Abstract
Plant growth and crop production can be greatly affected by common environmental stresses such as drought, high salinity and low temperatures. Gene expression is affected by several abiotic stresses. Stress-inducible genes are regulated by transcription factors and epigenetic mechanisms such as histone modifications. In this mini-review, we have explored the role of transcriptional adaptor ADA2b in Arabidopsis responses to abiotic stress. ADA2b is required for the expression of genes involved in abiotic stress either by controlling H3 and H4 acetylation in the case of salt stress or affecting nucleosome occupancy in low temperatures response.
Key words: drought, salt, low temperature, transcription regulation, transcriptional adaptors, histone acetylation, chromatin
Plant growth and crop production can be greatly affected by abiotic stresses. Abiotic stress includes a large number of stresses caused by complex enviromental conditions such as drought, freezing, salinity, high and low temperatures, UV light, heavy metals, flooding and hypoxia. According to Intergovermental Panel of Global Change (www.ipcc.ch), these stresses will increase in the near future as a result of climate change. For example, water supply is the single most limiting factor in many countries, including Mediterranean areas and drought stress is one of the main factors affecting plant productivity.1 Likewise, salt is a major constraint to global food crop production. It is estimated that 20% of all cultivated land and nearly half of irrigated land is salt-affected,2 reducing yields below the genetic potential. Low temperatures have a huge impact on the survival and geographical distribution of crops. Therefore, understanding plant responses to these abiotic stresses will lead to major advances in the area of crop productivity and tolerance to abiotic stresses.
Various abiotic stresses induce gene expression in many plants.3 Stress-inducible genes are regulated by multiple stress signals through the action of transcription factors and epigenetic mechanisms such as histone modifications and DNA methylation.4–6 The precise control of chromatin modification may play a central role in regulating gene expression in response to environmental cues because the switch between permissive and repressive chromatin allows alteration of gene expression in response to environmental changes. A number of studies have shown that plants are capable of adapting their growth and development to environment changes such as light, temperature, biotic and abiotic stresses through modulation of histone acetylation.7
Histone Acetylation and Abiotic Stress
Acetylation of the N-terminal tails of histones was one of the first chromatin modifications to be characterized and is generally correlated with increased accessibility and transcription of the associated DNA.8 The identification of the transcriptional regulator Gcn5 as a histone acetyltransferase gave rise to characterization of enzymes and their regulatory partners that form large multiprotein complexes which function to alter chromatin states.9 Gcn5 physically associates with Ada2 in several larger transcriptional coactivator complexes that are particularly well characterized in the yeast Saccharomyces cerevisiae.10,11 Similar to yeast and mammals, Arabidopsis GCN5 has been shown to acetylate H3 in vitro12,13 and global H3 acetylation is reduced in gcn5 mutant.14 More specifically, H3K14 and H3K27 acetylation are reduced at defined loci in gcn5 mutants.15 Also, in Arabidopsis, GCN5 interacts in vitro with the two homologs ADA2a and ADA2b.12,13 Arabidopsis ADA2b enhances the HAT activity of GCN5,12 which is also known in yeast.16 A subset of genes is regulated by the Arabidopsis ADA2b and GCN5, since expression of 5% of investigated 8,200 genes is changed in gcn5 and ada2b mutant leaves.17 As a result in Arabidopsis, gcn5 mutants exhibit pleiotropic developmental defects, including dwarfism, loss of apical dominance, aberrant meristem function in root and shoot, leaf and flower development, short petals and stamens, flower infertility and plant responses to abiotic stress.14,15,17–23 Chromatin immunoprecipitation coupled with a promoter chip revealed that about 40% of the promoters are associated with the Arabidopsis GCN5 protein.24 The GCN5 bromodomain is required for a small fraction (11%) of the AtGCN5 targets and their H3K14 acetylation.24
The Transcriptional Coactivator ADA2b and Abiotic Stress Responses
ADA2 (alteration/deficiency in activation 2) adaptor proteins are integral parts of GCN5-containing complexes. In Arabidopsis, two related ADA2 factors (ADA2a and ADA2b) have been identified.13 T-DNA mutations in ADA2b gene result in pleiotropic defects in development, some of which are similar to gcn5 phenotypes.17,22,25 ADA2b is also required to modulate histone acetylation in response to auxin.26 However, the related factor ADA2a does not appear to be required for plant development, since ada2a mutants are indistinguishable from wild-type plants.20 Moreover, expression of AtADA2a from a constitutive promoter fails to complement the ada2b-1 mutant phenotype, supporting the hypothesis that the two proteins target distinct sets of genes and may have different biochemical roles.20 The Drosophila genome also encodes two ADA2 proteins, which appear to be specific components of different GCN5-containing complexes.27,28 For instance, dADA2a is present in the ATAC (Ada2a-containing) complex,29,30 while dADA2b is present in the SAGA complex.31–33 However, contrary to what the situation is in Arabidopsis, both dADA2A and dADA2b are required for normal development.32 It remains to be learned whether AtADA2a and AtADA2b are part of different GCN5-containing complexes in Arabidopsis, which have different HAT activity and/or specificity.
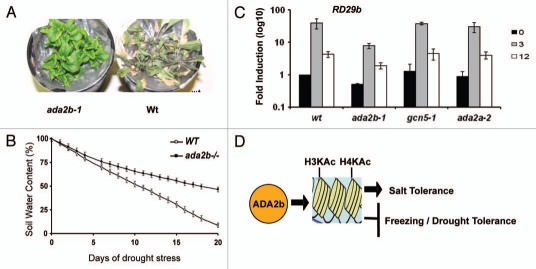
Chromatin regulation by histone acetylation is important for plant adaptation to environmental changes.34,35 GCN5 and ADA2b are involved in abiotic stress responses including low temperature and salt stress.17,18,20 The CBF (C-repeat binding factor) family of transcriptional activators function as master regulators of cold acclimation in Arabidopsis by stimulating the transcription of cold-responsive (COR) genes.36 COR gene expression is reduced upon exposure to low temperatures and delayed in both ada2b-1 and gcn5-1 mutants,17 indicating that CBFs might stimulate transcription through the recruitment of GCN5-containing complexes to the promoters of its target genes. The non-acclimated ada2b-1 mutant is more freezing tolerant,17,20 suggesting that ADA2b may repress the ability of plants to tolerate freezing by a mechanism independent of COR gene expression. To determine the response of the ada2b-1 mutant to water starvation, plants were grown in pots until the reproductive stage and then subjected to a 20 d water-starvation period (Fig. 1A). Ten days after water starvation, the wild-type plants showed a withering phenotype and after 20 d the leaves of wild-type plants were almost completely dry. In contrast ada2b-1 mutants were not wilting and survived this severe drought stress. The whole plant transpiration rate was reduced in ada2b-1 mutants in comparison to wild-type plants (Fig. 1B) suggesting that the drought tolerant arise from reduction of transpiration water loss probably through stomata closure. Although at the moment there is no evidence of rapid stomata closure of ada2b-1 mutants in response to drought stress, those mutants were hypersensitive to ABA20 a situation that could lead to rapid stomata closure. Moreover ada2b-1 mutants were hypersensitive to salt stress,18,20 suggesting that ADA2b is not only involved in abiotic stress responses but also its function is stress-dependent. Furthermore, abiotic stress responses are more pronounced in the ada2b mutant than gcn5 or ada2a mutants,20 suggesting that ADA2b is a key link of histone acetylation and transcription regulation upon abiotic stress.
Figure 1.
ADA2b is a key regulator of abiotic stress. (A) ada2b-1 mutants are drought tolerant; photo was taken 20 d after water starvation. (B) Whole plant transpiration of wild type (white square) and ada2b-1 (black square) mutant plants. Changes in soil water content during drought stress is indicated. (C) The effect of ADA2a, ADA2b and GCN5 on salt-induced RD29b expression. Wild-type Ws, ada2a-1, ada2a-2 and gcn5-1 plants were subjected to salt stress (100 mM NaCl) for various time as indicated. RNA were isolated from whole seedling and real-time Reverse Transcription PCR was performed using specific primers for RD29b. Fold induction of these genes after exposure to 100 mM NaCl were calculated by normalizing values from salt-treated plants with those from untreated samples. Values are presented on a logarithmic scale. For each mutant and wild-type plants, triplicate samples in each experiment were assayed twice. Three independent experiments were performed, error bars represent SE where n = 4. (D) Schematic representation of ADA2b function. ADA2b promotes histone acetyaltion in salt-induced genes and confers salt tolerance whereas it represses freezing and drought tolerant in Arabidopsis.
Indeed, the induction of COR and other stress related gene expression during salt treatment was reduced in ada2b-1 plants.18 These results indicate that ADA2b may act as a positive regulator of salt-induced gene expression. This ADA2b's action is distinct from that of ADA2a and GCN5, since in the ada2a-2 and gcn5-1 as well as gcn5-2 mutants the expression of the stress-inducible genes was not affected by high salinity stress (Fig. 1C).18,20,37
Histone modifications play a key role in gene expression and plant development under stress.5,34 In ada2b-1 plants the level of histone H3K9/K14 and H4 tetra-acetylation was reduced at the promoter and coding regions of the salt-induced genes18 which was correlated with reduced salt-induced gene expression, suggesting that ADA2b-mediated histone acetylation may facilitate the binding of transcription factors that have multiple saltresponsive modules to the promoter.34
When Arabidopsis is exposed to low temperature H3 acetylation increases and nucleosome occupancy decreases at COR promoters.19 H3 acetylation at COR promoters was stimulated upon cold acclimation in ada2b and gcn5 plants as in wild type plants, but the decrease in nucleosome occupancy was diminished. These data suggest that GCN5 and ADA2b are not required for cold-stimulated histone acetylation at COR gene promoters, but they are required for changes in nucleosome occupancy during cold acclimation.
SGF29 is another component of GCN5-containing complexes in yeast and there are two orthologs of yeast SGF29 in the Arabidopsis genome, designated as SGF29a and SGF29b.18 In root growth and seed germination assays, and in contrast to the ada2b salt responses, it was found that sgf29a mutants were more resistant to salt stress than their wild-type counterparts, suggesting that different components of HAT complexes may play a different role in plant abiotic stress responses.18
Conclusions and Future Perspectives
In summary ADA2b is implicated in several abiotic stresses. ADA2b is required for gene expression of genes involved in abiotic stress either by controlling H3 and H4 acetylation in the case of salt stress or affecting nucleosome occupancy in low temperatures response (Fig. 1D). The specificity of the ADA2b in chromatin function is believed to be mainly regulated by interaction with specific DNA-binding transcription factors. Therefore, identifying interaction between ADA2b and key transcription factors involved in abiotic stress will be needed to understand the chromatin regulation of stress-response gene expression. Moreover performing a genome-wide expression analysis of stress-induced genes in the ada2b mutants under different stress conditions will give rise to specific ADA2b target that could lead either to salt tolerance or to drought sensitivity.
Acknowledgments
We thank Prof. Frantisek Baluska for kindly inviting this mini review. We also thank Dr. Amy Hark, (Muhlenberg College, Allentown) for helpful discussion on the manuscript. Financial support by Greek Ministry of Education (EPEAEK, Pythagoras #21964), Greek General Secretary of Research and Technology (# 82337) and by funds from Aristotle University of Thessaloniki, School of Biology to K.V. is greatfully acknowledged.
References
- 1.Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 2.Ghassemi F, Jakeman AJ, Nix HA. Salinisation of land and water resources: Human causes, extent, management and case studies. Sydney, Australia: UNSW Press; 1995. p. 540. Wallingford, UK: CAB International. [Google Scholar]
- 3.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 5.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta. 2007;1769:295–307. doi: 10.1016/j.bbaexp.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdone L, Caserta M, Mauro ED. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- 9.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an ada complex and the saga (spt/ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 11.Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim Biophys Acta. 2006;1759:69–79. doi: 10.1016/j.bbaexp.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertrand C, Bergounioux C, Domenichini S, Delarue M, Zhou DX. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J Biol Chem. 2003;278:28246–28251. doi: 10.1074/jbc.M302787200. [DOI] [PubMed] [Google Scholar]
- 15.Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1 and TAF1/HAF2 interact to regulate histone acetylation required for liqht-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development and gene expression. Plant Cell. 2003;15:626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaldis A, Tsementzi D, Tanriverdi O, Vlachonasios KE. Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta. 2011;233:749–762. doi: 10.1007/s00425-010-1337-0. [DOI] [PubMed] [Google Scholar]
- 19.Pavangadkar K, Thomashow MF, Triezenberg SJ. Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant Mol Biol. 2010;74:183–200. doi: 10.1007/s11103-010-9665-9. [DOI] [PubMed] [Google Scholar]
- 20.Hark AT, Vlachonasios KE, Pavangadkar KA, Rao S, Gordon H, Adamakis ID, et al. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim Biophys Acta. 2009;1789:117–124. doi: 10.1016/j.bbagrm.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Cohen R, Schocken J, Kaldis A, Vlachonasios KE, Hark AT, McCain ER. The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta. 2009;230:1207–1221. doi: 10.1007/s00425-009-1012-5. [DOI] [PubMed] [Google Scholar]
- 22.Kornet N, Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21:1070–1079. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 24.Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, et al. Genomescale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 2008;56:493–504. doi: 10.1111/j.1365-313X.2008.03606.x. [DOI] [PubMed] [Google Scholar]
- 25.Sieberer T, Hauser M, Seifert GJ, Luschnig C. PROPORZ1 a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol. 2003;13:837–842. doi: 10.1016/S0960-9822(03)00327-0. [DOI] [PubMed] [Google Scholar]
- 26.Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, et al. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc Natl Acad Sci USA. 2010;107:10308–10313. doi: 10.1073/pnas.0913918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, et al. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase containing complexes. Mol Cell Biol. 2003;23:306–321. doi: 10.1128/MCB.23.1.306-21.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-19.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol. 2006;26:9413–9423. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-82.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciurciu A, Tombacz I, Popescu C, Boros I. GAL4 induces transcriptionally active puff in the absence of dSAGA- and ATAC-specific chromatin acetylation in the Drosophila melanogaster polytene chromosome. Chromosoma. 2009;118:513–526. doi: 10.1007/s00412-009-0215-7. [DOI] [PubMed] [Google Scholar]
- 32.Pankotai T, Komonyi O, Bodai L, Újfaludi Z, Muratoglu S, Ciurciu A, et al. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol. 2005;25:8215–8227. doi: 10.1128/MCB.25.18.8215-27.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol. 2004;24:8080–8089. doi: 10.1128/MCB.24.18.8080-9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JM, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010;33:604–611. doi: 10.1111/j.1365-3040.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- 35.Servet C, Silva NCE, Zhou DX. Histone Acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol Plant. 2010;3:670–677. doi: 10.1093/mp/ssq018. [DOI] [PubMed] [Google Scholar]
- 36.Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servet C, Benhamed M, Latrasse D, Kim W, Delarue M, Zhou DX. Characterization of a phosphatase 2C protein as an interacting partner of the histone acetyltransferase GCN5 in Arabidopsis. Biochim Biophys Acta. 2008;1779:376–382. doi: 10.1016/j.bbagrm.2008.04.007. [DOI] [PubMed] [Google Scholar]