Abstract
3′-end cleavage and subsequent polyadenylation are critical steps in mRNA maturation. The precise location where cleavage occurs [referred to as poly(A) site] is determined by a tripartite mechanism in which a A(A/U) UAAA hexamer, GU-rich downstream element and UGUA upstream element are recognized by the cleavage and polyadenylation factor (CPSF), cleavage stimulation factor (CstF) and cleavage factor Im (CFIm), respectively. CFIm is composed of a smaller 25 kDa subunit (CFIm25) and a larger 59, 68 or 72 kDa subunit. CFIm68 interacts with CFIm25 through its N-terminal RNA recognition motif (RRM). We recently solved the crystal structures of CFIm25 bound to RNA and of a complex of CFIm25, the RRM domain of CFIm68 and RNA. Our studies illustrated the molecular basis for UGUA recognition by the CFIm complex, suggested a possible mechanism for CFIm mediated alternative polyadenylation, and revealed potential links between CFIm and other mRNA processing factors, such as the 20 kDa subunit of the cap binding protein (CBP20), and the splicing regulator U2AF65.
Key words: cleavage factor, CPSF, CPSF6, CPSF7, protein-RNA complex, 3′ processing, splicing, mRNA processing, alternative polyadenylation, mRNA capping
Introduction
3′ processing of message RNA (mRNA) is an essential maturation step that increases the stability of mRNA, facilitates its export from the nucleus to the cytoplasm, and enhances translation efficiency.1 3′ end formation is a two-step process involving, first, endonucleolytic cleavage at a polyadenylation site [poly(A) site] followed by the addition of a polyadenine tail.2–5 Poly(A) site definition is accomplished through the recognition of specific cis-elements [referred to as poly(A) signal] located on the mRNA by their corresponding protein factors.4,5 Two of the well-studied poly(A) signals are the A(A/U)UAAA hexamer and downstream GU-rich element, which are bound by the cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) complexes, respectively.4,5 A third poly(A) signal consists of UGUA elements and was identified as the preferred binding site of Cleavage Factor Im (CFIm) by SELEX and biochemical analyses.6,7 The tripartite core protein-RNA complexes, together with Cleavage factor IIm serve as a platform to recruit other 3′ processing factors to modulate the efficiency of the cleavage and polyadenylation reaction.1–3,8
CFIm is a two-subunit complex, composed of a small 25 kDa (CFIm25) subunit and a larger 59/68/72 kDa subunit.9 CFIm25 is encoded by one gene, CPSF5, whereas two separate genes, CPSF6 and CPSF7, code for two isoforms of the large subunit, CFIm68 and CFIm59. The third isoform, CFIm72, is an alternatively spliced form of CFIm68.9–11 CFIm68 and CFIm59 both contain an N-terminal RRM domain, a central proline-rich region, and a C-terminal RS-like domain.10,11 The N-terminal RRM of CFIm59/68 mediates the interaction with CFIm25.11 Besides its fundamental role in UGUA-mediated poly(A) site recognition,6,7 CFIm has been shown to influence alternative poly(A) site selection,12–14 mRNA export,15,16 and mRNA splicing.17 The recently solved crystal structures of CFIm25-RNA and CFIm68 RRM-CFIm25-RNA complexes18 taken together with biochemical analyses shed light on the molecular mechanisms underpinning CFIm's specificity for UGUA elements and its role in alternative poly(A) site selection. The major findings resulting from these studies and their implications are discussed below.
CFIm Binds mRNA through the Collaboration of Two Distinct Protein Domains
A quick glance at the domain organization of the two subunits of CFIm might give the erroneous impression that CFIm68 is likely to be the subunit that recognizes UGUA sequence elements, because the RRM it contains is the most abundant single-stranded RNA binding domain in vertebrates.19,20 Furthermore, this motif interacts with RNA in a sequence-specific manner in a large number of instances.21,22 In contrast, CFIm25 possesses a Nudix hydrolase domain,23,24 a motif found in housekeeping enzymes which primarily hydrolyze (di)nucleotides.25,26 However, UV crosslinking11 and gel shift assays27 indicated that CFIm25 is capable of binding RNA. The CFIm68 RRM, on the other hand, enhances RNA binding mediated by CFIm25, but is not able to bind RNA by itself.11
Crystal structures of CFIm25 in complex with an RNA oligonucleotide containing a UGUA element unveiled the molecular basis for sequence specific recognition.27 Comparison with other Nudix proteins revealed that CFIm25 possesses a unique α-helix loop motif preceding its Nudix fold. This α-helix loop motif not only blocks the canonical hydrolase active site, but also provides a scaffold for CFIm25 to bind RNA.27 The UGUA element is recognized via a variety of hydrogen bonding interactions. U1 is mainly recognized by main chain atoms from Phe104, whereas U3 is recognized by the side chain of Arg63. In addition to the interaction with the side chain of Glu55, G2 forms an intramolecular Watson-Crick/sugar-edge base pair with A4.27 Moreover, Phe103 stacks with U1 and G2 to further stabilize the CFIm25-UGUA complex.27
To further investigate the role of the larger CFIm subunit in poly(A) site recognition, we solved a crystal structure of a CFIm68 RRM-CFIm25-RNA ternary complex.18 Consistent with previous observations, CFIm68 and CFIm25 forms a 2:2 heterotetramer.12 However, instead of forming a dimer, two CFIm68 RRM molecules bind to opposite sides of the CFIm25 homodimer.18 The CFIm68 RRM adopts the typical β1α1β2β3α2β4 architecture.22 The RRM contacts CFIm25 through the loops connecting β1/α1 and β2/β3, referred to as loop1 and loop3, respectively. Interactions mediated by loop1 are mainly hydrophobic, whereas loop3 residues participate in hydrogen bonding interactions, involving both side chain and main chain atoms. Mutagenesis analyses illustrated that only loop3 is critical for the CFIm complex formation.
Biochemical data demonstrated a critical role of CFIm68 in looping the intervening sequence between the two UGUA elements bound by the CFIm25 dimer. Two CFIm25 monomers are oriented in an anti-parallel orientation, so that the 5′ end of the two UGUA elements are facing each other. Hence, the intervening sequence needs to loop around to position both UGUA elements in the RNA binding pocket of CFIm25. Mutagenesis analyses attempting to sketch a low resolution RNA path have revealed that the CFIm68 RRM residues located in the clefts formed at CFIm68-CFIm25 interface are essential for RNA looping. A CFIm68 RRM quadruple variant bearing mutated residues in both clefts (W90A/W91A/N117A/R118A) nearly abrogated the RNA binding affinity of CFIm. Unlike other RRMs, which bind RNA across the β-sheet surface formed by the four anti-parallel β-strands,22 CFIm68 RRM is likely to direct the looping RNA beneath the β-sheet surface: Asp94, which is located at the bottom of the RRM, significantly reduced the RNA binding affinity of CFIm when mutated to alanine. Furthermore, an additional C-terminal α-helix (α3) immediately following the RRM blocks the surface of the β-sheet, which is the platform that usually binds RNAs.22 Also, in the CFIm68 RRM one of the three highly conserved aromatic residues that are responsible for RNA binding is replaced by a leucine (L128 on RNP1).22
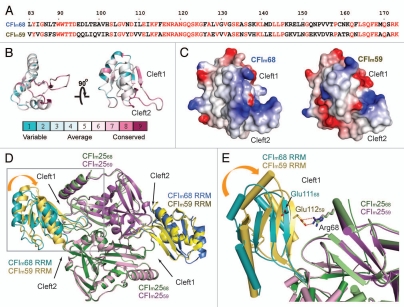
CFIm68, CFIm59 and their homologues were aligned using ClustalW28 and sequence conservation was calculated and mapped onto the CFIm68 RRM model using ConSurf (Fig. 2B).29 In agreement with the proposed RNA path based on the biochemical data, residues in the vicinity of clefts 1 and 2, and located at the bottom of the RRM manifest higher conservation than the rest of the RRM. Taken together these observations suggest that the RNA is unlikely to loop across the β-sheet surface.
Figure 2.
Sequence and structure comparison of CFIm68 and CFIm59. (A) Sequence alignment of the CFIm68 and CFIm59 RRM domains. Identical residues are highlighted in red. (B) Conservation of the CFIm68 RRM was calculated using the ConSurf server29 and displayed with PyMOL.53 (C) The electrostatic surface potential of the CFIm68 (left) and CFIm59 (right) RRM domains was calculated with Delphi58 and colored according to the electrostatic potential (blue, positive; red, negative). Models are in the same orientation as the model on the right in (B). (D) Superposition of the CFIm25/CFIm59 (PDB ID: 3N9U) complex with CFIm25/CFIm68 (3Q2S). Both complexes are shown as cartoon models. CFIm25 monomers complexed with CFIm59 (CFIm2559) are colored in purple and pink, whereas CFIm25 monomers in complex with CFIm68 (CFIm2568) are colored in green and light green. The CFIm59 RRMs are colored in gold and yellow and CFIm68 RRMs are colored in teal and blue. Cleft1 on the left side of the picture is narrower due to a movement in the RRM domain, indicated with the orange curved arrow. The region delineated by a gray rectangle is shown in more detail in (E). (E) A close-up view of cleft1. Arg68 of CFIm25 participates in a salt bridge with Glu112 of CFIm59 (Glu11259), but the same arginine residue is too far away to make contact with the analogous glutamate in CFIm68 (Glu11168). Residues are shown as stick models and colored according to the molecules they belong to. Ionic bonds are shown as red dashed lines. Helices are shown as cylinders for clarity.
A Potential RNA Looping-Mediated Regulation of Alternative Polyadenylation by CFIm
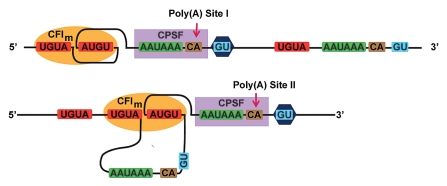
Pre-mRNA may be polyadenylated in several different ways due to the presence of multiple polyadenylation sites in the 3′-UTR.30 Deep sequencing and bioinformatics analyses have demonstrated the prevalence of alternative polyadenylation,8,31,32 which gives rise to mRNAs with 3′ UTR of various lengths30,33,34 and subsequently affects a variety of cellular events, such as gene silencing, tissue differentiation, and development.30,35 A recent report illustrated the involvement of CFIm in alternative polyadenylation in male germ cells.12–14 Moreover, knockdown of either CFIm25 or CFIm complex in Hela cell extracts led to a shift to the use of an upstream poly(A) site.12,13 The ability to loop the intervening sequence between two UGUA elements by CFIm68 provided a potential mechanism for CFIm to regulate alternative poly(A) site selection.18 Gel shift assays using UGUA-containing RNAs of various lengths for the intervening sequence showed that a minimum of 7 nucleotides (7 nt) is required for effective binding by the CFIm68 RRM-CFIm25 complex, whereas longer spacers (9 to 15 nt) enhanced the binding affinity. These data led to the hypothesis that CFIm68 RRM might not restrain the maximal length of the intervening RNA, and might therefore loop out an entire poly(A) site, including the AAUAAA hexamer and downstream GU rich element (Fig. 1). A similar RNA looping-mediated regulation mechanism has been proposed for the splicing regulator pyrimidine track binding protein (PTB).36 The antiparallel organization of the RRMs in PTB may allow the protein to loop out and exclude an entire exon from the mature mRNA.36 Interestingly enough, the 3-subunit cleavage stimulation factor complex (CstF) has been proposed to form a heterohexamer consisting of two copies of each subunits: CstF77, CstF64 and CstF50.3,37–39 Although CstF64 contains only one RRM domain, which has been shown to recognize GU rich element,40,41 CstF64 might achieve a similar RNA looping mechanism facilitated by the dimeric status of CstF complex and thereby influence the usage of an alternative poly(A) site. The hypothesis is consistent with the previous observation that a lower level of CstF64 in plasma B cells correlates with the use of alternative poly(A) sites as compared to pre-B cells.42 We speculate that RNA looping might be a general mechanism utilized by some 3′ processing factors to regulate polyadenylation.
Figure 1.
A model for how CFIm might facilitate alternative polyadenylation.
CFIm68 and CFIm59 Might Play Different Roles in 3′ Processing
CFIm68 and CFIm59 share a similar domain composition that is the hallmark of splicing regulator SR proteins,43 with a central proline-rich region flanked by an N-terminal RRM domain and a C-terminal RS-like domain. On the other hand, CFIm68 possesses an additional glycine-arginine rich (GAR) motif, which is missing in CFIm59. Interesting, CFIm68 has previously been shown to participate in the export of mRNA out of the nucleus and the GAR motif is responsible for the interaction with the mRNA export receptor NXF1/TAP.15,16 These data demonstrated a potentially different function for the two larger subunits. A recent study focused on post-translational modifications also shed light on the role of the multiple forms of the larger subunit.10 In the report, Martin and colleagues identified distinct methylation patterns of arginine residues in CFIm68 and CFIm59, and the different enzymes they are methylated by.10 The RS-like domain of both CFIm68 and CFIm59 are weakly methylated by PRMT1, a member of a family of protein arginine methyltransferases (PRMTs).10,44 The SH3 domain of PRMT2 was found to inter act with CFIm59, but not CFIm68.10,45 On the other hand, the PRMT5 complex only methylates CFIm68 within the GAR motif that is absent in CFIm59.10 The distinct methylation patterns of the two larger CFIm subunits suggested they might play different roles in mRNA 3′ processing, but the function of these modifications have yet to be determined.
Besides the crystal structure of the CFIm68 RRM-CFIm25 complex,18 a structure of a CFIm59 RRM-CFIm25 complex has also been solved recently (Structural Genomics Consortium, Karolinska Institute; PDB ID code 3N9U). Although different RRM constructs were used for crystallization, namely residues 13–235 for CFIm68 and 50–182 for CFIm59, both groups could observe interpretable electron density only for the RRM domain, i.e., residues 81–173 of CFIm68 and 82–177 of CFIm59. A structural comparison between the two complexes provides insight into the function of various CFIm isoforms (Fig. 2D). The overall protein architecture of CFIm59 RRM is very similar to that of CFIm68, with a typical RRM fold appended with a C-terminal helix positioned on top of the β-sheet. (RMSD 1.23 Å calculated on 93 Cα atoms) (Fig. 2D). In addition to the hydrophobic stacking interaction between Phe168 and Tyr127 and van der Waals contacts between Leu165 and Tyr85, which stabilize α3 as observed in CFIm68, Arg159 makes stacking and hydrogen bonding interactions with Phe168 and Gln167, respectively. These additional forces are not observed in CFIm68, since a threonine is located at the position corresponding to CFIm59 Arg159. Another interesting feature within α3 is the hydrogen bond between Ser166 and the main chain carbonyl of residue 162,59 (numbered as in CFIm59), which exists in both CFIm59 and CFIm68. Ser166 is subject to phosphorylation,46 which would disrupt the hydrogen bonding interaction with the main chain carbonyl of residue 161,68 and potentially destabilize helix α3. Interestingly, when this serine was mutated into an aspartate, a phosphate mimic, we observed a two-fold increase in the RNA binding affinity of the CFIm68/CFIm25 complex (data not shown). The potential role of Ser166 phosphorylation in the regulation of mRNA processing will need to be explored further.
Despite the fact that they crystallized in different space groups, the heterotetramer of CFIm59 RRM-CFIm25 is organized in the same manner as the CFI 68 RRM-CFIm25 complex, with two CFIm59 RRMs flanking the CFIm25 homodimer. In order to investigate the relationship between CFIm59 and CFIm68 in RNA binding, we inspected the sequence conservation (Fig. 2A and B) and electrostatics potential of the two RRM domains (Fig. 2C). Most of the surface charges are similar between CFIm59 and CFIm68. One notable difference is that the cleft 2 side of CFIm59 is more negatively charged than in CFIm68. The potential impact of the charge difference on RNA binding will require further experimental investigation.
Although the overall domain architecture of individual subunits are nearly identical between CFIm68 RRM-CFIm25 and CFIm59 RRM-CFIm25 complexes, a superposition of the entire heterotetramer revealed interesting differences (Fig. 2D and E). The large subunit of CFIm contacts the CFIm25 dimer through loop1 and loop3 of the RRM domain, and two clefts are formed at the RRM-CFIm25 interface. These clefts have been proposed to serve as the entry and exit paths for the mRNA bound by CFIm. In the CFIm68 RRM-CFIm25 complex, the exit clefts (designated as cleft2) are quite wide (∼20 Å) whereas the entrance clefts (designated as cleft1) are narrower (∼8 Å). Similarly, in the CFIm59 RRM-CFIm25 complex, cleft2 is open. While one cleft1 exhibits the same width as in the CFI 68 RRM-CFIm25 the other cleft1 is much narrower (∼4 Å), due to a ∼4° shift of the RRM domain which does not affect loop1 or loop3, which remain in the same position (Fig. 2D and E). As a consequence, a salt bridge is formed between Glu112 of CFIm59 and Arg68 of CFIm25 (Fig. 2E). The movement of the RRM domain suggests that CFIm68/CFIm25 and CFIm59/CFIm25 complexes might bind RNA in a different manner, since an RNA molecule is not expected to thread through a 4 Å cleft. We cannot rule out, however, that the RRM movement might be the result of crystal packing interactions. Structures of CFIm in complex with a long RNA containing two UGUA elements will be required to define the path of the intervening looping sequences and possibly shed light on the different roles that CFIm68 and CFIm59 play in RNA binding, in particular and mRNA processing in general.
CFIm may Bridge 3′ Processing with Other mRNA Processing Events
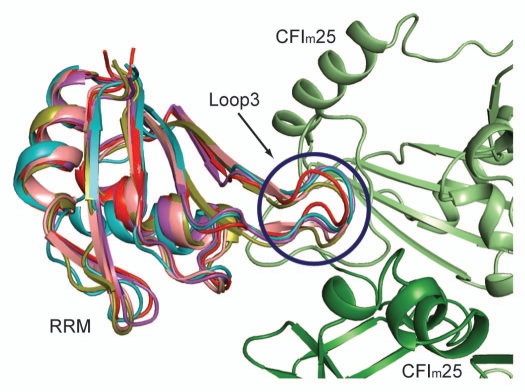
Whereas many studies have focused on elucidating the mechanism of individual steps in mRNA processing, emerging evidence suggests that all steps are intimately connected (reviewed in ref. 47). As an integral part of mRNA processing, 3′ processing is coupled to 5′ capping and splicing.48 An earlier study has identified a physical connection between the cap binding protein (CBP) complex with 3′ processing by showing that the 3′ processing complex was less stable when CBP was depleted from HeLa cell nucleus extract, while addition of recombinant CBP complex restored the 3′ cleavage efficiency.49 The crystal structure of CFIm68 RRM-CFIm25 complex and structural alignment analyses suggest that CFIm25 might be the 3′ processing factor that mediates the connection. CBP is a two-subunit complex composed of a larger 80 kDa (CBP80) and a smaller 20 kDa (CBP20) subunit, which binds to m7GpppG Cap through its RRM domain.50,51 A superposition of CBP20 and CFIm68 illustrated that not only the β-sheets and α-helices are superimposable, but more importantly, loop1 and loop3 aligned nearly perfectly (Fig. 3). As shown by mutagenesis, only loop3 is critical for the formation of CFIm complex. Considering that 4 out of 7 hydrogen bonding interactions mediated by loop3 are through main chain atoms, it is plausible that a protein, such as CBP20 could establish stable contacts with CFIm25, regardless of the amino acid sequence in loop3. Moreover, CBP20 interacts with CBP80 through loop2 and loop4, which are located on the opposite side of loop1 and loop3. This would allow CFIm25 to bind CBP20 without interfering with its binding partner, CBP80.
Figure 3.
Superposition of CFIm68 with RRMs harboring a loop3 of similar shape. The CFIm25 monomers are colored in green and light green. The CFIm68 RRM (PDB ID: 3Q2S18) is colored in teal, CBP20 (1H2V51) in red, U2AF65 (2G4B55) in olive, PAPBC (1CVJ54) in purple and Rna15 (2X1A40) in salmon.
In light of the CBP20-CFIm68 structural alignment, we performed a systematic search for homologous structures of the CFIm68 RRM domain using the DALI server.52 We aligned all the homologs with a Z score greater than 5 and inspected them manually using PyMOL.53 Among 598 homologs (This data set contains duplicates due to the presence of multiple chains in PDB files), only 4 proteins (duplicates were excluded) were found to have the same shape as loop1 and loop3 in CFIm68 (Fig. 3): the first RRM of cytoplasm poly(A) binding protein (PAPBC) (Z score = 12.5, PDB ID: 1CVJ),54 CBP20 (Z score = 11.8, PDB ID: 1H2V),51 Rna15, the yeast homolog of CstF64 (Z score = 10.9, PDB ID: 2X1A)40 and the second RRM of the splicing factor U2AF65 (Z score = 10.4, PDB ID: 2G4B).55 Identification of U2AF65 is intriguing, since the subunits of CFIm have been identified in purified human spliceosomes.56,57 In addition, specific interactions between CFIm and the splicing factor U2AF65 have been established experimentally.17 The sequence alignment comparison revealed a potential molecular mechanism for how CFIm25 and U2AF65 may bridge 3′ processing and splicing. Future efforts will be devoted to validate the direct interactions between CFI 25 and the identified protein factors and investigate the impact these interactions may have on mRNA processing.
Abbreviation
- CFIm
cleavage factor Im
- RRM
RNA recognition motif
Note
After this review was accepted for publication a paper describing a crystal structure of a CFIm complex was published:
Li H, Tong S, Li X, Shi H, Ying Z, Gao Y, Ge H, Niu L, Teng M. Structural basis of pre-mRNA recognition by the human cleavage factor I(m) complex. Cell Res 2011; 21:1039–51.
References
- 1.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Chan S, Martinez-Santibanez G. An up-close look at the pre-mRNA 3′-end processing complex. RNA Biol. 2009;6:522–525. doi: 10.4161/rna.6.5.9554. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahle E, Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 6.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 8.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rüegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 10.Martin G, Ostareck-Lederer A, Chari A, Neuenkirchen N, Dettwiler S, Blank D, et al. Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA. 2010;16:1646–1659. doi: 10.1261/rna.2164210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Yamamoto J, Chen Y, Aida M, Wada T, Handa H, et al. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells. 2010;15:1003–1013. doi: 10.1111/j.1365-2443.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 13.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78:472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- 15.Ruepp MD, Schümperli D, Barabino SML. mRNA 3′ end processing and more—multiple functions of mammalian cleavage factor I-68. Wiley Interdisciplinary Reviews—RNA. 2011;2:79–91. doi: 10.1002/wrna.35. [DOI] [PubMed] [Google Scholar]
- 16.Ruepp MD, Aringhieri C, Vivarelli S, Cardinale S, Paro S, Schümperli D, et al. Mammalian pre-mRNA 3′ end processing factor CFIm68 functions in mRNA export. Mol Biol Cell. 2009;20:5211–5223. doi: 10.1091/mbc.E09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Coseno M, Gilmartin GM, Doublié S. Crystal structure of a human cleavage factor CFIm25/CFIm68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–377. doi: 10.1016/j.str.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 20.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez C, Fisette JF, Chabot B, Allain FH. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–861. doi: 10.1038/nsmb.1814. [DOI] [PubMed] [Google Scholar]
- 22.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Tresaugues L, Stenmark P, Schuler H, Flodin S, Welin M, Nyman T, et al. The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins. 2008;73:1047–1052. doi: 10.1002/prot.22198. [DOI] [PubMed] [Google Scholar]
- 24.Coseno M, Martin G, Berger C, Gilmartin G, Keller W, Doublié S. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36:3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, et al. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Yang Q, Gilmartin GM, Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci USA. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Liu Y, Liu L, Liang C, Li QQ. Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics. 2008;179:167–176. doi: 10.1534/genetics.108.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdisciplinary Reviews: RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 36.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 37.Moreno-Morcillo M, Minvielle-Sebastia L, Mackereth C, Fribourg S. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA. 2011;17:412–418. doi: 10.1261/rna.2481011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 40.Pancevac C, Goldstone DC, Ramos A, Taylor IA. Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res. 2010;38:3119–3132. doi: 10.1093/nar/gkq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 43.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 46.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 47.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. Crystal structure of the human nuclear cap binding complex. Mol Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 51.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLano WL. “The PyMOL Molecular Graphics System”. San Carlos CA, USA: 2008. http://wwwpymolorg. [Google Scholar]
- 54.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 55.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YI, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 58.Nicholls A, Honig B. A rapid finite-difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J Compu Chem. 1991;12:435–445. [Google Scholar]