Abstract
The processes controlling glutamate receptor expression early in synaptogenesis are poorly understood. Here, we examine glutamate receptor (GluR) subunit mRNA expression and localization in Drosophila embryonic/larval neuromuscular junctions (NMJs). We show that postsynaptic GluR subunit gene expression is triggered by contact from the presynaptic nerve, approximately halfway through embryogenesis. After contact, GluRIIA and GluRIIB mRNA abundance rises quickly approximately 20-fold, then falls within a few hours back to very low levels. Protein abundance, however, gradually increases throughout development. At the same time that mRNA levels decrease following their initial spike, GluRIIA, GluRIIB and GluRIIC subunit mRNA aggregates become visible in the cytoplasm of postsynaptic muscle cells. These mRNA aggregates do not colocalize with eIF4E, but nevertheless presumably represent mRNP particles of unknown function. Multiplex FISH shows that different GluR subunit mRNAs are found in different mRNPs. GluRIIC mRNPs are most common, followed by GluRIIA and then GluRIIB mRNPs. GluR mRNP density is not increased near NMJs, for any subunit; if anything, GluR mRNP density is highest away from NMJs and near nuclei. These results reveal some of the earliest events in postsynaptic development and provide a foundation for future studies of GluR mRNA biology.
Key words: glutamate receptor, ribonucleoprotein particle, mRNA, Drosophila
Introduction
Most fast intercellular information transfer in the human brain occurs via glutamatergic synapses. The trafficking, clustering and modulation of glutamate receptor protein has been extensively studied. But almost nothing is known about glutamate receptor subunit gene expression during synapse development, or subsequent regulation of GluR subunit mRNAs.
The Drosophila embryonic/larval body wall neuromuscular junction (NMJ) is an easily accessible and genetically malleable model glutamatergic synapse that has provided many important insights into synapse formation and function. It is an ideal model for the study of GluR mRNA during synaptogenesis. However, the expression and localization of Drosophila GluR subunit mRNA in situ has not been characterized.
Drosophila embryogenesis takes 22–24 h at 25°C. Approximately 8–11 h after egg laying (AEL), myoblasts fuse to form syncytial body wall muscles while axons from motor neurons in the ventral nerve cord begin extending from the ventral nerve cord toward them. Body wall NMJ formation begins 10–13 h AEL, when growth cones on motor neuron axons begin contacting and exploring potential postsynaptic muscle targets. Over the next few hours, pre and postsynaptic proteins accumulate at sites of cell-cell contact. As soon as postsynaptic glutamate receptors accumulate, the synapse begins to function.
Postsynaptic glutamate receptor expression depends almost completely on cell-cell contact. Functional glutamate receptors can be detected electrophysiologically on nascent muscles before contact from the presynaptic motor nerve terminal, but there are very few receptors and they are distributed throughout the muscle surface.1 Within minutes of contact from an appropriate presynaptic partner, muscle glutamate receptors begin clustering at the site of cell-cell contact.1–3 New muscle receptors are then added rapidly to the forming NMJ during the latter half of embryogenesis (13–24 h AEL), and then more slowly throughout larval development (24–120 h AEL) as the NMJ grows.1 In the absence of innervation, muscles do not cluster receptors or express additional receptors.1,3 The intercellular signal that triggers postsynaptic receptor clustering and production remains unknown. The steps between reception of this signal and glutamate receptor protein production remain unknown.
Functional ionotropic glutamate receptors require four core protein subunits. Drosophila body wall muscles express five ionotropic glutamate receptor (GluR) subunits, called GluRIIA, GluRIIB, GluRIIC, GluRIID and GluRIIE.4–7 All receptors in the NMJ contain GluRIIC (also known as GluRIII), GluRIID and GluRIIE, plus either GluRIIA or GluRIIB. Thus, Drosophila embryonic/larval NMJs contain two types of glutamate receptor: A-type receptors containing GluRIIA+C+D+E, and B-type receptors containing GluRIIB+C+D+E.4,5,8 The availability of GluRIIA and GluRIIB subunits appears to be rate-limiting for formation of functional glutamate receptors, and the relative abundance of these two subunits determines the relative proportions of each glutamate receptor subtype in the synapse.9,10
Here, we describe for the first time the developmental changes and innervation-dependence of GluRIIA and GluRIIB mRNA expression, as well as the distribution and appearance of postsynaptic GluR subunit mRNA aggregates that likely represent glutamate receptor messenger ribonucleoprotein (mRNP) particles. These insights provide an important foundation for understanding how contact from pre-synaptic cells regulates synapse development, and how the composition and distribution of glutamate receptor mRNPs control glutamate receptor protein expression.
Results
Postsynaptic glutamate receptor production depends on contact between pre and postsynaptic cells. It is thus a contact-dependent form of differentiation. Previous studies used immunohistochemistry and electrophysiology to show that this differentiation involves clustering of functional glutamate receptors at the synapse.1,3 But where did these receptors come from? Cell-cell contact could trigger assembly of pre-synthesized protein subunits, translation of pre-transcribed mRNAs, or synthesis and subsequent translation of appropriate subunit mRNAs. To determine which, we used quantitative real-time RT-PCR and immunoblots (Fig. 1).
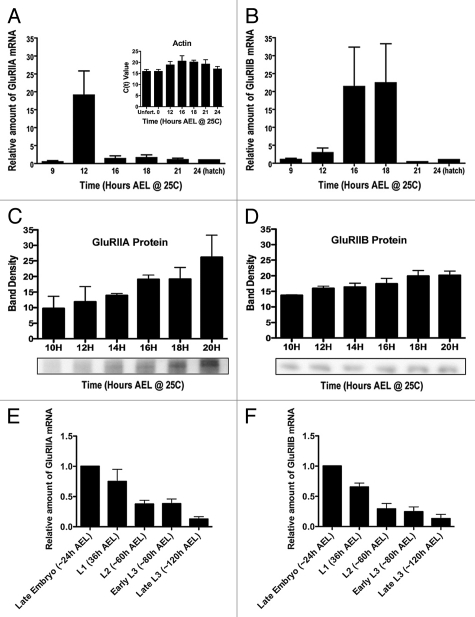
Figure 1.
Expression of Drosophila GluRIIA and GluRIIB mRNA and protein through embryonic and larval development. (A) Amount of GluRIIA mRNA in wildtype (Oregon R) embryos, measured using quantitative real time RT-PC R. Quantity is presented relative to the amount measured at 24 h after egg laying (AEL). N = 5–10 independent mRNA isolations and measurements per time point. Actin 5C mRNA was amplified and measured concurrently with GluRs as a control for mRNA isolation and amplification. Inset: Actin 5C raw C(t) values from unfertilized (unfert.) eggs laid by virgin females, and at various time points during embryogenesis. Note the lack of significant variation. (B) Amount of GluRIIB mRNA during embryogenesis, measured and quantified as described for GluRIIA. (C) Amount of GluRIIA protein, as measured by quantitative immunoblots. N = 3–8 independent protein isolations and measurements per time point. (D) Amount of GluRIIB protein, as measured by quantitative immunoblots. N = 3–8 independent protein isolations and measurements per time point. (E) Amount of GluRIIA mRNA in hatch-age (24 h AEL) embryos, first instar (L1), second instar (L2) and third instar (L3) larvae. N = 3 independent mRNA isolations and measurements per time point. (F) Amount of GluRIIB mRNA in hatch-age embryos and larvae, as described for (E). N = 3 independent mRNA isolations and measurements per time point.
GluRIIA and GluRIIB are rate-limiting for formation of glutamate receptors in the embryonic/larval NMJ; their rate of production and abundance determine the timing and types of glutamate receptors that are produced. Furthermore, quantitative real time RT-PCR can be used to measure abundance of GluRIIA and GluRIIB mRNA because GluRIIA and GluRIIB are expressed only in muscle.6,7 We used this method to quantify GluRIIA and GluRIIB mRNA abundance throughout embryonic and larval development (Fig. 1A).
As shown in Figure 1A, GluRIIA mRNA is expressed at very low but detectable levels before suddenly increasing approximately twenty-fold around 12 hours AEL. Surprisingly, GluRIIA mRNA abundance then falls back to the initial basal level within three hours and remains at that low level until the animal hatches (Fig. 1A). GluRIIB mRNA abundance is also initially very low, but like GluRIIA mRNA rises approximately twenty-fold before falling again to basal levels for the rest of embryogenesis (Fig. 1B). However, the peak of GluRIIB mRNA expression during embryogenesis is slightly delayed (to 16–18 h AEL), compared to that of GluRIIA.
To ensure the reliability of our measurements, actin 5C mRNA was isolated and amplified simultaneously for every GluRIIA or GluRIIB measurement, and used to correct for possible sample-to-sample variation in mRNA isolation and/or amplification.11 As shown in Figure 1A (inset), this variation was minimal, as actin 5C C(t) values were not statistically different across samples. The dramatic changes in GluRIIA and GluRIIB mRNA abundance therefore appear to be real. These mRNA spikes are also visible in data from the modENCODE Drosophila transcriptome project, which uses high-density whole genome tiling microarrays to identify all the transcriptionally active regions of the genome throughout embryogenesis.12 In agreement with our quantitative real-time PCR results, the MODENCODE data show qualitative peaks in GluRIIA and GluRIIB exon expression in mid embryogenesis, with GluRIIB exon expression peaking slightly later than GluRIIA (www.modencode.org/).
Although production of protein depends on the existence of mRNA, the abundance of GluRIIA and GluRIIB mRNA might not necessarily correlate with abundance of GluRIIA and GluRIIB protein. We therefore measured the amount of GluRIIA and GluRIIB protein during embryogenesis by quantitative immunoblotting (Fig. 1C and D). In contrast to the spikes of mRNA abundance, GluRIIA and GluRIIB protein increased gradually through embryogenesis, consistent with the gradual increases in abundance of clustered and functional NMJ glutamate receptors measured in previous studies.3,13
GluR mRNA and protein abundance are apparently disconnected during embryonic development, given that GluR mRNA abundance spikes midway through embryogenesis while protein abundance gradually increases. Is GluR mRNA abundance also disconnected from protein abundance during larval development? Drosophila larval NMJ development has been extensively studied, and it is well established that increasing numbers of glutamate receptors are added to the growing NMJ during larval development. Quantitative real time PCR, however, shows that GluRIIA and GluRIIB mRNA abundance falls gradually through larval development (Fig. 1E and F). This result is also supported by MODENCODE data.
How can GluR mRNA and protein abundance be disconnected? Glutamate receptor protein perdurance is very high: greater than 24 h in larvae.9,14 Therefore, it does not require much mRNA for protein to accumulate—even less as NMJ growth slows in larvae. The only time that a lot of GluR mRNA would be required is at the very start of synaptogenesis, when a large number of receptors must be synthesized de novo. Consistent with this, GluRIIA and GluRIIB mRNA abundance peaks during embryogenesis at about the same time that presynaptic nerves contact postsynaptic muscles, when the need for GluR protein production per unit time is highest. In other words, contact by the presynaptic neuron appears to trigger a burst of transcription such that the postsynaptic muscle is quickly flooded with glutamate receptor mRNA. Presumably, only a subset of the mRNA is subsequently required for translation. This subset is preserved, while the rest of the mRNA is degraded.
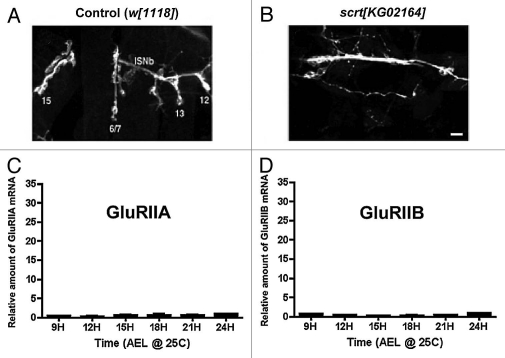
The first step in testing this hypothesis is determining whether GluRIIA and GluRIIB mRNA production in postsynaptic muscle does in fact depend on contact from the presynaptic nerve. To test this, we genetically manipulated pre and postsynaptic cell contact using scratch (scrt) mutants. Scratch is a predicted transcription factor expressed in neuronal precursor cells and required for proper neuronal differentiation.15 In a previous study,16 we showed that scrt[KG02164] mutants extend neuronal axons into the body wall musculature, but the neurons completely fail to form neuromuscular junctions onto muscles. Muscles in scrt[KG02164] mutants therefore develop without significant contact by presynaptic neurons, and there is no detectable expression of GluRIIA or GluRIIB in immunoblots (data not shown). If our hypothesis that postsynaptic GluRIIA and GluRIIB mRNA production is triggered by contact by the presynaptic nerve is correct, then the spikes in GluRIIA and GluRIIB mRNA production shown in Figures 1A and B should be absent from scrt[KG02164] mutant embryos.
Figure 2A shows a confocal micrograph of NMJs formed by intersegmental nerve branch B (ISNb) on ventral muscles 15, 6, 7, 13 and 12 in a single embryonic (∼22 h AEL) hemisegment. Figure 2B shows the same muscle field in a scrt[KG02164] mutant embryo. Note the presence of ISNb, but no NMJs. Figure 2C shows GluRIIA mRNA abundance at various time points during embryogenesis, as in Figure 1A, except in scrt[KG02164] embryos. Figure 2D shows the same thing for GluRIIB mRNA. The spikes in GluRIIA and GluRIIB mRNA abundance measured from wildtype embryos are completely absent.
Figure 2.
GluRIIA and GluRIIB mRNA expression in scratch mutant embryos. (A) Confocal micrograph of motor neuron terminals from intersegmental (ISN) nerve branch ISNb innervating ventral longitudinal muscles 15, 6/7, 13 & 12 in control (w[1118]) embryos (24 h AEL). Motor neuron terminals were visualized using fluorescently-conjugated anti-HRP antibodies. (B) Confocal micrograph as in (A), but showing ISNb in a scratch[KG02164] mutant embryo, where body wall NMJs do not form. Scale bar: 5 um. (C) GluRIIA mRNA abundance measured from scratch[KG02164] mutant embryos, using quantitative real time RT-PCR, as described for Figure 1A. (D) GluRIIB mRNA abundance measured from scratch[KG02164] mutant embryos, using quantitative real time RT-PC R, as described for Figure 1B. Note the lack of GluRIIA or GluRIIB mRNA expression.
We conclude based on these results, that GluRIIA and GluRIIB mRNA transcription is triggered in postsynaptic muscles by contact from the presynaptic nerve. GluRIIA and GluRIIB mRNA abundance rises quickly after cell-cell contact. GluR mRNA abundance drops off rapidly within a few hours of initial synapse formation, then more gradually throughout the rest of embryogenesis and larval development. Despite this drop off in mRNA abundance, GluRIIA and GluRIIB protein production proceeds steadily, leading to gradual accumulation of GluR protein. Presumably, a subset of the initially produced mRNA is preserved and preferentially used for translation. But where is this mRNA?
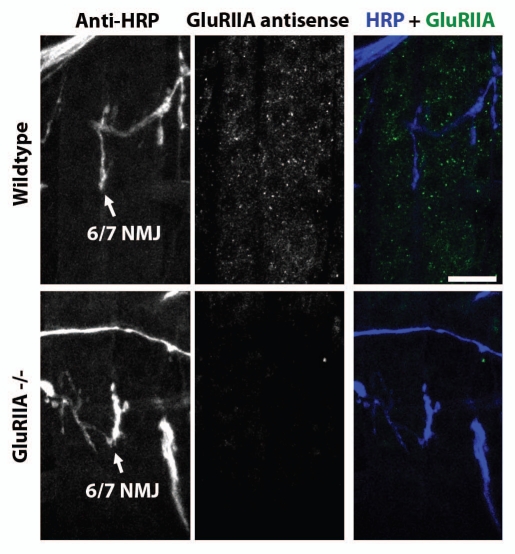
Figure 3 shows confocal micrographs of anti-GluRIIA mRNA Fluorescence In Situ Hybridization (FISH) in combination with immunohistochemistry in muscles of first instar larvae (∼40 h AEL). Specifically, the figure shows FISH and immunohistochemistry in ventral longitudinal muscles 6 and 7, and the NMJ formed in the cleft between the two muscles. Presynaptic axons and nerve terminals have been visualized using anti-HRP antibodies (left column and right column, blue). GluRIIA mRNA has been visualized using fluorescently-labeled GluRIIA mRNA antisense probes and FISH (middle column and right column, green). As shown, the GluRIIA mRNA appeared as small puncta distributed throughout the muscle.
Figure 3.
GluRIIA mRNA aggregates in first instar larval muscles. Parts show confocal micrographs of ventral longitudinal muscles 6 and 7 in one hemisegment of a first instar larvae. Left column shows NMJs in cleft of muscles 6 and 7, visualized using fluorescently-conjugated anti-HRP. Middle column shows anti-GluRIIA FISH signal. Right column shows merge of anti-HRP (blue) and anti-GluRIIA mRNA (green) signal. Top parts show micrographs from a wildtype larva. Bottom parts: As top, but from a GluRIIA[AD9]/Df(2L)Exel8016 mutant embryo, in which the GluRIIA gene is deleted. Scale bar: 15 um.
Several types of experiments confirmed that the puncta revealed by FISH really represented GluRIIA mRNA. First, we performed FISH using GluRIIA sense probes, which should not hybridize to the GluRIIA mRNA. As expected, FISH using sense probes showed no GluRIIA puncta (data not shown). Next, we used FISH to look for GluRIIA mRNA puncta in GluRIIA[AD9]/Df(2L) Exel8016 mutants (GluRIIA-/-), which contain deletions that completely remove the GluRIIA gene. As expected, GluRIIA puncta were not visible in GluRIIA-/- mutants (Fig. 3).
We also performed FISH under conditions where GluRIIA expression is increased. For example, we measured GluRIIA mRNA aggregate abundance after knockdown of Dicer1. Dicer1 is required for production of microRNAs, including microRNAs that suppress GluRIIA and GluRIIB mRNA abundance.10 When Dicer1 levels are reduced, GluRIIA expression is dramatically increased.10 Consistent with the idea that GluRIIA FISH puncta represent GluRIIA mRNA, we observed a large increase in the number of GluRIIA puncta after RNAi-mediated knockdown of muscle Dicer1.10
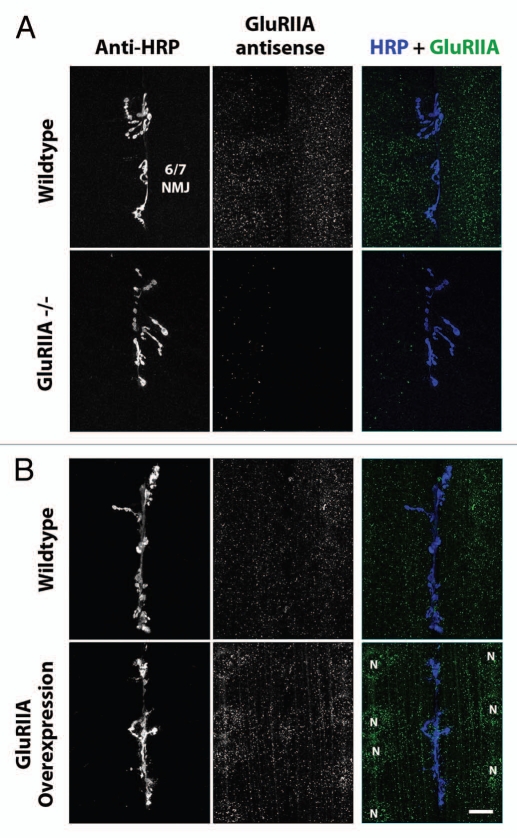
More directly, we overexpressed GluRIIA in third instar larvae (∼100 h AEL; Fig. 4B) using a muscle-specific Gal4 driver (24B Gal4) in combination with a UAS-GluRIIA full-length genomic transgene.14 The GluRIIA mRNA puncta in third instar larvae appeared similar to those in first instar larvae, and were also absent in GluRIIA null mutants (Fig. 4A). But when GluRIIA was overexpressed, the density of GluRIIA mRNA aggregates increased approximately 7-fold, compared to wildtype larvae or 24B-Gal4/+ controls (24B-Gal4 = 1.0 ± 0.4, N = 11; 24B-Gal4; UAS-GluRIIA = 6.9 ± 1.3, N = 13; p = 0.0005).
Figure 4.
GluRIIA mRNA aggregates in third instar larval muscles. (A) Top parts: Confocal micrographs of ventral longitudinal muscles 6 and 7 in one hemisegment of a wild type third instar larvae. Left column shows NMJs, visualized using fluorescently-conjugated anti-HRP. Middle column shows anti-GluRIIA FISH signal. Right column shows merge of anti-HRP (blue) and anti-GluRIIA mRNA (green) signal. Bottom parts: As top, but showing hemisegments from a GluRIIA[AD9]/Df(2L) Exel8016 mutant larva, in which the GluRIIA gene is deleted. (B) Top parts: Confocal micrographs of third instar ventral muscles 6 and 7, as in (A). Bottom parts: As top, but showing hemisegments from a [UAS-GluRIIA/+; 24BGal4/+] mutant larva, in which GluRIIA is overexpressed specifically in muscle cells. Muscle nuclei are labeled with letter ‘N’. Scale bar: 15 um.
Based on these results, we conclude that the GluRIIA FISH signal represents GluRIIA, and that the density of GluRIIA FISH puncta correlates with GluRIIA mRNA abundance.
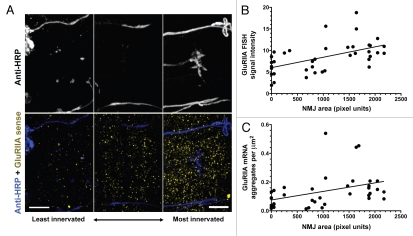
In Figures 1 and 2, we provided evidence that GluRIIA gene expression depends on muscle innervation. The presence of GluRIIA FISH puncta must therefore also be dependent on muscle innervation. To test this, we simultaneously visualized GluRIIA using FISH and motor axon terminals using anti-HRP antibodies, in homozygous prospero[17] mutant embryos. Prospero[17] mutants show delayed, highly variable, and/or absent body wall muscle innervation,1,3 and are therefore ideal for microscopically comparing GluRIIA puncta and degree of innervation (Fig. 5A). We quantified the GluRIIA FISH signal two ways. First (Fig. 5B), we measured total GluRIIA FISH fluorescence signal intensity (average pixel value over the entire visible muscle surface). This was compared to the NMJ size (area, measured in pixels). Consistent with the idea that muscle innervation triggers GluRIIA mRNA expression and GluRIIA mRNA expression correlates with GluRIIA FISH signal intensity, we observed a weakly linear (R2 = 0.29) but highly statistically significant (slope different from 0 at p = 0.0005) correlation between GluRIIA FISH signal intensity and NMJ size (Fig. 5B). We also measured GluRIIA mRNA puncta density by manually counting puncta and dividing by muscle area to calculate mRNA aggregate density (Fig. 5C). Similar to the results obtained by measuring FISH signal intensity, we observed a weakly linear (R2 = 0.15) but statistically significant (slope different from 0 at p = 0.02) correlation between GluRIIA mRNA aggregate density and NMJ size (Fig. 5C).
Figure 5.
GluRIIA mRNA aggregate density is proportional to NMJ size. (A) Confocal micrographs showing muscles 6 and 7 in three different homozygous prospero[17] mutant embryos, in which muscle innervation is delayed and variable. The examples are arranged with an example of a very poorly innervated hemisegment on the left, and a relatively normally-innervated hemisegment on the right. Top parts show anti-HRP signal. Bottom parts show anti-HRP signal (blue) and GluRIIA FISH signal (yellow). Scale bar: 10 um. (B) GluRIIA mRNA FISH signal intensity (average pixel intensity) versus NMJ size. (C) GluRIIA mRNA puncta density (aggregates per square micrometer) versus NMJ size. Lines represent best linear fit to all points.
FISH using whole undissected embryos showed GluRIIA mRNA punctae similar to those visible in dissected larval muscle (Figs. 3–4), but it was more difficult to determine the distribution and density of the mRNPs under these conditions. Qualitiatively, few/no GluRIIA mRNPs were visible in embryonic body wall muscle 6–9 h or 9–12 h AEL (before innervation), while mRNPs were clearly abundant at 12–15 h AEL, consistent with the idea that innervation triggers GluRIIA expression and mRNA aggregate formation. Sense controls, as expected, showed little/no FISH signal (data not shown).
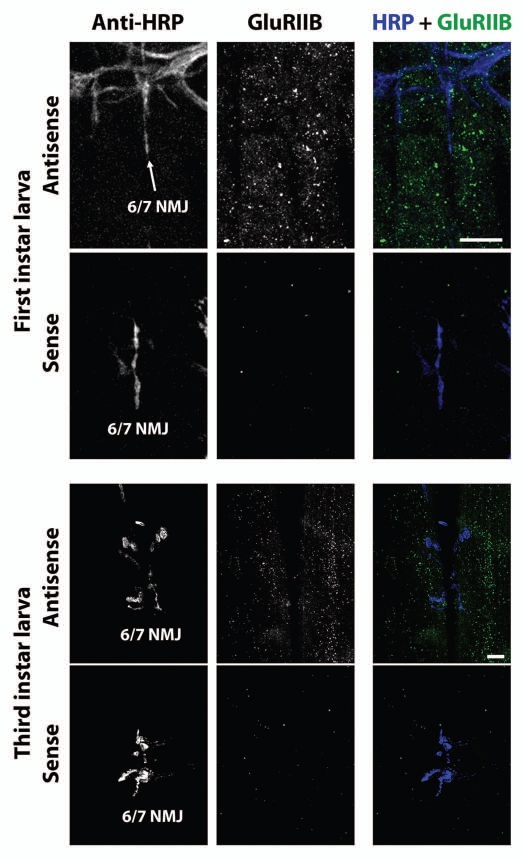
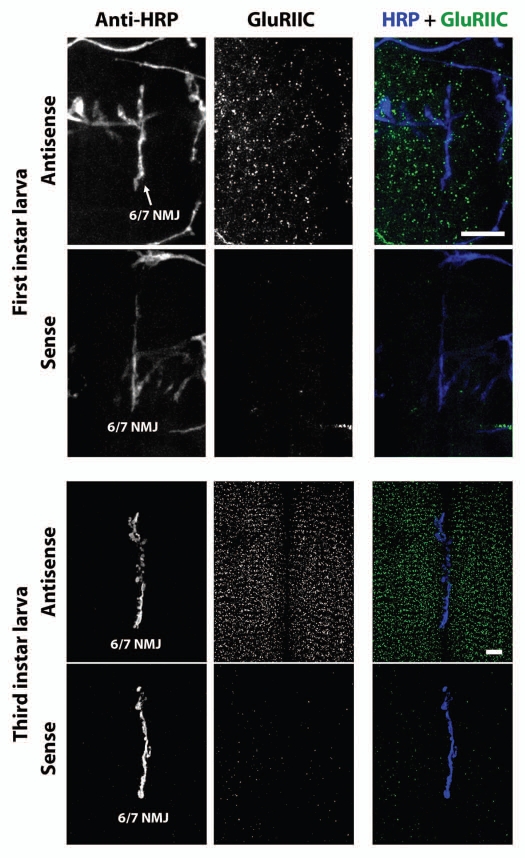
What about other GluR mRNAs? GluRIIA subunits are present in only a subset of Drosophila embryonic/larval body wall NMJ glutamate receptors. Other receptors contain GluRIIB, and both GluRIIA and GluRIIB-containing receptors contain GluRIIC. We therefore visualized GluRIIB and GluRIIC mRNA using FISH. Figure 6 shows confocal micrographs of GluRIIB FISH in ventral longitudinal muscles of first instar (top six parts) and third instar (bottom six parts) larvae. Figure 7 shows confocal micrographs of GluRIIC FISH in ventral longitudinal muscles of first instar (top six parts) and third instar (bottom six parts) larvae. As with GluRIIA, FISH against GluRIIB and GluRIIC revealed what appear to be small mRNA aggregates distributed throughout the muscle cells. Sense probe controls also showed no signal.
Figure 6.
GluRIIB mRNA aggregates in first and third instar larval muscles. Top six parts: Confocal micrographs of first instar NMJs and muscles as in Figure 3, except using antisense probes against GluRIIB instead of GluRIIA. Bottom parts show (lack of) FISH signal resulting from use of sense (negative control) probes. Lower six parts: Confocal micrographs of third instar NMJs and muscles as in Figure 4, except using antisense probes against GluRIIB instead of GluRIIA. Bottom parts show (lack of) FISH signal resulting from use of sense (negative control) probes. Scale bars: 15 um.
Figure 7.
GluRIIC mRNA aggregates in first and third instar larval muscles. Parts are as described for Figure 6, except probes for GluRIIC mRNA were used. Scale bars: 15 um.
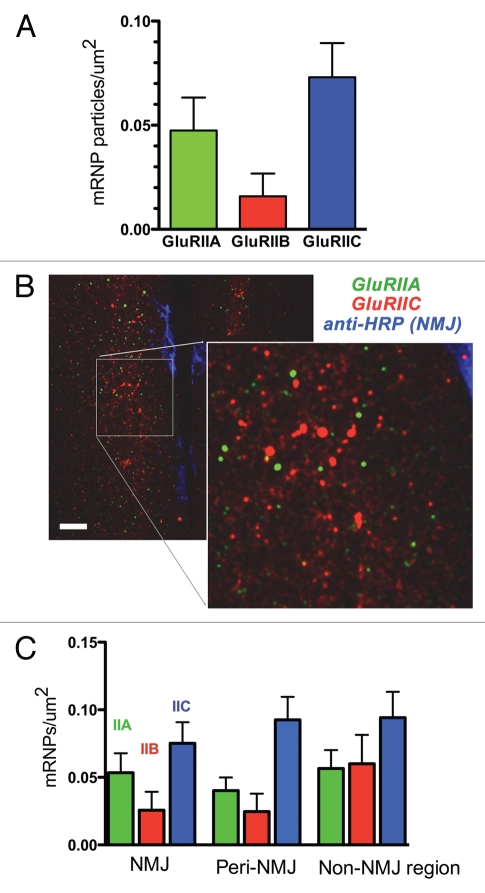
However, close inspection and comparison of GluRIIA, GluRIIB and GluRIIC mRNA aggregates revealed some differences. Primarily, the density of GluRIIC mRNA aggregates appeared to be noticeably increased, relative to GluRIIA or GluRIIB. This is interesting because Drosophila embryonic/larval body wall muscles require approximately double the amount of GluRIIC protein compared to GluRIIA or GluRIIB protein. In other words, the density of GluR mRNA aggregates appears to be proportional to the cells ‘need’ for that particular GluR protein. We quantified this by manually counting the number of mRNA aggregates for each GluR in first instar ventral longitudinal muscles (as described for GluRIIA in Fig. 5), and plotting the relative mRNA aggregate density (Fig. 8A). The number of GluRIIC mRNA aggregates was approximately the same as the number of GluRIIA and GluRIIB mRNA aggregates combined (Fig. 8A), consistent with the fact that all NMJ glutamate receptors contain GluRIIC, plus either GluRIIA or GluRIIC. The number of GluRIIA mRNA aggregates was also approximately double the number of GluRIIB aggregates (Fig. 8A), consistent with the fact that receptors with GluRIIA subunits predominate during embryogenesis and early larval development.10
Figure 8.
Different GluR mRNAs are associated with different mRNA aggregates. (A) mRNA aggregate density in third instar larval muscles 6 and 7, for GluRIIA, GluRIIB and GluRIIC. Values are ‘background-subtracted’ such that the number of punctae in sense controls is zero. (B) Confocal micrographs of third instar ventral longitudinal muscles 6 and 7, after multiplex FISH and simultaneous immunohistochemistry to visualize the NMJ. GluRIIA mRNA aggregates are green; GluRIIC mRNA aggregates are red, and the NMJ is blue. Note that the GluRIIA and GluRIIC mRNA aggregates vary in density and do not overlap. Scale bar: 15 um. (C) GluRIIA, GluRIIB and GluRIIC mRNA aggregate density in muscles 6 and 7, separated according to distance from the NMJ. ‘NMJ’ density was measured in the area delimited by anti-HRP staining. ‘Peri-NMJ’ density was measured outside the anti-HRP staining but within 10 um of the NMJ. ‘Non-NMJ region’ density was measured in muscle areas greater than 10 um from the NMJ. N = 9–14 animals per measurement.
If the density of GluR mRNA aggregates differs between GluRIIA, GluRIIB and GluRIIC, then GluRIIA, GluRIIB and GluRIIC mRNAs cannot all be found in the same aggregates. There must be separate aggregates for each GluR mRNA. To test this explicitly, we performed multiplex FISH to simultaneously visualize GluRIIA and GluRIIC mRNA aggregates, in combination with immunohistochemistry (Fig. 8B). As expected given the quantitative differences in mRNA aggregate density, GluRIIA and GluRIIC mRNA aggregates were physically segregated (Fig. 8B).
Finally, we quantified mRNA aggregate density relative to the NMJ (Fig. 8C). Many important postsynaptic proteins (including CaMKII, calmodulin, PCP4, dendrin, neurogranin, TrkA, TrkB, NMDAR1, GluR2, GluR5 and GlyR A2) are thought to be locally translated in dendrites,17–21 and there is evidence that GluRIIA may be preferentially translated near NMJs.22,23 However, it's unclear whether GluR mRNAs are preferentially localized near NMJs. A previous study by Currie et al. (1995) suggests that GluRIIA is not localized.24 However, the methods utilized in that study were not suitable for quantification, and did not permit visualization of mRNA aggregates or NMJs. We therefore quantified GluRIIA, GluRIIB and GluRIIC mRNA aggregate density relative to the NMJ, using simultaneous FISH and immunohistochemistry. Specifically, we used the anti-HRP signal (which delineates the NMJ) to quantify GluR mRNP density within three muscle regions: “NMJ” (defined by HRP signal), “Peri-NMJ” (within 10 um of any part of the NMJ) and “extra-NMJ” (muscle area outside the area defined by Peri-NMJ region). As shown (Fig. 8C), and consistent with the results described for each individual GluR mRNA (Figs. 3–7), there was no increase in mRNA aggregate density near NMJs for any of the three GluR mRNAs examined. If anything, there was a slight tendency for GluR mRNP density to be highest farther away from the NMJ, although this trend was not statistically significant. Qualitatively, most GluRIIA mRNA aggregates tended to surround nuclei when GluRIIA was overexpressed (Fig. 4B).
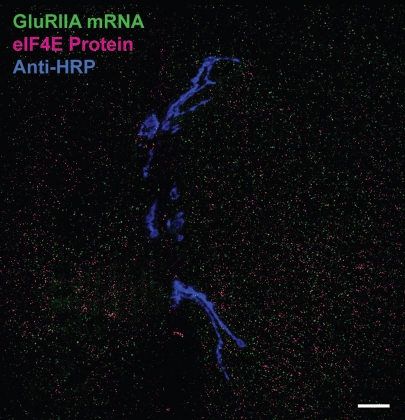
Increasing evidence suggests that mRNA in vivo is continuously associated with a shifting cast of proteins that control mRNA editing, trafficking, translation and stability. These mRNA and proteins often aggregate as so-called ‘messenger ribonucleoprotein (mRNP) particles. The GluR mRNA aggregates that we observed in confocal micrographs using FISH are similar to previously described mRNP particles.25 Furthermore, the density of GluR mRNA aggregates appears proportional to protein need, suggesting that the GluR mRNA aggregates we describe here might represent translating mRNPs. Indeed, GluRIIA mRNA has previously been suggested to be associated with the eukaryotic translation initiation factor eIF4E near Drosophila third instar NMJs.22 The increased resolution of FISH, along with the ability to perform simultaneous immunohistochemistry to visualize eIF4E and NMJs, allows us to test directly whether GluR mRNA is colocalized with eIF4E near NMJs. As shown in Figure 9, eIF4E is, like GluRIIA mRNA, distributed in a punctate pattern throughout muscle cells. However, the nanoscale resolution of FISH and confocal microscopy shows clearly that eIF4E is not significantly colocalized with GluRIIA mRNA aggregates (Fig. 9). We confirmed the lack of colocalization quantitatively by calculating two different background-corrected colocalization measures: Pearson's correlation coefficient and Mander's overlap coefficient.26 Pearson's coefficient for GluRIIA and eIF4E was −0.635 ± 0.038 (N = 7), and the Mander's overlap coefficient was 0.033 ± 0.006 (N = 7). Both values are very low, suggesting no significant overlap. However, to confirm this we rotated the eIF4E channel 90 degrees clockwise relative to the GluRIIA FISH channel using Photoshop and re-measured the GluRIIA/eIF4E overlap coefficients. Pearson's coefficient for GluRIIA and rotated eIF4E was −0.659 ± 0.035 (N = 7), and the Mander's overlap coefficient was 0.016 ± 0.004 (n = 7). The lack of any significant difference confirms that any overlap is essentially coincidental. However, this does not mean that GluRIIA mRNA does not associate with eIF4E at all (see discussion).
Figure 9.
GluRIIA mRNA aggregates do not colocalize with eIF4E protein. Shown is a confocal micrograph of ventral longitudinal muscles 6 and 7 in a hemisegment of a third instar larva. The NMJ has been labeled with anti-HRP antibodies (blue). GluRIIA mRNA aggregates have been labeled with GluRIIA antisense probes (green). eIF4E has been labeled with anti-eIF4E antibodies. Note the lack of overlap between red and green. Results were similar for seven hemisegments examined. Colocalization coefficient statistics are presented in the text.
Discussion
Here, we have shown that GluR subunit gene expression depends on contact between pre and postsynaptic cells. In response to cell-cell contact, GluR subunit mRNA abundance increases rapidly, but then drops off again to very low levels within a few hours and continues to fall throughout larval development. At the same time that overall GluR mRNA levels decrease during embryogenesis, GluR mRNA aggregates appear throughout the postsynaptic muscle cell cytoplasm. Different GluR mRNAs are not colocalized, but seem to form separate aggregates whose density is proportional to amount of protein required by the cell. eIF4E protein does not appear to be a component of the GluR mRNA aggregates.
It is reasonable that contact between the pre and postsynaptic cells turns on glutamate receptor subunit gene expression, given the huge increase in postsynaptic receptor protein that contact triggers.1,3 However, this was not a forgone conclusion. It was equally reasonable for GluR subunit gene expression to be a normal part of muscle development following myoblast fusion, or for contact to trigger translation of pre-existing mRNAs. The signaling cascade that mediates contact-dependent postsynaptic glutamate receptor expression remains unknown. We now know that the pathway ends in muscle nuclei and triggers a large but transient burst of GluR mRNA production. This will facilitate genetic screens for pathway components.
It is interesting that GluR mRNA aggregate density correlates with the amount of protein required. All glutamate receptors in the Drosophila NMJ contain a GluRIIC subunit, plus either GluRIIA or GluRIIB, with GluRIIA being dominant. The number of GluRIIC mRNA aggregates is approximately equal to the number of GluRIIA and GluRIIB mRNA aggregates combined, and there are more GluRIIA aggregates than GluRIIB aggregates. This suggests that the GluR mRNA aggregates we describe here may be involved in translation. However, eIF4E does not appear to be a component of GluR mRNA aggregates (at least for GluRIIA). Since mRNA is only associated with eIF4E while being actively translated, the GluR mRNA aggregates are probably not translating, but rather might serve to preserve or traffick GluR mRNA in preparation for translation.
Our data shows that GluR mRNA aggregates are not preferentially localized near NMJs. These results are consistent with a previous study by Currie et al. that used In Situ Hybridization with DIG labeled RNA probes against GluRIIA followed by colorimetric detection and light microscopy.24 Although the techniques of Currie et al. could not reveal the small GluR mRNA aggregates that we observed, they definitely showed that GluRIIA was distributed throughout muscles, consistent with our results. A subsequent study by Sigrist et al. using similar techniques focused on GluRIIA mRNA near NMJs, but did not explicitly claim that GluRIIA mRNA was found exclusively at NMJs.27 We cannot determine whether or not GluR mRNA may be preferentially translated near NMJs.
If GluR mRNA or any preferentially translated subset of GluR mRNA were localized near NMJs, it would have to be trafficked there. The fact that GluR mRNA aggregates are distributed throughout muscle does not mean that they are not trafficked in interesting ways. Indeed, GluRIIA overexpression led to accumulation of GluRIIA mRNA aggregates near nuclei, possibly due to overload of unknown transport systems. Unfortunately FISH uses fixed tissue and therefore we could not observe GluR mRNA aggregate movement. We tried tagging and visualizing GluRIIA transcripts using the ‘MS2/MCP-GFP system’,28,29 which allows live motion tracking of mRNA down to single mRNA level.29–31 However, unlike FISH, the MS2/MCP-GFP system does not allow visualization native mRNA. Rather, one or more MS2 ‘stem-loop’ sequences are inserted into a transgenic mRNA of interest. This confers a specific 3D structure that can be bound by GFP-tagged ‘MS2 coat binding proteins’ (MCPs) expressed in the same cells. We synthesized a 9X repeat of MS2 sequence with multiple cloning sites and inserted this MS2 sequence into a GluRIIA transgene capable of rescuing loss of GluRIIA in vivo.14 After verifying that this tagged mRNA was indeed being produced and not being degraded, flies carrying the GluRIIA-MS2 transgene were crossed to flies expressing MCP-GFP. The resulting GluRIIA-MS2;MCP-GFP embryos and larvae were then examined. Unfortunately, most of the MCP-GFP was nuclear, consistent with the fact that MCP-GFP contains a default nuclear localization signal and the idea that MCP-GFP was unable to efficiently associate with GluRIIA-MS2. A small amount of GFP was visible in the cytoplasm, but only in rare instances did we see punctae mimicking the distribution of native GluRIIA mRNA. Since FISH represents the ‘gold standard’ for visualization of native mRNA, we concluded that GluRIIA mRNA aggregation is probably disrupted by incorporation of MS2 or association with MCP-GFP, and results involving indirect tagging of transgenic GluRIIA should be viewed with caution. Although it might be possible to optimize relative expression of GluRIIA-MS2 and MCP-GFP such that tagged GFP behaves similar to native mRNA, it should be noted that the MS2/MCP system has been previously used only for highly expressed mRNAs like nanos, gurken and bicoid,28,32 whereas GluRIIA is expressed at relatively low levels.
The best way to determine the function of GluR mRNA aggregates is to disrupt them and see what happens to mRNA stability and GluR protein production. Toward this end, we have begun biochemically isolating and proteomically identifying Drosophila GluR mRNA-associated proteins. Our results confirm that specific proteins are associated with GluR mRNAs. These proteins include highly conserved but previously unnamed proteins representing novel protein families. Disruption of these proteins causes dramatic loss of GluR protein, consistent with the idea that the GluR mRNA aggregates described here represent GluR messenger ribonucleoprotein (mRNP) particles. GluR mRNPs have not been previously described.
Consistent with our multiplex FISH results (Fig. 9), our proteomic screen did not identify eIF4E. This is not surprising. eIF4E associates with mRNA only during active translation. Given the stability of GluR protein (>24 h) and relatively low demand for new protein after initial synaptogenesis, it's likely that GluR mRNAs are not being actively translated most of the time. Instead, they appear to be sequestered in aggregates for utilization as needed.
In summary, we have presented the first description of GluR mRNA aggregates in relation to each other and the glutamatergic synapse they support. Despite intense interest in synapse formation, receptor trafficking and receptor localization, relatively little interest has been paid to various aspects of glutamate receptor subunit gene expression. Gene expression encompasses many processes, including nuclear transcription and transcript processing (capping, splicing, editing, polyadenylation, etc., and finally nuclear export), through cytoplasmic mRNA trafficking, sequestration, translation and eventual degradation.33 All of these processes are mediated and/or regulated by mRNPs,34 which are visible as aggregates, or ‘granules’ in the nucleus or cytoplasm. We propose that the GluR mRNA aggregates described here are bona fide GluR mRNP particles, probably required for GluR mRNA stability and/or translation, and that studying their composition and function will lead to important insights concerning glutamate receptor gene expression and nervous system development.
Materials and Methods
Genetics.
Flies were maintained on standard cornmeal agar food. Embryos and larvae were raised on apple-juice agar plates supplemented with yeast paste. GluRIIA mRNA was overexpressed using a UAS-full length GluRIIA genomic transgene previously shown capable of driving substantial increases in GluRIIA-containing receptor abundance,14 in combination with the mesoderm-specific Gal4 driver 24B. UAS-GluRIIA flies were gifts from Stephan Sigrist (Freie Universität Berlin, Berlin, Germany). GluRIIA deletion mutants were gifts from Aaron DiAntonio (Washington University, St. Louis MO). Other fly strains were obtained from the Bloomington Indiana Drosophila stock center (flystocks.bio.indiana.edu/).
Real-time RT PCR.
Quantitative real time RT-PCR to quantify GluR mRNA abundance was performed as previously described in references 4, 10, 16 and 35. First, total mRNA was extracted from whole embryos or larvae using standard Trizol (Invitrogen) extraction.36 Next, mRNA was reverse transcribed using oligo dT primers and a SuperScript III RT Kit (Invitrogen). Real-time PCR (Opticon 2; MJ Research) was then performed concurrently for GluRIIA, GluRIIB and Actin-5C using target-specific primers and SYBR green (Biorad) for amplicon detection and quantitation. Relative mRNA levels were quantified using the method detailed by Horz et al. (2004). For this method, the cycle threshold (C(t)) values for GluRIIA, GluRIIB and actin 5C were first determined for each sample. C(t) values were then corrected for mRNA isolation and amplification using the formula: ΔC(t)sample = C(t)target − C(t)actin 5C. These corrected sample ΔC(t) values were then referenced to C(t) values from concurrently-run age-specified wild-type control samples (the “calibrator”) to determine the relative amount of target mRNA in the test sample. The formula used for determining relative target levels was: ΔΔC(t)sample = ΔC(t)sample − ΔC(t)calibrator. The amount of target for each sample is then reported as a ‘fold difference’ relative to the calibrator using: 2−ΔΔC(t). Essentially, actin 5C serves as an internal control for sample-to-sample variations in mRNA isolation and PCR amplification, while the calibrator provides a “standard” quantity of mRNA abundance to which other samples (time points, mutants, etc.,) can be compared. Other aspects of the calculation correct for the fact that PCR product is exponentially (rather than linearly) related to the amount of starting template.
For embryo collection and mRNA isolation (Figs. 1 and 2), cages with Oregon R or scrt[KG02164] flies were set up over yeast-supplemented apple juice & agar plates. Plates were changed every hour. Once embryos reached the desired age, they were homogenized in Trizol for mRNA isolation and extraction. 100 embryos were used per sample. For larval mRNA measurements, animals were allowed to grow on apple juice and agar plates supplemented with yeast paste until they reached the appropriate stage. They were then homogenized in Trizol for mRNA isolation and extraction.
Unfertilized eggs laid by virgin females showed no real-time RT-PCR signal for either GluRIIA or GluRIIB. GluRIIA and GluRIIB null mutants also showed no real-time RT-PCR signal for either GluRIIA or GluRIIB.10
Immunoblots.
Immuno (western) blots were performed as previously described in references 35 and 37. We controlled for variation in protein extraction and isolation by measuring protein concentration in every sample by Bradford Assay, then loading each lane with 50 µg of protein. Measurements GluR protein quantity are therefore relative to total protein quantity rather than any particular protein, which could vary from genotype to genotype. Protein quantity was then measured from the bands using Quantity One software (BioRad).
FISH probe constructs.
cDNAs for GluRIIA and GluRIIB were gifts from Stephan Sigrist; GluRIIC cDNA (RE 65796) was obtained from the Drosophila Genomics Resource Center.
Fluorescence in situ hybridization (FISH) and immunohistochemistry.
RNA probes were synthesized in vitro from full-length target cDNAs, using standard methods appropriate to the cDNA vector and orientation (e.g., SP6 RNA polymerase for vectors containing an SP6 promoter at the 3′ end to generate an antisense probe). After synthesis, probes were fragmented by alkaline hydrolysis for better penetration of tissues38 and precipitated using standard methods. In most cases, the probe was labeled with digoxigenin-UTP (DIG-UTP; Roche) during transcription and then eventually visualized using anti-DIG antibodies (monoclonal mouse, Jackson Immuno). In some cases, the synthesis reaction included an amine-modified UTP that could be directly labeled with a fluorophore (“FISH tag” kit; Invitrogen) eliminating the need for secondary detection by antibodies. In situ hybridization was then performed using standard methods similar to those described previously in references 22, 39 and 40. In brief, manually filleted first or third instar larvae were fixed for 1 h in 4% paraformaldehyde in phosphate-buffered saline (PBS), and then permeabilized by incubating in 0.1% tween-20 in PBS for an hour, followed by pre-hybridization in [50% deionized formamide (Sigma), 4x Saline Sodium Citrate buffer (Sigma), 1x Denhardt's Solution (Invitrogen), 0.25 mg/ml yeast tRNA (Invitrogen), 0.25 mg/ml sheared salmon sperm DNA (Ambion), 0.05 mg/ml heparin (Sigma), 0.5% tween-20 in DEPC water] for 1 to 4 h at room temperature. Samples were incubated with RNA probe (∼1 ng/µl) in hybridization buffer (same as pre-hybridization buffer with the addition of 2.5% Dextran Sulfate) at 55°C overnight. Samples were then washed for 24 h in wash buffer [50% Deionized Formamide, 2x Saline Sodium Citrate buffer, 0.5% tween-20 in DEPC water] and then stained with antibodies.
Antibodies used were: anti-eIF4E antibodies to visualize eIF4E protein, anti-HRP antibodies to visualize NMJs, and/or anti-DIG antibodies to visualize mRNA. Antibodies were obtained from Jackson Immunoresearch.35 Except for anti-eIF4E antibodies, which were a generous gift from Paul Lasko (McGill University, Montreal Canada). For simultaneous visualization of multiple different GluR mRNAs, mRNA probes were directly tagged with fluorophores during probe synthesis; a different color was chosen for each probe.
Imaging.
Imaging was done using a laser scanning confocal system (Fluoview FV500; Olympus) with a 40x/1.3 NA UPlan-FIuar or a 60x/1.4 NA Plan-Apochromat objective lens (Olympus) using the sequential scanning mode to avoid any ‘bleed through’ artifacts. These were then converted to maximum-intensity Z-projections using ImageJ. Each GluRIIA/B/C FISH experiment was always performed in parallel with a negative control (sense probe or mutant).
Quantification of mRNP density from FISH images.
To calculate GluR mRNA aggregate density, GluRIIA/B/C mRNA aggregates in each FISH image were counted manually, and this number was divided by muscle area. Density was measured from both muscle 6 and muscle 7 in each image, and averaged to generate each value. Fluorescence signal intensity represents ‘mean pixel intensity’, measured using Adobe Photoshop.
Colocalization.
Colocalization of eIF4E immunoreactivity and GluRIIA mRNA FISH signal was quantified using Colocalizer Express 1.6.1 software (Colocalization Research Software), per procedures described in reference 26.
Acknowledgments
We would like to thank Pei-San Ng for help with the western blots, Kaiyun Chen for help with first instar dissections, Paul Lasko for the eIF4E antibodies, Aaron DiAntonio for GluRIIA [AD9] flies, Stephan Sigrist for GluRIIA and GluRIIB FISH probe plasmids and the UAS-genomic GluRIIA flies, the Drosophila Genomics Resource Center (DGRC) for the GluRIIC cDNA clone, and the Bloomington Drosophila Stock Center for other fly lines. We are also grateful to the University of Illinois at Chicago Research Resource Center (RRC), which provided essential reagents and DNA services.
References
- 1.Broadie K, Bate M. Innervation directs receptor synthesis and localization in Drosophila embryo synaptogenesis [see comments] Nature. 1993;361:350–353. doi: 10.1038/361350a0. [DOI] [PubMed] [Google Scholar]
- 2.Featherstone DE, Rushton E, Broadie K. Developmental regulation of glutamate receptor field size by nonvesicular glutamate release. Nat Neurosci. 2002;5:141–146. doi: 10.1038/nn789. [DOI] [PubMed] [Google Scholar]
- 3.Chen K, Featherstone DE. Discs-large (DLG) is clustered by presynaptic innervation and regulates postsynaptic glutamate receptor subunit composition in Drosophila. BMC Biol. 2005;3:1. doi: 10.1186/1741-7007-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Featherstone DE. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J Neurosci. 2005;25:3199–3208. doi: 10.1523/JNEUROSCI.4201-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin G, Schwarz T, Kittel RJ, Schmid A, Rasse TM, Kappei D, et al. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J Neurosci. 2005;25:3209–3218. doi: 10.1523/JNEUROSCI.4194-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster CM, Ultsch A, Schloss P, Cox JA, Schmitt B, Betz H. Molecular cloning of an invertebrate glutamate receptor subunit expressed in Drosophila muscle. Science. 1991;254:112–114. doi: 10.1126/science.1681587. [DOI] [PubMed] [Google Scholar]
- 7.Petersen SA, Fetter RD, Noordermeer JN, Goodman CS, DiAntonio A. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 1997;19:1237–1248. doi: 10.1016/s0896-6273(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 8.Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci. 2004;24:1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid A, Hallermann S, Kittel RJ, Khorramshahi O, Frölich AM, Quentin C. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat Neurosci. 2008;11:659–666. doi: 10.1038/nn.2122. [DOI] [PubMed] [Google Scholar]
- 10.Karr J, Vagin V, Chen K, Ganesan S, Olenkina O, Gvozdev V. Regulation of glutamate receptor subunit availability by microRNAs. J Cell Biol. 2009;185:685–697. doi: 10.1083/jcb.200902062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horz HP, et al. Monitoring microbial populations using real-time qPCR on the MJ research Opticon 2 system. MJ Research Application Note. 2004:3. [Google Scholar]
- 12.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broadie K, Bate M. Innervation directs receptor synthesis and localization in Drosophila embryo synaptogenesis. Nature. 1993;361:350–353. doi: 10.1038/361350a0. [DOI] [PubMed] [Google Scholar]
- 14.Rasse TM, Fouquet W, Schmid A, Kittel RJ, Mertel S, Sigrist CB, et al. Glutamate receptor dynamics organizing synapse formation in vivo. Nat Neurosci. 2005;8:898–905. doi: 10.1038/nn1484. [DOI] [PubMed] [Google Scholar]
- 15.Roark M, Sturtevant MA, Emery J, Vaessin H, Grell E, Bier E. scratch, a pan-neural gene encoding a zinc finger protein related to snail, promotes neuronal development. Genes Dev. 1995;9:2384–2398. doi: 10.1101/gad.9.19.2384. [DOI] [PubMed] [Google Scholar]
- 16.Liebl FL, Werner KM, Sheng Q, Karr JE, McCabe BD, Featherstone DE. Genome-wide P-element screen for Drosophila synaptogenesis mutants. J Neurobiol. 2006;66:332–347. doi: 10.1002/neu.20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol. 2005;64:116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- 18.Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- 19.Pfeiffer BE, Huber KM. Current advances in local protein synthesis and synaptic plasticity. J Neurosci. 2006;26:7147–7150. doi: 10.1523/JNEUROSCI.1797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bramham CR. Local protein synthesis, actin dynamics and LTP consolidation. Curr Opin Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 22.Sigrist SJ, Thiel PR, Reiff DF, Lachance PE, Lasko P, Schuster CM. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 2000;405:1062–1065. doi: 10.1038/35016598. [DOI] [PubMed] [Google Scholar]
- 23.Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Currie DA, Truman JW, Burden SJ. Drosophila glutamate receptor RNA expression in embryonic and larval muscle fibers. Dev Dyn. 1995;203:311–316. doi: 10.1002/aja.1002030303. [DOI] [PubMed] [Google Scholar]
- 25.Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, et al. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinchuk V, Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Current protocols in cell biology/editorial board, Bonifacino JS, [et al.] 2008;4:19. doi: 10.1002/0471143030.cb0419s39. [DOI] [PubMed] [Google Scholar]
- 27.Sigrist SJ, Thiel PR, Reiff DF, Schuster CM. The postsynaptic glutamate receptor subunit DGluR-IIA mediates long-term plasticity in Drosophila. J Neurosci. 2002;22:7362–7372. doi: 10.1523/JNEUROSCI.22-17-07362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 29.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 30.Singer RH. RNA localization: visualization in real-time. Curr Biol. 2003;13:673–675. doi: 10.1016/s0960-9822(03)00605-5. [DOI] [PubMed] [Google Scholar]
- 31.Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- 32.Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 34.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebl FL, Chen K, Karr J, Sheng Q, Featherstone DE. Increased synaptic microtubules and altered synapse development in Drosophila sec8 mutants. BMC Biol. 2005;3:27. doi: 10.1186/1741-7007-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts DB. Drosophila: A Practical Approach. Oxford: Oxford University Press; 1998. p. 2. [Google Scholar]
- 37.Chen K, Augustin H, Featherstone DE. Effect of ambient extracellular glutamate on Drosophila glutamate receptor trafficking and function. J Comp Physiol A. 2008;195:21–29. doi: 10.1007/s00359-008-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 39.Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, -beta and -gamma during rat embryonic development. Endocrinology. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 40.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:88. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]