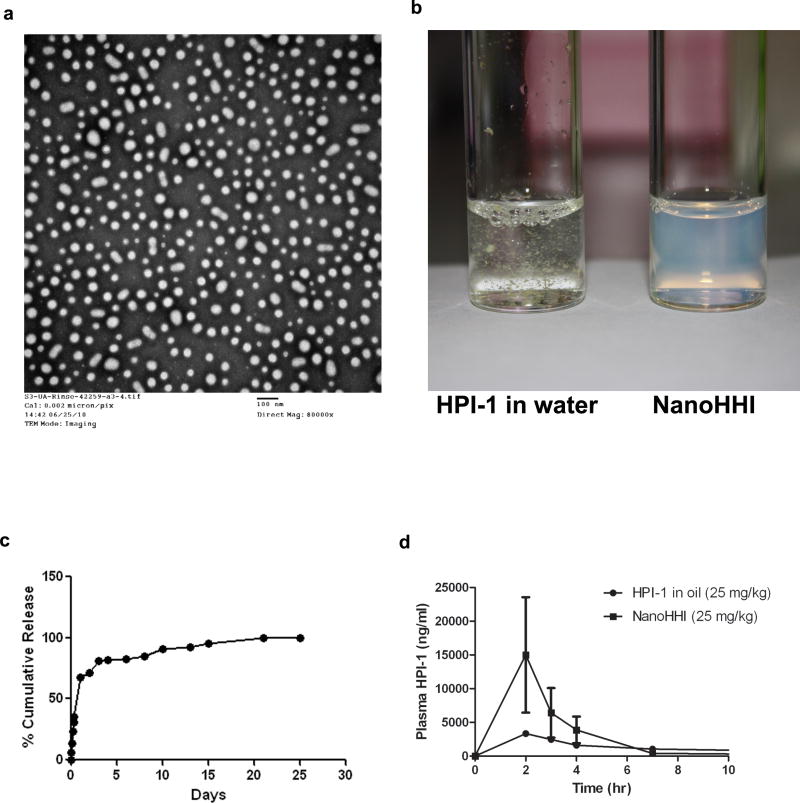
Transmission electron microscopy (Figure 1a) and dynamic light scattering (DLS) studies (data not shown) confirmed that NanoHHI has an average diameter of 60nm. In contrast to the extremely poor aqueous solubility of free HPI-1, NanoHHI was well dispersed to form a uniform suspension in aqueous media (Figure 1b). HPI-1 was loaded efficiently in the void PLGA-PEG nanoparticles (2% w/w loading; 90% encapsulation efficiency). The in vitro release kinetics of HPI-1 from NanoHHI was assessed over an approximately four week period by dialysis method, using 50% v/v ethanol as release medium to ensure sink conditions due to the nearly complete insolubility of free HPI-1 in aqueous media (Figure 1c). A small burst release in the initial 24 hours was observed (cumulative ~30%), followed by a relatively slow release over 3 days (cumulative ~70%), with complete drug being released by day 25.
Figure 1. Physicochemical characterization and pharmacokinetics of NanoHHI.
(a) Transmission electron micrograph (TEM) pictures of NanoHHI, demonstrating uniformly dispersed nanoparticles with an average size diameter of <100nM. The scale bar on lower right corner equals 100nM.
(b) NanoHHI (right) is a uniform suspension in aqueous media while parental HPI-1 is essentially insoluble, with flakes of drug clearly visible in the suspension. Equivalent amounts of HPI-1 were used in both instances.
(c) In vitro release kinetics of HPI-1 from NanoHHI, performed at 37°C. The Y-axis charts the % cumulative release and X-axis denotes time in days.
(d) Pharmacokinetic disposition of parenteral NanoHHI administered as a single dose (25 mg/kg) compared to that of “free” HPI-1 (25 mg/kg) in corn oil. The experiments were performed in non-tumor bearing CD1 mice, with three mice per cohort. Plasma HPI-1 levels were assessed using LC-MS/MS.
Pharmacokinetics of parenteral NanoHHI compared to free HPI-1 and brain distribution of NanoHHI
We compared the bioavailability of NanoHHI to free HPI-1 dissolved in corn oil, following administration of a single equivalent dose of either formulation through the intraperitoneal (i.p.) route in non-tumor-bearing mice (Figure 1d). Relevant pharmacokinetic parameters, including Cmax, Tmax, and area under the curve (AUC) are tabulated in Supplementary Table 1. There was an appreciable difference in the bioavailability of i.p. HPI-1 between the free drug and “nano” formulation, with NanoHHI showing an AUC0-∞ value twice that of free HPI-1 in corn oil (52 ± 45 μg*h/mL versus 26 ± 12 μg*h/mL). We also attempted to compare the pharmacokinetics of intravenous (i.v.) NanoHHI versus free HPI-1 dissolved in 40% ethanol as an excipient; however the mice receiving i.v. ethanolic HPI-1 succumbed within 12 minutes of injection, likely from the excipient-related toxicity. The AUC0-∞ value of i.v. NanoHHI alone was 268 ± 98 μg*h/mL (Supplementary Figure 1a and Supplementary Table 1). A more pronounced improvement in bioavailability was observed after oral administration of 30 mg/kg HP!-1 equivalents, with the NanoHHI AUC0-∞ value being four-times higher than that of free HPI-1 in corn oil (17 ± 31 μg*h/mL versus 4.2 ± 1.4 μg*h/mL) (Supplementary Figure 1b and Supplementary Table 1). Thus, we were able to establish the feasibility of administering NanoHHI through multiple routes, with a two-to-four hold improved AUC values for the two modes (i.p. and oral) where an equimolar comparison could be made versus free HPI-1.
A potential clinical application of Gli-targeted Hh agents will be in the setting of medulloblastomas that have acquired secondary mutations in SMO; thus, establishing tractable delivery of systemic NanoHHI past the blood brain barrier (BBB) is paramount in that respect. Following single dose i.v. administration of NanoHHI, HPI-1 was readily detectable in brain tissue at 3.9±2.1 μg/g after 10 minutes, and 1.4±0.4μg/g at 30 minutes after injection (Supplementary Figure 1c).
NanoHHI inhibits the in vivo growth of murine medulloblastoma allografts harboring Smo antagonist-resistant binding site mutation
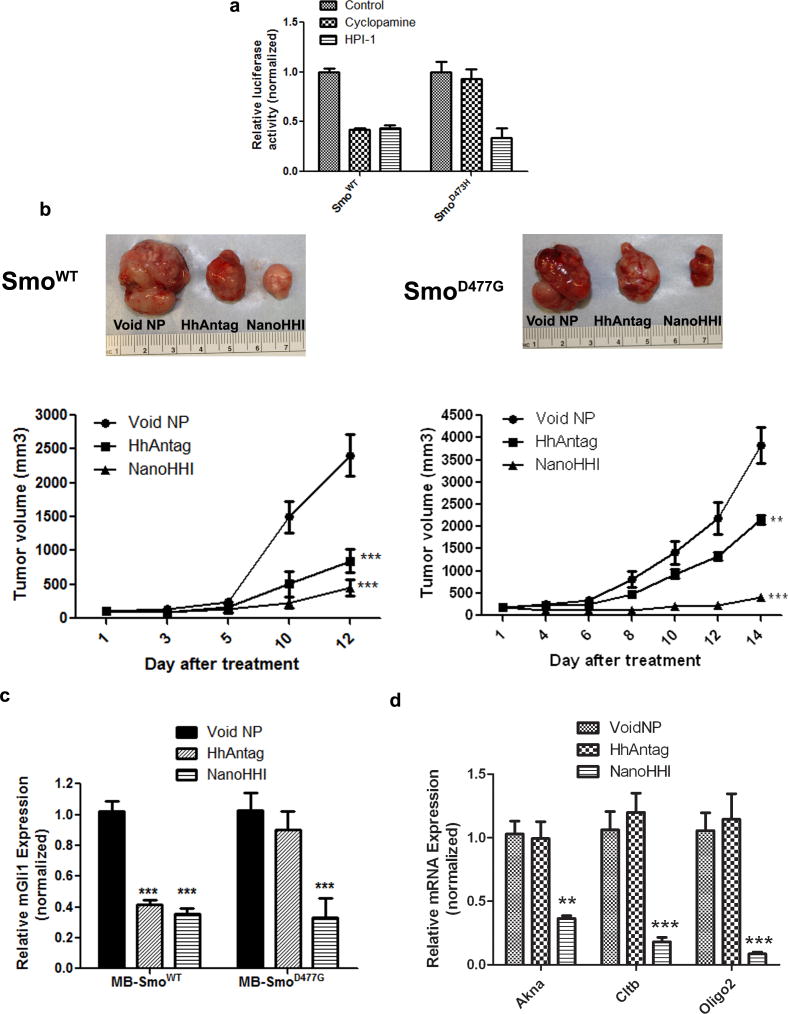
In vitro Hh reporter assays were conducted using HEK-293 cells expressing either wild type pRK5-SmoWT, or the Smo antagonist resistant binding site mutant pRK5-SmoD473H construct (Figure 2a). While both HPI-1 and cyclopamine were able to significantly inhibit reporter activity in the setting of wild-type Smo, only HPI-1 resulted in significant downregulation of reporter activity in HEK-293 cells expressing the mutant SmoD473H. These results confirmed the ability of our indigenously synthesized free HPI-1 to bypass the Smo binding site mutation in vitro.
Figure 2. NanoHHI inhibits the in vivo growth of murine medulloblastoma allografts selected for acquired resistance to Smo inhibitors.
(a) HPI-1 can block Hh signaling in HEK-293 cells expressing human Smo (HsSmo) with a binding site mutation. HEK-293 cells stably expressing either wild type HsSmo (SmoWT) or mutant HsSmoD473H were transiently transfected with Gli1-binding site firefly luciferase reporter, and exposed to cyclopamine or HPI-1. Renilla luciferase was used as a control for transfection efficiency. Both cyclopamine and HPI-1 can block Hh signaling in HEK-293 cells expressing HsSmoWT (left); in contrast, only HPI-1 blocks Hh signaling in HEK-293 cells expressing HsSmoD473H, while cyclopamine, which is unable to bind to mutant Smo receptor has minimal effect (right). All luciferase assays were performed in triplicate, and the Y-axis denotes relative luciferase units normalized to Renilla.
(b) Murine medulloblastomas arising in Ptch+/−; Trp53−/− mice, and harboring either murine SmoWT (MmSmoWT) or an acquired MmSmoD477G mutation, were allografted subcutaneously in athymic mice, and treated with either HhAntag or NanoHHI. Void PLGA-PEG nanoparticles were used as vehicle control. In allografts expressing wild type MmSmo, both HhAntag and NanoHHI demonstrate comparable effects in terms of tumor growth inhibition on day 12 (the experiment had to be terminated due to the rapid increase in growth of vehicle control allografts) (left). In contrast, in allografts expressing the MmSmoD477G binding site mutation, NanoHHI demonstrates a markedly better efficacy than HhAntag in growth inhibition (right, P<0.01 for HhAntag and P<0.001 for NanoHHI). Representative post-treatment allografts are illustrated for each cohort above the growth curves.
(c) Murine Gli1 (MmGli1) mRNA levels were assessed following treatment in both cohorts, as a measure of Hh pathway inhibition. Mirroring the growth phenotypes, both HhAntag and NanoHHI significantly downregulate MmGli1 levels in MmSmoWT allografts, while only NanoHHI significantly downregulates MmGli1 expression in MmSmoD477G expressing allografts. The Y-axis represents relative expression normalized to housekeeping control.
(d) In medulloblastoma allografts expressing the MmSmoD477G mutation, NanoHHI significantly downregulates bona fide Gli1 target genes, including Akna, Cltb, and Olig2, compared to HhAntag-treated allografts (expression is normalized to the void polymer control treatment arm).
We then proceeded to in vivo assays using the NanoHHI formulation, comparing it to the potent and selective Smo antagonist, HhAntag. In subcutaneous medulloblastoma allografts derived from SmoWT; Ptch+/−; Trp53−/− tumors, both NanoHHI and HhAntag inhibited in vivo growth to a comparable degree (Figure 2b, left). In contrast, in allografts derived from SmoD477G; Ptch+/−; Trp53−/− tumors, there was a marked difference in efficacy between NanoHHI and HhAntag (Figure 2b, right). Although even HhAntag had a significant negative impact on allograft growth (P<0.01), NanoHHI essentially “flattened” the tumor growth curve (P<0.001). The partial response seen with HhAntag in the setting of a binding site mutation may reflect residual binding capability to the mutant Smo receptor, and/or off-target effects, an effect not observed with the clinical grade compound (14). In treated allografts, qRT-PCR for MmGli1 transcripts mirrored the efficacy data (Figure 2c), with significant MmGli1 downregulation observed in both treatment arms for SmoWT; Ptch+/−; Trp53−/− tumors (P<0.001), but only in the NanoHHI arm for SmoD477G; Ptch+/−; Trp53−/− tumors (P<0.001) (all of the transcript results were normalized to levels observed in allografts treated with void polymer). Since HPI-1 is a direct inhibitor of Gli function and acts downstream of Smo, we evaluated the impact of NanoHHI or HhAntag treatment in a panel of recently described Gli-target genes in murine medulloblastoma, including Akna, Cltb and Olig2 (Figure 2d) (22). For these analyses, only the SmoD477G; Ptch+/−; Trp53−/− allografts were assessed, and confirmed the ability of NanoHHI, but not the targeted Smo inhibitor, to significantly block bona fide Gli target gene expression in the treated tumors. In this study, we observed no significant alterations of body weight in any of the NanoHHI treatment arms, while the HhAntag-treated mice demonstrated ~10% body weight loss at the end of treatment (Supplementary Figure 1d).
NanoHHI inhibits the growth of orthotopic pancreatic cancer xenografts in combination with gemcitabine
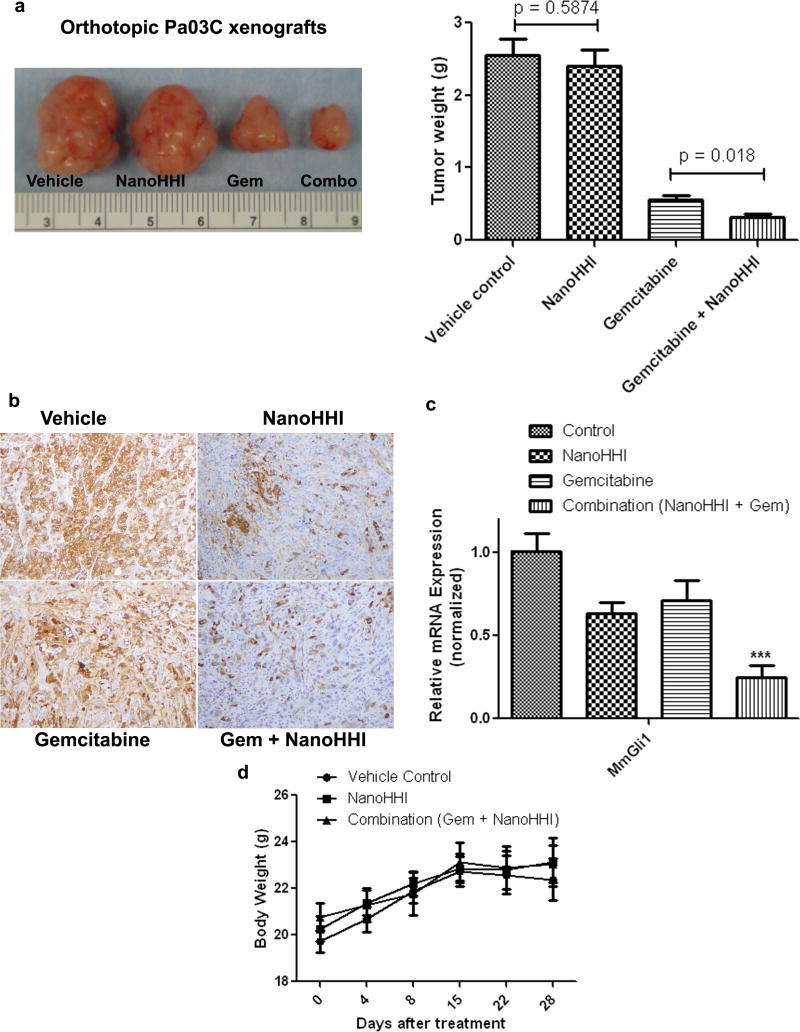
Gorlin syndrome tumors with cell autonomous Hh signaling comprise only a minor fraction of cases in the universe of solid cancers, with ligand-dependent Hh signaling being the most common mechanism of activation by far. In this latter instance, much of the signaling is paracrine in nature, and the non-neoplastic stromal cells may be significantly less likely to acquire secondary mutational hits during therapy. Nonetheless, given the demonstrable efficacy of Hh blockade in preclinical models of ligand dependent cancers, we explored the in vivo effects of NanoHHI in an orthotopic human pancreatic cancer xenograft model. In Pa03C xenografts, we found that NanoHHI monotherapy had no significant effect on orthotopic tumor growth (P=0.58), mirroring what has been reported with cyclopamine and other Smo antagonists (18, 23). In contrast, NanoHHI in combination with gemcitabine significantly improved tumor growth inhibition over gemcitabine alone (P =0.018) (Figure 3a). We have previously demonstrated that cyclopamine and other Smo antagonists deplete the “primary” tumor of cells expressing aldehyde dehydrogenase (ALDH), a credentialed marker of tumor initiating cells (i.e., “cancer stem cells”) (24, 25), even when there is no impact on gross tumor volume (18, 23). We confirmed that NanoHHI, either as a single agent, or in combination with gemcitabine, can cause a marked decrease in ALDH-expressing cells within the orthotopic Pa03C xenograft (Figure 3b). Further, we also confirmed that stroma-derived MmGli1 was significantly reduced in both NanoHHI single-agent (P<0.05) and combination therapy arms (P<0.001) (Figure 3c), consistent with inhibition of Hh signaling in the murine stromal compartment. In contrast, gemcitabine alone had no significant impact on MmGli1 levels. We also examined the levels of HsCMYC, a key transcriptional regulator in pancreatic cancer, and found no significant effects of NanoHHI compared to void nanoparticles (notably, gemcitabine resulted in significant downregulation of expression in both single agent and combination therapy arms) (Supplementary Figure 2a). The expression of HsHES1, a Notch pathway target gene, however, was significantly downregulated by NanoHHI, consistent with the recent data on non-canonical regulation of HES1 expression by Hh signaling (26). Finally, we examined the levels of MmNestin, a marker of neo-endothelial cells in the murine stroma (27), and found significant downregulation in both single-agent and combination therapy arms, consistent with the known anti-angiogenic effects of Hh blockade in tumors.
Figure 3. NanoHHI and gemcitabine blocks the growth of orthotopic pancreatic cancer xenografts with paracrine Hh signaling.
(a) Orthotopic human pancreatic cancer Pa03C xenografts were generated in athymic mice by surgical orthotopic implantation, and treated with vehicle control, NanoHHI, gemcitabine, or the combination. At four weeks of therapy, tumor weights were assessed at necropsy, and demonstrated a significant reduction in weight (in grams) for the combination arm compared to gemcitabine alone (P=0.018). Expectedly, NanoHHI monotherapy had minimal impact on primary tumor weight. Representative post-treatment xenografts are illustrated for each cohort (Left)
(b) NanoHHI depletes aldehyde dehydrogenase (ALDH) expressing cells in Pa03C xenografts. ALDH expression in neoplastic cells was assessed by immunohistochemistry on representative xenografts from each of the four arms (as indicated). Marked reduction in ALDH expressing cells is observed in both NanoHHI monotherapy and combination treatment arms.
(c) Relative expression of stroma-specific murine Gli1 (MmGi1) were assessed in the treated pancreatic cancer Pa03C xenografts. Significant downregulation of MmGli1 was observed in both NanoHHI monotherapy (P<0.05) and combination arms (P<0.001), consistent with downregulation of Hh signaling in the stromal compartment.
(d) Body weight measurements were obtained at five time points in mice bearing orthotopic Pa03C xenografts that were treated with vehicle, NanoHHI or the combination of NanoHHI and gemcitabine over a four week time period. No significant body weight losses are encountered in either NanoHHI arm.
Minimal systemic toxicity of NanoHHI in mice
The preclinical and emerging clinical utilization of small molecule Hh blockade has shown the surprising resilience of non-neoplastic Hh-dependent compartments (e.g., bone marrow) to this class of agents (28). Thus, there appears to be a “therapeutic window,” wherein depriving Hh signals to cancer cells themselves, or to the microenvironment, can inhibit tumor growth without major systemic deleterious effects. To determine whether a similar safety profile might exist for NanoHHI, particularly since the active pharmaceutical ingredient has a distinct target (Gli) from the currently studied Smo antagonists, we performed a panel of laboratory assays on NanoHHI-administered mice. In both the medulloblastoma allograft and the orthotopic pancreatic cancer xenograft experiments, which lasted for a period of two to four weeks, we observed no loss of body weight in any of the NanoHHI treatment arms (see Supplementary Figure 1d and Figure 3d). We also examined the histopathology of major viscera in NanoHHI treated mice, which did not reveal any microscopic abnormalities (data not shown). Finally, we examined a panel of laboratory parameters, including hematological (RBC, WBC, and platelet counts and hemoglobin), renal and liver function tests in NanoHHI mice compared to vehicle treated animals (Supplementary Figure 3), and observed no significant differences in any of the results.
Discussion
Aberrant activation of the Hh pathway is observed in many solid and hematological cancers, generating considerable excitement about targeted inhibition of this pathway. As recent data have shown, patients whose tumors harbor mutational loss of Patched can experience dramatic responses to a class of orally bioavailable Smo antagonists, of which GDC-0449 is farthest along in clinical development (29, 30). Nonetheless, several mechanisms of secondary resistance to Hh blockade in tumors have emerged recently, most notably the acquisition of somatic mutations of Smo, which render the receptor incapable of binding to GDC-0449 and other antagonists of this class (12, 13). The acquisition of secondary resistance to targeted therapies is certainly not unprecedented, with chronic myelogenous leukemia being one of the better known examples where this phenomenon occurs in response to imatinib (31).
There is an urgent impetus for finding alternative targets in the Hh pathway, particularly those that function downstream of Smo. The Gli transcription factors represent ideal targets for this purpose, in light of their role as the final intracellular mediator of Hh signal transduction, both ligand-dependent and independent. Given that Gli1 can be either primarily amplified (for example, in gliomas, from which this gene inherits its eponymous designation) (32), or secondarily amplified/overexpressed in the setting of Hh inhibitor therapy (13), a pharmacological strategy targeting Gli could have tangible benefits. Recent small molecule screens have indeed identified such lead candidates, of which a panel of four Hh pathway inhibitors (HPIs) has been extensively characterized in vitro vis-à-vis their mechanisms of action (15). Of these, HPI-1 is a potent inhibitor of Gli function, including exogenously expressed Gli1 in cells, underscoring its potential as an agent of choice in cancers that acquire resistance to Smo antagonists. A major pitfall of the parental HPI-1 compound is its lipophilicity, which makes systemic delivery and adequate bioavailability a challenge.
In this manuscript, we present the initial characterization of a novel nanoparticle formulation of HPI-1 using the polymer PLGA as a backbone. PLGA has been extensively used as a vehicle for enabling systemic delivery of therapeutic agents that otherwise require noxious excipients like cremophor or ethanol, and has been conferred a GRAS status by the USFDA (16, 33). Although the active pharmaceutical ingredient (API) in NanoHHI is identical to that of free HPI-1, we have demonstrated the improved systemic bioavailability of the nanoformulation compared to the free drug upon both oral and parenteral administration in mice. In addition, we have documented the ability of i.v. NanoHHI to cross the BBB, resulting in detectable levels of API in the brain. In this study, we establish the ability of NanoHHI to bypass secondary resistance to “conventional” potent and selective Smo antagonists in a medulloblastoma allograft model. We also shown that in preclinical models of pancreatic cancer, wherein the major component of Hh signaling is paracrine and acquired mutational resistance is potentially less of an issue, NanoHHI has therapeutic efficacy comparable to what has been reported for cyclopamine and related Smo antagonists (18, 23). Although we did not explicitly examine the efficacy of NanoHHI in a model of Gli overexpression (such as in a glioblastoma with GLI1 amplification), data presented in the Hyman et al study suggests that NanoHHI should demonstrate activity in this setting, based on its ability to block Gli function at multiple levels (15). Whether NanoHHI also acts in acquired Smo inhibitor resistance secondary to other mechanisms like activation of the PI3-kinase pathway remains to be seen.
In conclusion, we present a novel polymer nanoparticle formulation of a potent Hh inhibitor, NanoHHI, which demonstrates in vivo ability to circumvent acquired mutational resistance to the commonly used clinical Smo antagonists. In light of its demonstrable efficacy in preclinical allograft and xenograft models of both ligand independent and ligand dependent Hh signaling, and its ability to cross the BBB, NanoHHI has the potential to be utilized across a wide spectrum of tumor types. Future studies will expand the application of NanoHHI into the areas of authochthonous and orthotopic mouse models, including those generated by stereotactic injection within the central nervous system. The oral bioavailability of NanoHHI also provides an avenue for exploring a role for Hh-targeted chemoprevention in cognate systems.
Supplementary Material
References
- 1.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nature reviews. 2003;3:903–11. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M, Maitra A. The hedgehog pathway and pancreatic cancer. The New England journal of medicine. 2009;361:2094–6. doi: 10.1056/NEJMcibr0905857. [DOI] [PubMed] [Google Scholar]
- 3.Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–7. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 5.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 7.Lam CW, Xie J, To KF, Ng HK, Lee KC, Yuen NW, et al. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18:833–6. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 8.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–81. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 9.Theunissen JW, de Sauvage FJ. Paracrine Hedgehog signaling in cancer. Cancer research. 2009;69:6007–10. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- 10.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–10. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends in cell biology. 2007;17:438–47. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–4. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer research. 2011;71:435–44. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 15.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106:14132–7. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu JM, Wang X, Marin-Muller C, Wang H, Lin PH, Yao Q, et al. Current advances in research and clinical applications of PLGA-based nanotechnology. Expert Rev Mol Diagn. 2009;9:325–41. doi: 10.1586/erm.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6:229–40. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer research. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263:1600–3. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 21.Shaikh J, Ankola DD, Beniwal V, Singh D, Kumar MN. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur J Pharm Sci. 2009;37:223–30. doi: 10.1016/j.ejps.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, et al. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci U S A. 2011;107:9736–41. doi: 10.1073/pnas.1004602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, et al. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725–35. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimeno A, Feldmann G, Suarez-Gauthier A, Rasheed Z, Solomon A, Zou GM, et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol Cancer Ther. 2009;8:310–4. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. Journal of the National Cancer Institute. 2010;102:340–51. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 27.Teranishi N, Naito Z, Ishiwata T, Tanaka N, Furukawa K, Seya T, et al. Identification of neovasculature using nestin in colorectal cancer. Int J Oncol. 2007;30:593–603. [PubMed] [Google Scholar]
- 28.Bisht S, Brossart P, Maitra A, Feldmann G. Agents targeting the Hedgehog pathway for pancreatic cancer treatment. Curr Opin Investig Drugs. 2011;11:1387–98. [PubMed] [Google Scholar]
- 29.De Smaele E, Ferretti E, Gulino A. Vismodegib, a small-molecule inhibitor of the hedgehog pathway for the treatment of advanced cancers. Curr Opin Investig Drugs. 2011;11:707–18. [PubMed] [Google Scholar]
- 30.Von Hoff DD, Lorusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, et al. Inhibition of the hedgehog pathway in advanced Basal-cell carcinoma. The New England journal of medicine. 2009;361:1164–72. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 31.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 32.Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–3. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 33.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–47. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.