Abstract
Hfq-binding small RNAs (sRNAs) are critical regulators that form limited base-pairing interactions with target mRNAs in bacteria. These sRNAs have been linked to diverse environmental responses, yet little is known how Hfq-binding sRNAs participate in the regulatory networks associated with each response. We recently described how the Hfq-binding sRNA Spot 42 in Escherichia coli contributes to catabolite repression, a regulatory phenomenon that allows bacteria to consume some carbon sources over others. Spot 42 base pairs with numerous mRNAs encoding enzymes in central and secondary metabolism, redox balancing, and the uptake and consumption of non-preferred carbon sources. Many of the corresponding genes are transcriptionally activated by the Spot 42-repressor CRP, forming a regulatory circuit called a multi-output feedforward loop. We found that this loop influences both the steady-state levels and dynamics of gene regulation. In this article, we discuss how the CRP-Spot 42 feedforward loop is integrated into encompassing networks and how this loop may benefit enteric bacteria facing uncertain and changing nutrient conditions.
Key words: catabolite repression, Escherichia coli, feedforward loop, glucose, Hfq, regulatory circuit, small RNA, Spot 42
Background
Bacterial survival depends on extracting and consuming available carbon and energy sources from the environment. When multiple carbon sources are present, bacteria can selectively consume carbon sources that are metabolized more efficiently while ignoring other non-preferred carbon sources. This phenomenon, termed catabolite repression, has been studied extensively for over 60 years and has laid the groundwork for our current understanding of metabolic regulation in bacteria.1 Catabolite repression represents a collection of regulatory mechanisms that block the uptake of non-preferred carbon sources and inhibit the expression of unneeded transporter and catabolism genes. In many cases, transcription of transporter and catabolism genes requires the global transcription regulator CRP. In the absence of a preferred carbon source, bacteria synthesize the secondary messenger cAMP that binds and activates CRP. Once bound to cAMP, CRP activates or represses the expression of hundreds of metabolic genes, where many of the genes activated by CRP are responsible for the uptake and catabolism of non-preferred carbon sources.
Of the handful of genes repressed by CRP, one gene (spf) encodes the small RNA (sRNA) Spot 42. This 109-nt sRNA is found throughout enteric bacteria and its sequence is highly conserved. Despite its discovery over 30 years ago, the biological function of Spot 42 was a mystery until 2002, when the Valentin-Hansen group revealed that Spot 42 acts as an Hfqbinding sRNA.2 These sRNAs rely on the RNA chaperone Hfq to form limited basepairing interactions with target mRNAs and thereby alter mRNA stability and translational efficiency.3 The Valentin-Hansen group identified one Spot 42 target, the galK gene encoding galactokinase.4 Galactokinase phosphorylates galactose in the first enzymatic step in the consumption of this non-preferred carbon source. Since most Hfq-binding sRNAs regulate the expression of multiple genes and catabolite repression affects numerous carbon sources besides galactose, we investigated whether Spot 42 plays a broader role in this well-studied phenomenon.
Spot 42 Regulates Numerous Metabolic Genes
Microarray analysis following transient sRNA overexpression has proven to be a dependable technique for the genome-wide identification of targets of Hfq-binding sRNAs.5 Employing this technique, we identified 16 different genes—including galK—that in three independent experiments were modulated at least two-fold following Spot 42 overexpression.6 Many of these genes appear to be direct targets of Spot 42 based on results from base-pairing prediction algorithms, translational reporter fusions and mutational analyses.6 Almost all of the identified genes play roles in central and secondary metabolism, redox balancing or antioxidant biosynthesis. Intriguingly, most of the genes repressed by Spot 42 encode transporters and catabolic enzymes of diverse non-preferred carbon sources. Besides galactose, implicated carbon sources are L-fucose, D-xylose, L-lactic acid, N-acetylneuraminic acid and D-sorbitol. Lowering the cutoff to 1.5-fold regulation in the microarray analyses revealed genes involved in the consumption of other non-preferred carbon sources such as maltose (malX). Some genes may only be regulated at the level of translation, leaving open the possibility that Spot 42 also targets genes besides those we identified.
Since the identified transporter and catabolism genes are repressed by Spot 42, we hypothesized that Spot 42 overexpression would limit growth on the associated carbon sources. As anticipated, high levels of Spot 42 reduced growth on L-fucose, D-xylose, L-lactic acid, N-acetylneuraminic acid and D-sorbitol but not on the unrelated carbon source glycerol or in nutrient-rich LB media. The strength of the growth phenotype varied with the carbon source, suggesting that Spot 42 exerts different amounts of control over each catabolic pathway.
CRP and Spot 42 Participate in a Multi-Output Feedforward Loop
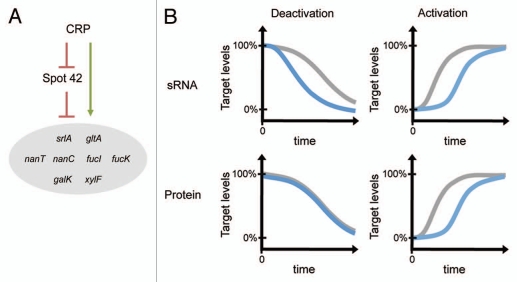
From a network perspective, Hfq-binding sRNAs are thought to act as signal transmitters, where most sRNAs appear to be induced by stress signals and modulate the expression of downstream genes important in the stress response. However, accumulating examples show that sRNAs also function in complex genetic circuits, including feedback loops, feedforward loops and other regulatory configurations.7 To explore the extent to which Spot 42 participates in regulatory circuits, we examined how the identified gene targets are controlled at the transcriptional level. Strikingly, over half of the genes repressed by Spot 42 appear to be transcriptionally activated by Spot 42's repressor CRP. The accompanying regulatory circuit can be called a multi-output coherent feedforward loop (Fig. 1A). Here, CRP activates the expression of multiple target genes directly at the transcriptional level and indirectly through repression of Spot 42. The identification of this regulatory circuit raised the question: does Spot 42 contribute to the regulation of these genes beyond what CRP already provides? Fortunately, a tremendous body of work from Alon and colleagues has begun to address this question for feedforward loops only composed of transcription regulators.8–11
Figure 1.
CRP and Spot 42 participate in a multi-output, coherent feedforward loop. (A) Depiction of the CRP-Spot 42 feedforward loop with known target genes. CRP activates target genes directly at the transcriptional level and indirectly by repressing Spot 42. (B) Feedforward loops containing base-pairing sRNAs or protein transcription regulators may confer different regulatory dynamics. The observed regulatory dynamics (blue) are depicted for the CRP-Spot 42 feedforward loop (sRNA) and an equivalent loop where Spot 42 is replaced with a protein transcription repressor (Protein). The regulatory dynamics of direct regulation (gray) are included as a reference. For each plot, a sustained activating or deactivating signal is applied at time 0.
Transcriptional coherent feedforward loops have been reported to behave differently in comparison to direct regulation in three ways: (1) processing multiple signals, (2) altering steady-state levels and (3) altering dynamics. Since CRP is the only known regulator of Spot 42, we focused on the last two potential differences. We used srlA and fucI, two target genes that appear to be regulated by the CRP-Spot 42 feedforward loop, to assess how Spot 42 alters the regulation of genes already modulated directly by CRP.
Notable differences between the full CRP-Spot 42 feedforward loop and direct regulation by CRP were observed under both steady-state and dynamic conditions. Under steady-state conditions, Spot 42 further repressed target protein levels in the presence of glucose. Under dynamic conditions, Spot 42 delayed the increase in target protein levels following loop activation and accelerated the decline in target protein levels following loop deactivation (Fig. 1B). The delay following loop activation was predicted for transcriptional feedforward loops with the same configuration as the CRP-Spot 42 loop.9 Following loop deactivation, however, the transcriptional feedforward loop displayed the same dynamics as direct regulation.9 In contrast, we found that the CRP-Spot 42 loop accelerated the decline in protein levels following loop deactivation. The difference between the dynamics of the CRP-Spot 42 feedforward loop and the equivalent transcriptional feedforward loop was intriguing, since it suggests that base-pairing sRNAs and protein transcription regulators can confer different network behaviors.
It is worth noting other possible explanations for the observed dynamics. For instance, there could be an unidentified regulator that affects the expression of CRP, Spot 42 or the target genes. Another possibility is that CRP activity fluctuates following the appearance of glucose.12 How these factors contributed to the observed behavior of the CRP-Spot 42 feedforward loop remains to be seen. To determine the true properties of the CRP-Spot 42 feedforward loop, a synthetic gene regulatory network could be constructed with the same regulatory configuration. Since this synthetic network would be insulated from other cellular processes, the dynamics of the loop could be reexamined to say definitively whether feedforward loops with sRNAs behave differently than transcriptional feedforward loops.
The CRP-Spot 42 Feedforward Loop Operates within the Context of Sugar Consumption Operons
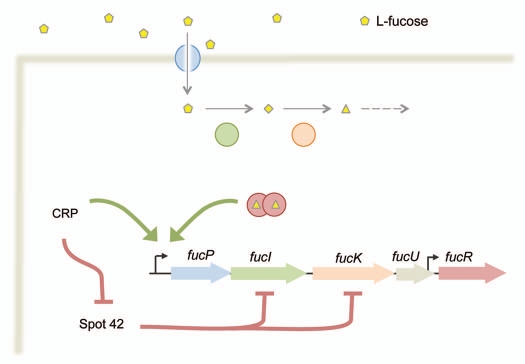
How does the CRP-Spot 42 feedforward loop connect to other regulatory networks? Of the eight genes that appear to be regulated by CRP and Spot 42, seven are encoded in operons responsible for the uptake and consumption of non-preferred carbon sources. For instance, two targets, fucI and fucK, are encoded in the fucPI-KUR operon responsible for the uptake and catabolism of L-fucose (Fig. 2). The last gene in the operon encodes a transcription regulator that induces transcription of the operon in the presence of the FucK product L-fuculose-1-phosphate.13,14 Thus, CRP and Spot 42 target an operon that responds to the operon's cognate sugar in addition to glucose.
Figure 2.
The CRP-Spot 42 feedforward loop interfaces with multiple sugar consumption operons. The fucPIKUR operon is shown as one example. FucP transports L-fucose into the cell, FucI isomerizes L-fucose to L-fuculose, FucK phosphorylates L-fuculose and FucR activates the transcription of the fucPIKUR operon in the presence of L-fuculose-1-phosphate. Not shown is FucU, which interconverts L-fucose between its α and β anomers. The fucPIKUR operon is transcriptionally regulated by CRP and FucR. The fucR gene also is transcribed from a weak promoter in fucU.28 Spot 42 represses the expression of fucI and fucK, which may limit the intracellular accumulation of L-fuculose-1-phosphate and subsequent activation of the fucPIKUR operon in the presence of both L-fucose and glucose.
How do these types of sugar consumption operons respond to both glucose and their cognate sugar? By monitoring a transcriptional readout of different sugar consumption operons, Kaplan and coworkers showed that the response is operon-specific. This can be illustrated for the fucP and galE promoters.15 In glucose-fed cells, the fucP promoter was induced only in the presence of both L-fucose and cAMP. In contrast, the galE promoter was induced in the presence of only D-galactose and showed optimal induction at intermediate concentrations of cAMP.
Sugar consumption operons also have been found to display complex responses at the single-cell level. A prototypical example is the lacZYA operon, which is repressed by glucose and induced by allolactose, a catabolic intermediate derived from lactose. Allolactose binds the LacI repressor and relieves repression of the operon. Glucose import inhibits lactose transport by the LacY permease and partially reduces transcription of the lacZYA operon through deactivation of CRP.16
Early studies of the lacZYA operon utilized the non-hydrolyzable inducer TMG.17,18 In the absence of glucose, the operon showed an “all-or-none” response to TMG, where individual cells were either uninduced or fully induced. This behavior was attributed to positive feedback from the inducing sugar upregulating the expression of the transporter, which transports more inducing sugar into the cell. In the presence of glucose, the “all-or-none” transition shifted to higher TMG concentrations.19 Intriguingly, the TMG concentrations associated with this transition depended on the history of the cells, a phenomenon called hysteresis.19 Here, the TMG concentrations associated with the “all-or-none” transition were lower for initially induced cells than for initially uninduced cells.
Ozbudak and coworkers demonstrated that the “all-or-none” response of the lac-ZYA operon is sensitive to the properties of the regulatory network.19 Introduction of additional binding sites for LacI or use of the cell-permeable inducer IPTG or the hydrolyzable sugar lactose resulted in a graded, unimodal response. Kaplan and coworkers also observed unimodal induction of the araBAD promoter with the hydrolyzable sugar arabinose, although others have reported an “all-or-none” response from the same promoter in other E. coli strains.15,20,21 Therefore, the stability and cell permeability of the inducing sugar as well as strain-specific factors may be important for an “all-or-none” response in sugar consumption networks.
Given the complex responses of sugar consumption operons to glucose and the inducing sugar, how might Spot 42 contribute to these responses? Most of the Spot 42 targets that we identified encode sugar transporters (e.g., srlA) or enzymes that synthesize the inducing molecule (e.g., fucI, fucK). By repressing these genes, Spot 42 may help limit the induction of sugar consumption operons in the presence of glucose and the inducing sugar. However, other Spot 42 targets encode enzymes that break down the inducing molecule (e.g., galK). For these genes, Spot 42 could help improve the induction of sugar consumption operons in the presence of glucose and the inducing sugar. Therefore, depending on the operon, Spot 42 conceivably could limit or improve the induction of sugar consumption operons by the inducing sugar.
What are the Physiological Contributions of Spot 42?
Of the observed and potential contributions of the CRP-Spot 42 feedforward loop, which of these contributions (if any) account for the strong conservation of Spot 42 across enteric bacteria? Although we do not yet have a complete picture of the impact of the loop on the inducibility of targeted sugar consumption operons, we can speculate on how the observed steady-state and dynamic behaviors might benefit cells. Under steady-state conditions, reducing the levels of target proteins may free metabolic resources that can be used toward cell growth and the consumption of the preferred carbon source glucose. Under dynamic conditions, the accelerated loss and delayed accumulation of target proteins may bias cells toward the consumption of glucose. When glucose appears, cells with Spot 42 can more quickly focus on glucose consumption, while, when glucose disappears, cells delay scavenging for nonpreferred carbon sources in case glucose reappears. However, it is also plausible that the steady-state and dynamic contributions of the CRP-Spot 42 feedforward loop provide no evolutionary advantage to cells. Spot 42 instead may have been selected for other reasons and the steady-state or dynamic behavior of the feedforward loop are proverbial spandrels of evolution.22 For instance, the principal role of Spot 42 may be regulating genes in central metabolism, redox balancing and oxidative protection, which may optimize metabolism during the consumption of a preferred carbon source. How then can we determine whether enteric bacteria care about the CRP-Spot 42 feedforward loop and, if so, what properties of the loop are most critical?
One way to pin down the physiological contributions of the CRP-Spot 42 feedforward loop is to identify growth phenotypes in a Spot 42-deletion (Δspf) strain. Hatfull and Joyce found that one Δspf strain of E. coli (CM4722) grew more slowly in comparison to the wild type E. coli strain on solid media at 23°C and in liquid L broth at 23°C and 42°C.23 However, no obvious growth defects were observed for a few different carbon sources and for other strains of E. coli.23 Based on our dynamics data, a Δspf growth phenotype may only be observed under changing nutrient conditions. In addition, our steady-state data suggest that growth phenotypes may appear under specific media conditions (e.g., glucose minimal media) and are subtle enough to require growth competition assays.
While we can envision various experimental conditions that might reveal a Δspf growth phenotype, arguably the best conditions to test reflect the bacteria's natural environment. Enteric bacteria reside in the gut of most animals but also can be found in environments outside their host, such as on vegetation and in sand, soil and water. While still relatively uninformed about the survival of enteric bacteria outside of their host, we are learning more about the lifestyle of enteric bacteria in the gut. Commensal E. coli, the predominant facultative anaerobe in the gastrointestinal tracts of mammals, colonizes the mucosal lining of the intestines and can consume 11 different carbon sources derived from mucus.24 Of the seven carbon sources that appear to be important in colonization, three (N-acetylneuraminic acid, L-fucose and D-ribose) are linked to Spot 42.25 Specifically, we found that Spot 42 represses the expression of transporters and catabolic enzymes associated with N-acetylneuraminic acid and L-fucose, while Autieri and coworkers showed that D-ribose consumption requires the L-fucose catabolic genes also targeted by Spot 42.26 Since consumption of the remaining carbon sources also may be influenced by Spot 42 expression, this sRNA may be an important regulator in enteric bacteria inhabiting the gut.
Assuming CRP is the only regulator of Spot 42, commensal E. coli would need to consume a preferred carbon source to lower cAMP levels and induce Spot 42 expression. Consumption of gluconate, a degradation product of mucus and the major carbon source used by E. coli during gut colonization, reduces cAMP levels almost as efficiently as glucose.25,27 Once gluconate is depleted, the other carbon sources become important, suggesting that Spot 42 could play a role in carbon source selection as bacteria initiate and maintain colonies in the mucosal lining. It will be interesting to see how gluconate consumption affects Spot 42 expression and what the resulting impact is on the consumption of other carbon sources during gut colonization.
Conclusions
The Hfq-binding sRNA Spot 42 is implanted firmly in the regulatory architecture of catabolite repression. Spot 42 regulates numerous genes in metabolism and functions as a co-regulator with CRP, the global regulator of catabolite repression. As part of the CRP-Spot 42 feedforward loop, Spot 42 alters the steady-state and dynamic behavior of CRP regulation and may influence inducibility of targeted sugar consumption operons. Based on all of these contributions of Spot 42, it will be intriguing to see how and why Spot 42 confers a selective advantage to cells. Since Spot 42 is highly conserved across enteric bacteria, Spot 42 and the CRP-Spot 42 feedforward loop are likely to be critical for the survival of enteric bacteria in their natural environments. By continuing to explore the regulatory role of Spot 42, we will expand our understanding of the participation of sRNAs in regulatory networks and, in turn, how sRNAs contribute to bacterial adaptation to harsh and changing environments.
Acknowledgments
Work carried out in the laboratory of G.S. is supported by the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. C.B. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
References
- 1.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 2.Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 3.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Beisel CL, Storz G. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 9.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 11.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paigen K. Phenomenon of transient repression in Escherichia coli. J Bacteriol. 1966;91:1201–1209. doi: 10.1128/jb.91.3.1201-1209.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkus JM, Mortlock RP. Isolation of a mutation resulting in constitutive synthesis of L-fucose catabolic enzymes. J Bacteriol. 1986;165:710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YM, Zhu Y, Lin EC. The organization of the fuc regulon specifying L-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet. 1987;210:331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan S, Bren A, Zaslaver A, Dekel E, Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol Cell. 2008;29:786–792. doi: 10.1016/j.molcel.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn M, Horibata K. Inhibition by glucose of the induced synthesis of the β-galactoside-enzyme system of Escherichia coli. Analysis of maintenance. J Bacteriol. 1959;78:601–612. doi: 10.1128/jb.78.5.601-612.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick A, Weiner M. Enzyme Induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 20.Khlebnikov A, Risa O, Skaug T, Carrier TA, Keasling JD. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J Bacteriol. 2000;182:7029–7034. doi: 10.1128/jb.182.24.7029-7034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 23.Hatfull GF, Joyce CM. Deletion of the spf (Spot 42 RNA) gene of Escherichia coli. J Bacteriol. 1986;166:746–750. doi: 10.1128/jb.166.3.746-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peekhaus N, Conway T. What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun. 2007;75:5465–5475. doi: 10.1128/IAI.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein W, Rothman-Denes LB, Hesse J. Adenosine 3′,5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]