Abstract
Helicases are motor proteins that can transiently catalyze the unwinding of energetically stable duplex DNA or RNA molecules by using ATP hydrolysis as the source of energy. Many helicases share a core region of highly conserved sequence motifs, and belong to the rapidly growing DEAD-box protein family. Pea DNA helicase 45 (PDH45), that exhibits striking homology with eukaryotic translation initiation factor 4A (eIF4A), contains ATP-dependent DNA and RNA helicase, DNA-dependent ATPase and ATP-binding activities. The transcript of the PDH45 gene was reported to be upregulated in pea plant in response to high salinity, cold stress, abscisic acid (ABA), dehydration and early wounding. The first direct evidence that overexpression of PDH45 confers salinity stress tolerance without yield loss has also been reported. A promoter analysis of PDH45 gene has not been reported thus far. The cis-regulatory elements present on promoter region of the gene act as binding sites for RNA polymerase and transcription factors and control the regulation of gene expression. Here we report the promoter of the PDH45 gene that contains stress-responsive cis-regulatory elements which may be responsible for regulating the expression of PDH45 under abiotic stress conditions.
Key words: gene expression, PDH45 promoter, plant eIF4A, plant helicase, salinity stress, stress responsive cis-regulatory elements
A full-length cDNA (1,630 bp) encoding PDH45 was cloned (Accession number Y17186) from pea cDNA library as described earlier in reference 1. The cDNA is consisted of an open reading frame (ORF) of 1,224 bp, a 5′ untranslated region (UTR) of 78 bp and a 3′ UTR of 328 bp including a 18 bp poly(A) tail. The PDH45 transcript was reported to be upregulated in response to NaCl (200 mM), cold (4°C), dehydration, ABA and early wounding stresses.2 The PDH45 overexpression driven by a constitutive cauliflower mosaic virus-35S promoter in tobacco plants has been shown to confer salinity tolerance.2 T1-transgenic plants grow normally and set viable seeds without yield penalty under salinity stress.2 The exact mechanism of PDH45-mediated tolerance to salinity stress is not understood. A promoter of PDH45 gene has not been reported so far. However, recently, a promoter sequence of a salinity stress-induced pea MCM6 DNA helicase3,4 has been reported to contain some stress responsive cis-regulatory elements.5 To check whether the stress-regulated cis-regulatory elements are present in the promoter of PDH45 gene, the promoter has also been isolated. For this first the pea genomic DNA library was prepared and then the PDH45 promoter isolated as described below:
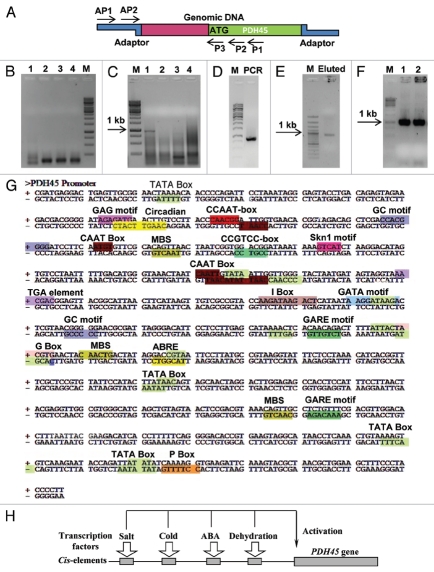
For isolation of PDH45 promoter, the genomic DNA library was made by digesting genomic DNA with different restriction enzymes [EcoRV (Library 1), DraI (Library 2), PvuII (Libray 3), StulI (Library 4)] in separate tubes, these enzymes will generate blunt ends of the genomic DNA. After digestion, the blunt end DNA was ligated to BD Genome Walker Adaptor which possesses the complimentary sequences of primers (AP1 and AP2). The ligated DNA is used for the isolation of promoters by amplification using gene specific primers (P1 and P2). Schematic representation of DNA walking technique used for PDH45 promoter isolation is described in Figure 1A.
Figure 1.
Isolation of PDH45 promoter by PCR. (A) Schematic representation of DNA walking technique used for PDH45 promoter isolation. (B) Primary PCR of PDH45 promoter with P1 and AP1 primers using genomic library as template (lane 1–4). (C) Nested PCR of PDH45 promoter with nested primer P2 and AP2, first PCR used as a template. (D) Second-nested PCR using only PCR product of library one as a template with nested P3 and AP2 primers. (E) The eluted fragment from the secondary PCR. (F) The fragment was cloned in TOPO vector and colonies were checked with vector specific primers. (G) A schematic representation showing various cis-regulatory elements presents in the genomic region upstream of the 5′-UTR sequence of the PDH45 gene as determined by PlantCARE program. TATA-box and CAAT sequences and various cis-acting elements are shown. (H) A hypothetical model representing the regulation of expression of stress-induced PDH45 gene under stress by cis-regulatory elements in the promoter region.
PDH45 promoter isolation was done with two PCRs. The first PCR was performed by using AP1 primer (5′-GTA ATA CGA CTC ACT ATA GGG C-3′) and gene specific primer, P1 (5′-CGA AAC CGT ATG CGT AGA TTC CAC GCA GCA AAT C-3′). Four DNA genomic libraries were used as a template for the first PCR. The PCR products were resolved on a 1% agarose gel. The results show that PCR from library No. 1 (Fig. 1B, lanes 1) have little smear and in other libraries no bands appeared (Fig. 1B, lanes 2 to 4). Later on, the Primary PCR product was diluted 10 times and used as a template for nested PCR with nested primers AP2 (5′-ACT ATA GGG CAC GCG TGG T-3′) and gene specific nested primer P2 (5′-CCC CAT CCC TTC GAA GGT CGC AAT CGC-3′). The PCR product of library No 1 showed a very specific band with approximate size of 1 kb (Fig. 1C, lane 1). The other library did not show any specific band (Fig. 1C, lanes 2–4). Figure 1D shows PCR with second-nested gene specific primer, P3 (5′-CGC AAT CGC CTT CAC ACC TTC CGT CGT CT-3′) and AP2 that confirmed right amplification of secondary PCR by giving almost same size band (distance between P2 and P3 primer is 58 nucleotides). The specific band from nested PCR later was eluted from the gel (Fig. 1D) and found to be pure as checked on gel (Fig. 1E). This eluted DNA fragment (promoter) was cloned in TOPO TA cloning vector (pCR®2.1-TOPO®) and checked with vector specific primers (Fig. 1F). Sequence analysis for identification of cis-regulatory elements followed by using PLACE and PlantCARE database. The sequence of promoter of PDH45 gene along with the different cis-regulatory elements is shown in Figure 1G. Sequence analysis of this clone showed the overlapping with 350 bp in 5′ end of PDH45 gene.
The promoter is the sequence where all transcription factors and RNA polymerases can bind and regulate the expression of the gene. Transcription factors will recognize the specific DNA sequence, which is known as cis-regulatory elements and regulate the gene expression in different conditions. A hypothetical model representing the regulation of expression of stress-induced PDH45 gene under stress by cis-regulatory elements in the promoter region is shown in Figure 1H. Promoter of PDH45 gene contains many cis-regulatory elements including stress related cis-acting elements. The putative functions of only some of these important cis-regulatory elements which are involved in stresses are described below:
ABRE: cis-acting element (TACGGTC) involved in the ABA responsiveness and drought tolerance.6
CCAAT BOX: Common sequence found in the 5′-non-coding regions of eukaryotic genes; “CCAAT box” found in the promoter of heat shock protein genes, located immediately upstream from the most distal HSE of the promoter; “CCAAT box” act cooperatively with HSEs to increase the heat shock promoter activity.7,8 It is recently identified that it helps to regulate flowering in Arabidopsis.9
MBS: MYB binding site found in the promoters of the dehydration-responsive gene rd22 and many other genes in Arabidopsis. It is also related to ABA signaling. The recognition sequence is YAACKG (Y = C/T; K = G/T).10
G-box: G-box cis-element (CACGTG) exists in many gene promoters and has critical effects on plant development,11 hormone responses,12,13 and fungal infections.14
GARE-motif: This sequence was identified as a gibberellin (GA) response element (GARE).15 The GARE motif in turn is necessary for gibberellin induction of LEAFY16,17 to promote flowering thus hinting at a role of GARE as a factor in GA-regulated signaling pathways that depend on cGMP as second messenger.
TGA-element: The identification of putative auxin response elements is based mainly on conservation of similar sequence elements found in a variety of genes induced by auxin. Most of these putative auxin response elements have not been tested for functionality, and their direct role in response to auxin remains to be demonstrated.18
Stress responsive genes can be expressed either through an ABA-dependent or ABA-independent pathway.19 The earlier results showed that PDH45 followed ABA-dependent pathways in abiotic stress.2 The PDH45 transcript results indicated that the stress response was specific to salinity and cold stress related pathways.2 Recently, an improved program (PromPredict) for promoter prediction which is based on DNA free energy has been described in reference 20. The PromPredict Web server (nucleix.mbu.iisc.ernet.in/prompredict/prompredict.html) can be used to predict promoter regions in the input sequence.20 To check whether the stress related cis-regulatory elements were present in the gene, the promoter of PDH45 was analyzed. The sequence analysis showed the presence of various stress related cis-regulatory elements including salt, cold, ABA and dehydration. The hypothetical model in Figure 1H shows that probably some transcription factors bind to the salt, cold, ABA and dehydration related cis-regulatory elements and activate the PDH45 gene transcription. Further work is needed to understand the presence of many cis-regulatory elements in the promoter region of PDH45. The validation of PDH45 promoter in vivo by using transgenic methods under stress conditions still needs to be studied. However, from these predicted cis-regulatory elements and together with expression profile we can briefly predict that these cis-elements may be involved in stress response of PDH45 gene.
Acknowledgments
Work on DNA replication and plant abiotic stress tolerance in N.T.'s laboratory is partially supported by Department of Biotechnology (DBT), Government of India. We are thankful to Miss Neha Vaid for her help in preparation of the manuscript.
References
- 1.Pham XH, Reddy MK, Ehtesham NZ, Matta B, Tuteja N. A DNA helicase from Pisum sativum is homologous to translation initiation factor and stimulates topoisomerase I activity. Plant J. 2000;24:219–229. doi: 10.1046/j.1365-313x.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 2.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran NQ, Dang HQ, Tuteja R, Tuteja N. A single subunit MCM6 from pea forms homohexamer and functions as DNA helicase. Plant Mol Biol. 2010;74:327–336. doi: 10.1007/s11103-010-9675-7. [DOI] [PubMed] [Google Scholar]
- 4.Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol. 2011;76:19–34. doi: 10.1007/s11103-011-9758-0. [DOI] [PubMed] [Google Scholar]
- 5.Dang HQ, Tran NQ, Tuteja R, Tuteja N. Promoter of a salinity and cold stress-induced MCM6 DNA helicase from pea. Plant Signal Behav. 2011:6. doi: 10.4161/psb.6.7.15502. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis DNA Encoding Two Desiccation-Responsive rd29 Genes. Plant Physiol. 1993;101:1119–1120. doi: 10.1104/pp.101.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haralampidis K, Milioni D, Rigas S, Hatzopoulos P. Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol. 2002;129:1138–1149. doi: 10.1104/pp.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieping M, Schöffl F. Synergistic effect of upstream sequences, CCAAT box elements and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- 9.Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum K, Wienand U, Meier I. Reduction of G-box binding factor DNA binding activity, but not G-box binding factor abundance, causes the downregulation of RBCS2 expression during early tomato fruit development. FEBS Lett. 1999;454:95–99. doi: 10.1016/S0014-5793(99)00784-X. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa H, Ohmiya K, Hattori T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996;9:217–227. doi: 10.1046/j.1365-313X.1996.09020217.x. [DOI] [PubMed] [Google Scholar]
- 13.Nantel A, Quatrano RS. Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem. 1996;271:31296–31305. doi: 10.1074/jbc.271.49.31296. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JTACF. an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/S0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 15.Skriver K, Olsen FL, Rogers JC, Mundy J. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA. 1991;88:7266–7270. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, et al. GAMYB-like genes, flowering and gibberellin signaling in Arabidopsis. Plant Physiol. 2001;127:1682–1693. doi: 10.1104/pp.010442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blázquez MA, Weigel D. Integration of oral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- 18.Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuteja N. Abscisic acid and abiotic stress signalling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morey C, Mookherjee S, Rajasekaran G, Bansal M. DNA Free Energy-Based Promoter Prediction and Comparative Analysis of Arabidopsis and Rice Genomes. Plant Physiol. 2011;156:1300–1315. doi: 10.1104/pp.110.167809. [DOI] [PMC free article] [PubMed] [Google Scholar]