Abstract
The mitogen-activated protein (MAP) kinase cascades are important signaling components that mediate various biological pathways in all eukaryotic cells. In our recent publication,1 we identified AtMPK4 as one of the downstream targets of AtMKK6 that is required for executing male-specific meiotic cytokinesis. Here we provide evidence that another target, AtMPK13, is developmentally co-expressed with AtMKK6 in Arabidopsis, and both AtMPK13 and AtMKK6 display high Promoter::GUS activity in the primary root tips and at the lateral root primordia. Partial suppression of either AtMKK6 or AtMPK13 expression significantly reduces the number of lateral roots in the transgenic lines, suggesting that the AtMKK6-AtMPK13 module positively regulates lateral root formation.
Key words: MAP kinase modules, lateral root, RNAi, developmental specificity, pericycle
The Proposed AtMKK6-AtMPK13 Module
The dual-specificity MAP kinase kinases reside in the center of the MAP kinase signaling module and transmit signals by interacting with and phosphorylating MAP kinases on threonine and tyrosine residues of the conserved-TXY-motif. All plant phyla examined seem to possess fewer MKKs than either MAPKKKs or MPKs,2 suggesting a common theme that one MKK might be capable of phosphorylating several MPKs. In fact, Arabidopsis makes efficient use of MKKs by recruiting different MPKs to the same MKK in different biological contexts.3 Among the ten Arabidopsis MPK kinases, AtMKK6 has long been proposed to specifically regulate phragmoplast expansion during cytokinesis, with AtMPK13 as its downstream target, based on homology between AtMPK13 and NtNRK1 in tobacco.4 NtNRK1/NtNTF6, as well as its alfalfa ortholog, MsMMK3, has been functionally characterized, and both of them are regulated in a cell cycle dependent manner.5–7 Consistent with such a model, heterologouslyexpressed AtMKK6 was shown to activate AtMPK13 in yeast.8 Meanwhile, AtMKK6 was also shown to phosphorylate AtMPK18 when transiently co-expressed with this AtMPK in Nicotiana benthamiana leaves.9 In a large scale protein microarray screen, AtMKK6 was reported to activate AtMPK3, AtMPK6 and AtMPK8, but in this study only 10 AtMPKs were assayed, and these did not include AtMPK13.10 A directed yeast two-hybrid assay using the full set of AtMKKs and AtMPKs revealed that AtMKK6 can interact with AtMPK4, AtMPK11, AtMPK6 and AtMPK13.11 Recently, AtMPK4 was demonstrated to serve as a target of AtMKK6 in processing somatic cytokinesis in the root,12 as well as in male-specific meiotic cytokinesis.1 By contrast, no cytokinesis defect was observed for the homozygous mpk13 T-DNA insertion mutant (SALK_130193).12 However, two variants of the AtMPK13 mRNA are expressed in Arabidopsis and this insertion event disrupts only the long coding sequence (CDS) of AtMPK13, and not the short CDS.1 Therefore, a clear picture of the biological function of the proposed AtMKK6-AtMPK13 cascade is still lacking.
AtMKK6 and AtMPK13 are Co-Expressed Developmentally
A comprehensive co-transcriptional analysis of 114 Arabidopsis MAP kinases was conducted earlier, using publicly available microarray data.13 In this study, AtMPK4, AtMPK13 and AtMPK20 are the MPKs whose expression is most closely correlated with that of AtMKK6 during plant development, and together they were assigned to the “shoot apex” group. In addition, AtMKK6 and AtMPK13 expression patterns are associated with mitotic activity, as are two MAPKKKs, AtMAPKKK2 and AtMAPKKK12. In silico analysis using the AtGenExpress Visualization Tool (AVT) supported the idea that AtMPK13 is co-expressed with AtMKK6 during different developmental stages (jsp.weigelworld.org/expviz/expviz.jsp?experiment=development&normalization=absolute&probesetcsv=At5g56580%2Cat1g07880&action=Run). We confirmed this co-expression pattern experimentally by RT-PCR analysis, using RNA samples from 15 different plant tissues/organs, where we compared the AtMKK6 and ATMPK13 patterns to the ubiquitous expression patterns of AtMPK4 and AtMPK6, and the restricted pattern of AtMPK11 (Fig. 1A).
Figure 1.
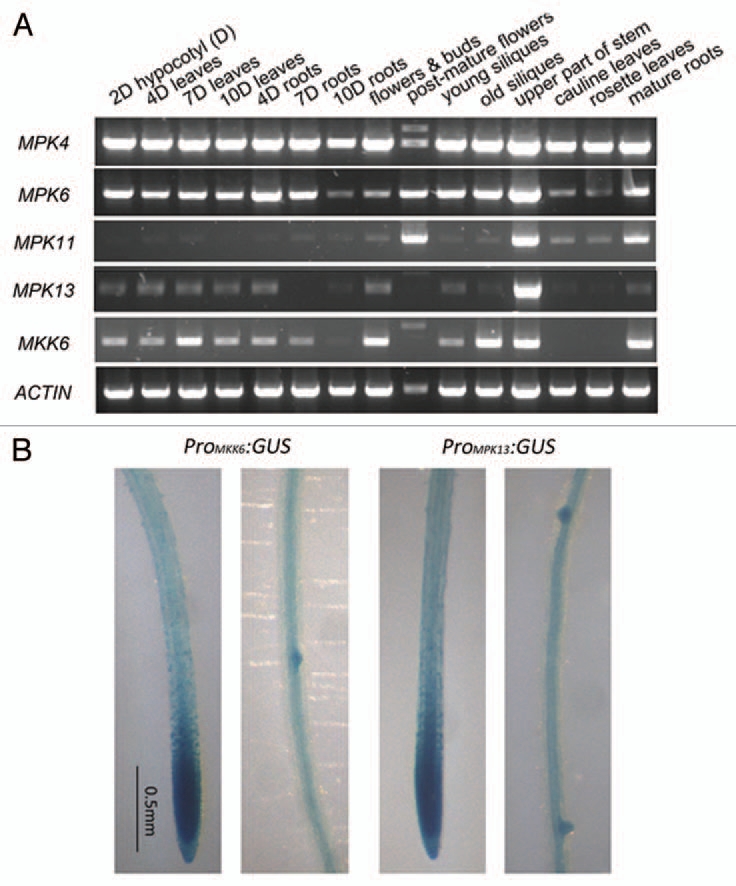
Expression patterns of AtMKK6 and related AtMPKs. (A) Comparison of expression patterns of AtMKK6 and AtMPK4, AtMPK6, AtMPK11 and AtMPK13 by RT-PCR. The (D) after “2D hypocotyls” indicates that the seedlings were germinated and grown in darkness before sample collection. Slower mobility bands observed in the “post-mature flowers” sample are from the genomic DNA fragment. (B) Histochemical analysis of PromoterMKK6::GUS and PromoterMPK13::GUS lines.
To obtain a more fine-grained view of these gene expression patterns, we examined ProMKK6::GUS and ProMPK13::GUS transgenic lines. The result showed that both AtMKK6 promoter activity and AtMPK13 promoter activity were concentrated in the root tip and in the sites of incipient lateral root emergence (Fig. 1B). In addition, GUS activity was only observed within the stele and pericycle cells, but not in the cortex or the epidermal cells. Taken together, these data suggest that the expression pattern of AtMPK13 is broadly similar to that of AtMKK6, and provide initial insight into the relevant biological function.
Conditional Knockdown Alleles of AtMKK6 and AtMPK13 Produce Fewer Lateral Roots
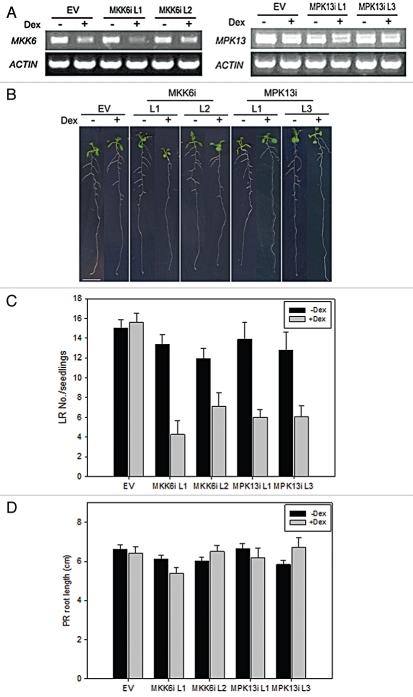
Since the AtMKK6 knockout allele is severely stressed and sterile, and there is no null knockout allele of MPK13,1 we generated dexamethasone (Dex)-inducible RNA interference lines for each kinase. Multiple independent T3 transgenic lines were selected that showed reduced accumulation of either AtMKK6 mRNA or AtMPK13 mRNA, respectively, 24 h after Dex-treatment (Fig. 2A).
Figure 2.
Phenotypic analysis of the Dex-inducible AtMKK6 and AtMPK13 RNAi lines. (A) RT-PCR examination of AtMKK6 and AtMPK13 transcripts in the AtMKK6 RNAi lines and the AtMPK13 RNAi lines, respectively, 24 h after Dex-treatment. Both transcript variants for AtMPK13 can be seen in the PCR products. (B) Lateral root reduction phenotype of the RNAi lines. Seedlings were germinated and grown on ½ MS for 5 days before being transferred to ½ MS plates with or without 1 µM dexamethasone. (C) Quantitative measurement of the lateral root (LR) number in the RNAi transgenic lines. (D) Quantitative measurement of the primary root (PR) length of the RNAi lines.
When germinated on ½ MS media and then transferred to Dex-containing plates 5 d after germination (DAG), both the AtMKK6 RNAi and AtMPK13 RNAi genotypes produced far fewer lateral roots (LRs) following Dex induction than did the untreated plants, while Dex treatment of pTA7002 empty vector lines grown in the same manner had no effect on LR formation (Fig. 2B). Quantitative analysis indicated that the reduction in LR formation was statistically significant (Fig. 2C). In contrast, primary root (PR) growth of AtMKK6 RNAi or AtMPK13 RNAi genotypes was largely unaffected by Dex induction under these growth conditions, indicating that partial suppression of either AtMKK6 or AtMPK13 results specifically in suppression of LR formation (Fig. 2D). Interestingly, the residual LRs formed in the Dex-treated transgenic plants are mostly located at the basal region of the PRs (Fig. 2B). Since these plants were transferred to Dexcontaining medium 5 DAG, when the seedlings had already formed their initial lateral root primordia (LRP), this distribution of LRs in the older seedlings suggests that LRP formed earlier (“pre-Dex”) can continue to develop into mature lateral roots, whereas Dex treatment, and the associated suppression of AtMPK13 or AtMKK6 expression, block the formation of new LRP.
Taken together, our results suggest that the AtMKK6-AtMPK13 module is required for initiating and/or sustaining pericycle cell division during lateral root initiation in Arabidopsis. The observation that partial suppression of either AtMKK6 or AtMPK13 gene expression resulted in formation of far fewer lateral roots without affecting primary root growth, provides evidence for this developmental specificity.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zeng Q, Chen JG, Ellis BE. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011;67:895–906. doi: 10.1111/j.1365-313X.2011.04642.x. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Ann Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 3.Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010;15:106–113. doi: 10.1016/j.tplants.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 2003;17:1055–1067. doi: 10.1101/gad.1071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderini O, Glab N, Bergounioux C, Heberle-Bors E, Wilson C. A novel tobacco mitogen-activated protein (MAP) kinase kinase, NtMEK1, activates the cell cycle-regulat ed p43Ntf6 MAP kinase. J Biol Chem. 2001;276:18139–18145. doi: 10.1074/jbc.M010621200. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi Y, Soyano T, Sasabe M, Machida Y. A MAP kinase cascade that controls plant cytokinesis. J Biol Chem. 2004;136:127–132. doi: 10.1093/jb/mvh118. [DOI] [PubMed] [Google Scholar]
- 7.Bögre L, Calderini O, Binarova P, Mattauch M, Till S, Kiegerl S, et al. A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell. 1999;11:101–113. [PMC free article] [PubMed] [Google Scholar]
- 8.Melikant B, Giuliani C, Halbmayer-Watzina S, Limmongkon A, Heberle-Bors E, Wilson C. The Arabidopsis thaliana MEK AtMKK6 activates the MAP kinase AtMPK13. FEBS Lett. 2004;576:5–8. doi: 10.1016/j.febslet.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 9.Hua ZM, Yang X, Fromm ME. Activation of the NaCland drought-induced RD29A and RD29B promoters by constitutively active Arabidopsis MAPKK or MAPK proteins. Plant Cell Environ. 2006;29:1761–1770. doi: 10.1111/j.1365-3040.2006.01552.x. [DOI] [PubMed] [Google Scholar]
- 10.Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, et al. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23:80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JS, Huh KW, Bhargava A, Ellis BE. Comprehensive analysis of protein-protein interactions between Arabidopsis MAPKs and MAPK kinases helps define potential MAPK signalling modules. Plant Signal Behav. 2008;3:1037–1041. doi: 10.4161/psb.3.12.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosetsu K, Matsunaga S, Nakagami H, Colcombet J, Sasabe M, Soyano T, et al. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell. 2010;22:3778–3790. doi: 10.1105/tpc.110.077164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menges M, Doczi R, Okresz L, Morandini P, Mizzi L, Soloviev M, et al. Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 2008;179:643–662. doi: 10.1111/j.1469-8137.2008.02552.x. [DOI] [PubMed] [Google Scholar]