Abstract
The primary function of the piRNA pathway is to repress the expression and transposition of transposable elements. However, the piRNA pathway has additional biological and developmental functions. These functions are either a consequence of transposon regulation, or they result from direct roles of transposable elements in chromosome structure and gene regulation through piRNAs. Recent data have extended the functions of transposable elements in gene regulation, revealing a trans-acting role of transposable element piRNAs in the control of gene expression. Over the last few years, extensive studies on the piRNA pathway have rapidly increased our understanding of the relationships between transposable elements and the host genome, and of the essential role of transposable elements in biological and developmental processes.
Key words: deadenylation, Drosophila, early development, gene regulation, mRNA decay, oogenesis, piRNA, RNA silencing, translational control, transposable elements
Introduction
Transposable elements are repeated sequences that are able to move and replicate within genomes. They represent a substantial proportion of the genomes in most eukaryotic species, making up as much as 45% of the human genome,1 and about 20% of the Drosophila genome.2 Their parasitic behavior that allows their maintenance within genomes has led to transposable elements being regarded as selfish junk DNA. However, and not surprisingly, since their very discovery in the 1940s, transposable elements were also suspected by reseachers in the field, to have essential functions in gene regulation and in genome dynamics.3 Transposable elements are now being recognized as a driving force in genome evolution (reviewed in refs. 4 and 5). Major ways by which transposable elements can affect genome dynamics include the creation of gene regulation modules through their insertion within, or in close proximity of genes, the increase of genome diversity by promoting genome rearrangements, and the domestication of transposases as a source of DNA-binding domain proteins.5 Other data also suggest the transposable element origin of several microRNAs (miRNAs).6,7 Transposable elements are important structural components of centromeres in many organisms, and of telomeres in Drosophila. Mammalian LINE retrotransposons play an important role in X chromosome inactivation by promoting heterochromatinization, and more recently, LINE activity was shown to contribute to X inactivation in mouse embryonic stem cells.8
Nevertheless, given that such an important part of the genomes is composed of transposable elements, genome integrity could not be maintained without a tight control of transposition. In particular, repressing transposition is specifically important in the germline, which transfers the genetic information from one generation to the next. Recent data have shown that the silencing of transposable elements in the germline depends on Piwi-interacting RNAs (piRNAs), via a mechanism that is similar to RNA interference or RNA silencing involving miRNAs or small interfering RNAs (siRNA) (reviewed in refs. 9–11). piRNAs interact with specific Argonaute proteins and are used as guides for target mRNA recognition and silencing. piRNAs differ from miRNAs and siRNAs in that they are not produced by Dicer enzymes and do not require the formation of stable double-stranded RNA intermediates for their biogenesis.
piRNA biogenesis and function in silencing transposable elements have been extensively studied in the (Drosophila) ovary, and the resulting data have been reviewed recently.12–17 Here I focus on the developmental functions of piRNAs and transposable elements, discussing recent data in the (Drosophila) model that support further examples of the developmental functions of the piRNA pathway.
The piRNA Pathway—A Summary
piRNAs are a specific class of 24- to 30-nucleotide-long non-coding RNAs bound to specific Argonaute proteins of the Piwi clade, Piwi, Aubergine (Aub) and Ago3. They largely derive from transposable elements dispersed in the genome and from clusters—called piRNA clusters—composed of repeated sequences and transposon remnants that are localized in pericentromeric and subtelomeric regions.18–22 Recent studies have established that piRNAs form two distinct groups depending on where they are produced in the (Drosophila) ovary, either in somatic follicle cells, or in germline cells.
Among the three Piwi-type Argonaute proteins, Piwi alone is expressed in the somatic follicle cells that surround the germline cells in the ovary. In these cells, the piRNAs are produced from piRNA clusters transcribed from a single DNA strand, such as the flamenco locus,23,24 and are antisense to transposon mRNAs. Accumulation of these piRNAs depends on, in addition to Piwi, the RNA helicase Armitage, the Tudor domain containing RNA helicase Yb and the putative nuclease Zucchini.25–28 The mechanisms by which these piRNAs repress transposable element expression are not yet known, however they may involve transcriptional silencing since Piwi is mostly nuclear and interacts with Heterochromatin Protein 1a (HP1a).29
The remaining two Piwi-type Argonaute proteins Aub and Ago3 are specific to the germline cells in the ovary. piRNA production in these cells depends on Piwi, Aub and Ago3, and most piRNAs are generated from piRNA clusters transcribed from both genomic strands. Piwi and Aub preferentially bind to piRNAs that are antisense to transposon mRNAs, the most abundant piRNAs, whereas Ago3 preferentially binds to those in the sense strand. Sequence analysis of germline piRNAs led to the conceptualization of the ping-pong model of piRNA amplification in which antisense piRNAs from piRNA clusters, bound mostly to Aub, target transposon mRNA to produce sense strand cleavage products, cleaved by Aub. These then associate with Ago3 to become sense piRNAs after 3′ trimming. Ago3 loaded with sense piRNAs targets and cleaves complementary piRNA precursors from clusters, thus initiating the production of more antisense piRNAs. In this way, the system amplifies the production of piRNAs and contributes to transposon silencing by mRNA cleavage.20–22 Primary piRNAs originating from clusters are required to initiate this amplification cycle and although the mechanism behind their production is not yet known, it might resemble that used in piRNA biogenesis in follicle cells, involving cleavage by Zucchini.
Role of the piRNA Pathway in Axis Specification of the Oocyte
In addition to the Piwi-family Argonaute proteins, several other proteins are involved in piRNA biogenesis and function. These include the RNA helicases Armitage,30 Spindle-E19 and Vasa,31–33 the putative nucleases Squash and Zucchini,34 and the homolog of HP1, Rhino.35 Mutations in the genes encoding these proteins derepress transposable elements and reduce the amounts of piRNAs present in ovaries.33,35
These mutants, including piwi, aub and ago3 show defects in germline development. One of these defects is the mislocalization within the oocyte of major determinants of polarity axes: the mRNAs encoding the TGFa homolog Gurken and the pole plasm determinant Oskar.30,34,35 Affected microtubule network organization is responsible for the mislocalization of these mRNAs which in turn leads to ventralized eggs with reduced or lacking dorsal appendages. Importantly, these defects are in part rescued by mutations in the gene encoding Chk2, a kinase involved in DNA damage signaling.34–37 This prompted the analysis of DNA damage in piRNA pathway mutants. Indeed, an early defect in these mutants is an increase of DNA breaks in germline cells in the germarium, that persist in later stages. These data led to propose a model in which the lack of transposon repression in mutants of the piRNA pathway results in DNA breaks due to high levels of transposition. This in turn induces the DNA damage checkpoint and triggers Chk2-dependent alterations of microtubule organization, leading to defects in oocyte patterning.14,15,34–37 However, the hypothesis that DNA breaks in the germarium of piRNA pathway mutants come from transposition of transposable elements has yet to be tested.
This data suggest that the axis specification defects observed in several mutants of piRNA pathway components are in fact a consequence of transposable element deregulation.
It should be noted, however, that for a number of piRNA pathway mutants — zucchini, spindle-E and piwi— patterning defects are not suppressed by chk2 mutations.34 This suggests that these genes have additional effects that should be independent of the DNA damage checkpoint activity.
Piwi has other functions in germline stem cell biology. All germline cells derive from two to three germline stem cells, that are localized at the anterior-most region of the germarium. Maintenance of these stem cells in part depends on their interactions with somatic niche cells (reviewed in ref. 38). Clonal analysis of piwi mutants has shown that piwi is required in somatic niche cells for germline stem cell maintenance, and that it also has an intrinsic function in germline stem cells for their division.39,40 The molecular basis of these developmental functions remains undetermined, specifically, whether these defects are linked to piRNA biogenesis and/or regulation of transposition.
Gene Regulation by piRNAs
In many systems, the majority of piRNAs do not map to transposons and although the function of most of these piRNAs is not yet known, they could be involved in gene regulation through sequence complementarity to specific genes (reviewed in refs. 15 and 17).
Two examples of direct gene regulation by piRNAs have been reported in Drosophila testes. In fact, the piRNAs that regulate two genes, Stellate and vasa, are the most abundant in Drosophila testes.41,42 Expression of the Stellate locus in testes is repressed by Suppressor of Stellate [Su(Ste)] repeats which produce piRNAs complementary to Stellate. Deletions of Su(Ste) repeats lead to elimination of Su(Ste) piRNAs, Stellate mRNA overexpression and accumulation of Stellate protein in crystals causing male sterility.43,44 Most piRNA pathway components (Aub, Spindle-E, Armitage, Squash, Zucchini and Ago3) are involved in the silencing of the Stellate locus.34,36,44,45 Analysis of the silencing mechanism involved revealed increased Stellate mRNA (but not pre-mRNA) levels and, to a significantly higher extent, increased Stellate protein levels in Su(Ste) deletions or aub mutants.46 This indicates that silencing of Stellate involves mRNA degradation and potentially translational repression.
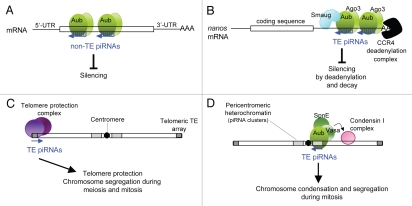
The second most abundant piRNAs in Drosophila testes, called AT-chX (−1 and −2), derive from short repeated sequences on chromosome X. These piRNAs are complementary to vasa mRNA, with three or four mismatches 41. Vasa protein levels are increased in aub and ago3 mutant testes,41,45 and production of AT-chX piRNAs is affected in most piRNA pathway mutants.42,45 In vitro analyses have shown that Aub complexes isolated from testes can cleave in vitro synthesized vasa mRNA, suggesting that vasa silencing could involve mRNA slicing.41 However, the mechanism of silencing has not yet been addressed in vivo (Fig. 1A).
Figure 1.
Role of the piRNA pathway in gene and chromosome regulation. Models of the mechanisms of regulation involving piRNAs. A and B: trans-effect of piRNAs on mRNAs leading to gene regulation. C and D: cis-effect of transposable elements and of potential interacting piRNAs involved in chromosome structure and function. A complete chromosome is represented; it remains unknown whether piRNAs interact with nascent transcripts in these cases. TE, transposable elements; SpnE, Spindle-E. See text for details.
Direct gene regulation by piRNAs might also occur in Drosophila ovaries. In normal ovaries, Oskar protein synthesis is repressed in early oocytes (stages 1 to 6), before Oskar accumulation at the posterior pole starting in stage 8 oocytes.47–49 In aub, armitage, spindle-E, squash and zucchini mutant ovaries, Oskar protein synthesis is derepressed in early oocytes.30,34 This defect has not been reported to be rescued by chk2 mutations and could reflect a direct regulation of oskar mRNA by the piRNA pathway.34
Recent data have shown that a number of piRNAs are produced by the 3′-untranslated region (3′-UTR) of genes in somatic follicle cells of ovaries.50,51 One of the genes that produce piRNAs is the transcription factor traffic jam (tj). tj piRNAs have been suggested to regulate fasIII mRNA which encodes a cell adhesion molecule. This was inferred from increased fasIII mRNA levels observed in mutant testes for tj and piwi, and from the complementarity between tj piRNAs and regions within the fasIII pre-mRNA.50 Potential target sites of tj piRNAs lie in the large first intron of fasIII, not in the mature mRNA, and the mechanism of silencing is not known. Because tj mRNA is sliced into piRNAs, this in turn may also potentially regulate the levels of Tj protein.17
Direct Role of piRNAs from Transposable Elements in Gene Regulation
piRNAs are maternally loaded into embryos52 and are thought to serve as primary piRNAs in the primordial germ cells at the posterior pole of the embryo, to initiate piRNA amplification in the future adult. We recently addressed the role of piRNAs in the bulk of the early embryo.
During early embryogenesis, maternally loaded mRNAs are degraded at the maternal-to-zygotic transition, contributing to the switch from maternal to zygotic control of gene expression.53 In Drosophila, this maternal mRNA degradation depends on the Smaug (Smg) RNA-binding protein expressed before zygotic transcription54 and on a miRNA cluster expressed zygotically.55 Using mRNA encoding the embryonic posterior morphogen Nanos (Nos) as a paradigm to study maternal mRNA decay, we found that nos mRNA degradation depends on its deadenylation by the CCR4-NOT deadenylation complex.56,57 This results from the recruitment of the CCR4-NOT complex onto nos mRNA by Smg.58
Deadenylation by the CCR4 deadenylase is also a major mechanism of RNA silencing by the miRNA pathway.59–62 The CCR4 deadenylation complex is recruited to specific mRNA targets by a complex containing a miRNA, Argonaute, GW182 and Cytoplasmic Poly(A) Binding Protein (PABPC).62–65 While analyzing the role of the RNA silencing pathways in the deadenylation of nos mRNA, we found that the piRNA pathway has a strong effect as several mutants of the pathway showed affected nos deadenylation in embryos. This effect on nos is direct since Smg and the CCR4 deadenylase are in complex with Aub and Ago3 in embryos, and nos mRNA is in complex with Aub protein.66,67 Strikingly, the data suggest that this regulation depends on piRNAs produced by two transposable elements, called 412 and roo. These piRNAs show complementarity with nos 3′-UTR and the deadenylation of nos transgenes is affected following deletion of the target sites of these piRNAs. These data led to a model in which piRNAs guide the interaction of Aub and Ago3 with nos mRNA and help recruit the CCR4 deadenylase leading to nos mRNA decay during the first hours of embryogenesis66 (Fig. 1B). This regulation of nos by the piRNA pathway is essential for anterior-posterior patterning of the embryo. Embryos harboring mutations in piRNA pathway components, or bearing transgenes that lack piRNA target sites, show defects in head development due to ectopic accumulation of Nos protein.
This represents the first example of gene regulation by piRNAs originating from transposable elements, and sheds light on a novel molecular mechanism by which transposable elements act in trans to regulate gene expression and developmental processes.
Interestingly, deadenylation by CCR4 has also been described to occur on transcripts from transposable elements and is thought to depend on piRNAs.68 Therefore a model similar to that described for nos mRNA could be envisaged, in which transposable element transcripts would undergo deadenylation and decay, following CCR4 recruitment by piRNAs and Aub. Whether or not mRNA slicing and the recruitment of deadenylation/degradation enzymes can occur simultaneously on the same mRNA, and what determines the choice between these two pathways, are important questions to address in the future.
While nos mRNA accumulates in the bulk of embryos of aub mutant mothers due to defective deadenylation and decay, these embryos show a complete lack of nos mRNA accumulation at the posterior pole and do not form primordial germline cells.66,69,70 These defects at the posterior pole result in part from earlier defects in oogenesis, specifically, the lack of oskar mRNA localization and translation at the posterior pole of the oocyte. As mentioned above, defects in oocyte patterning arise from the activation of the Chk2 checkpoint kinase in aub mutants. However, the lack of nos mRNA localization at the posterior pole of the embryo is only weakly rescued in chk2 aub double mutants,66 and a direct role for Aub in the localization of nos mRNA at the posterior pole has been proposed.67 It is intriguing to speculate that this function of Aub might also depend on its interaction with nos mRNA via the 412 and roo piRNAs, which in this case would be involved in the stabilization/localization of nos mRNA at the posterior pole, a role opposite to their role in nos deadenylation in the bulk of the embryo.
Role of the piRNA Pathway in Telomere Protection and in Chromosome Condensation During Mitosis
Transposable elements are important structural components of centromeres and telomeres and recent data have highlighted the role of piRNAs in this structural function. In Drosophila, telomeres are protected by specific transposable elements HeT-A and TART, the copy number of which depends on aub and spindle-E.71 It was shown recently that aub and armitage are required for the assembly of the telomere protection complex onto HeT-A transposon. This assembly is proposed to involve a specific population of piRNAs that match HeT-A and TART and that would recruit the protection complex to the telomeres72 (Fig. 1C). This mechanism is essential to prevent telomere fusion and chromosome segregation defects during meiosis and mitoses in the embryo.
In another study, Aub and Spindle-E were shown to form a complex with Vasa in germline cells in the germarium. This complex appears to have a function in chromosome condensation and segregation during mitosis through its direct role in the deposition of condensin I complex onto mitotic chromosomes.73 aub, spindle-E and vasa mutants show a delay in the prometaphase-metaphase transition and defects in chromosome segregation in dividing germline cells in the germarium. The complex was suggested to associate with pericentromeric regions through piRNAs complementary to piRNA clusters (Fig. 1D), although this was not addressed directly.
Concluding Remarks
The discovery of the piRNA pathway as the essential regulator of transposable elements has expedited our understanding of the relationships between transposable elements and the host genome. Initial studies concentrated on deciphering the mecanisms of defense developed by the host genome against transposition. However, further analyses of the piRNA pathway have rapidly deepened our understanding of how transposable elements are intimately linked to essential cellular and developmental functions. Transposable elements affect these functions by two mechanisms, either cis in which transposable elements provide structural or regulatory elements that can recruit piRNAs and/or protein complexes, or trans in which transposable elements produce piRNAs capable of interacting with gene sequences. An important question is whether this latter trans-regulation is widespread. An example in Leishmania shows that all copies of an extinct transposon are inserted within the 3′-UTR of genes leading to reduced stability of the mRNAs.74 This could correspond to an evolutionary intermediate of gene regulation by transposable element piRNAs.
Genes of the piRNA pathway have also been implicated in several biological functions in somatic tissues, including heterochromatin formation, long-term memory and suppression of phenotypic variation (canalization).75–77 These data suggest that piRNAs might be involved in many additional biological and developmental functions that are yet to be discovered.
Acknowledgments
I am grateful to Catherine Papin, Isabelle Busseau, Alain Pélisson, Séverine Chambeyron and Stéphane Ronsseray for helpful discussions and suggestions on the manuscript. Work in the Simonelig lab is supported by the CNRS UPR1142 and by grants from ANR Blanche (ANR-06-BLAN-0343, ANR-2010-BLAN-1201 01), FRM (Equipe FRM 2007 and Projets Innovants ING20101221078) and ARC Libre 2009 (N° 3192).
Abbreviation
- abbreviations
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 3.McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 9.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 10.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin AA, Hannon GJ, Brennecke J. The PiwipiRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 14.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 15.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell. 2010;19:687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Senti KA, Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 20.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 21.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 22.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. Embo J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 2010;286:3789–3797. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 31.Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germ-line of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 32.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004;131:4895–4905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- 39.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 41.Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16:2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 44.Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 2009;37:3254–3263. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- 48.Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development. 1995;121:3723–3732. doi: 10.1242/dev.121.11.3723. [DOI] [PubMed] [Google Scholar]
- 49.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 50.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 51.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, et al. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 54.Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 56.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 59.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Dev Biol. 2010;349:46–52. doi: 10.1016/j.ydbio.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 70.Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 71.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- 74.Bringaud F, Muller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 76.Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2010;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]