Abstract
For over two decades now, it is known that the nodule symbiosis between legume plants and nitrogen fixing rhizobium bacteria is set in motion by the bacterial signal molecule named nodulation (Nod) factor.1 Upon Nod factor perception a signaling cascade is activated that is also essential for endomycorrhizal symbiosis (Fig. 1). This suggests that rhizobium co-opted the evolutionary far more ancient mycorrhizal signaling pathway in order to establish an endosymbiotic interaction with legumes.2 As arbuscular mycorrhizal fungi of the Glomeromycota phylum can establish a symbiosis with the vast majority of land plants, it is most probable that this signaling cascade is wide spread in the plant kingdom.3 However, Nod factor perception generally is considered to be unique to legumes. Two recent breakthroughs on the evolutionary origin of rhizobium Nod factor signaling demonstrate that this is not the case.4,5 The purification of Nod factor-like molecules excreted by the mycorrhizal fungus Glomus intraradices and the role of the LysM-type Nod factor receptor PaNFP in the non-legume Parasponia andersonii provide novel understanding on the evolution of rhizobial Nod factor signaling.
Key words: parasponia, legumes, rhizobium, mycorrhizae, Nod factor
Rhizobium Nod Factors and Responses in Legumes
Elucidation of the Nod factor structure was a major step in the molecular approach to unravel the signaling pathway in legumes that is essential for the establishment of rhizobium symbiosis. Rhizobial Nod factor molecules are lipochito-oligosaccharides (LCOs) consisting of three to five N-acetyl-glucosamines that on the amino group of the non-reducing glucosamine are acylated with a fatty acid of 16–20 C-atoms in length (C16 to C20). Furthermore, species specific substitutions can be present on the terminal glucosamines, thereby determining specific recognition of Nod factor structure by the legume host plants.6 Examples of such modifications are glycosylation, sulfation, acetylation and methylation, for which the particular rhizobium species harbours specific nodulation (nod) genes.7,8 Therefore, it is generally assumed that the perception of Nod factors by legume host plants has co-evolved with their corresponding rhizobial symbionts. Some rhizobium species however, produce a diverse mixture of Nod factors often resulting in a large range of host plants.9 Of such broad host range rhizobium species, Sinorhizobium sp. NGR234 is iconic, as it is well studied at a molecular and genetic level.10 This species not only nodulates hundreds of legume species, but also Parasponia; the only non-legume genus able to establish a similar symbiosis with rhizobium.9,11
Nod factors are sufficient to trigger all symbiotic responses essential to develop fully differentiated nodules on legume roots.12,13 Furthermore, Nod factor signaling is imperative for intracellular bacterial infection.14 This infection process is set in motion through changes in growing root hairs resulting in root hair curling around Nod factor secreting rhizobia. These captured bacteria can initiate the formation of intracellular infection threads. Such infection threads are surrounded by a plant derived membrane and contain files of clonally propagating rhizobia. Within infection threads the rhizobia remain cell wall bound. Infection threads grow to the base of epidermal cells. Simultaneously, activation of mitosis in cortical cells below the infected root hair cells results in a nodule primordium. As infection threads reach newly formed nodule primordia the rhizobia are released into the nodule cells. Release from infection threads occurs at specific sites that do not contain a cell wall. Since the budding off of rhizobium from the infection thread have a droplet-like appearance, these regions in the infection threads are known as “unwalled infection droplets.” In this endocytotic-like process, rhizobia become surrounded by a plant membrane. Such unit, of one or a few rhizobia surrounded by a plant membrane, is named symbiosome. Symbiosomes act like transient nitrogen fixing organelles. Thereby the surrounding symbiosome membrane acts as an interface between both partners enabling exchange of nutrients. Along the whole infection process, from early responses in root hairs down to the formation of symbiosomes, Nod factor signaling is important.
In some more basal legume species symbiosome formation does not occur.15,16 Instead rhizobia remain in intracellular thread-like structures; so-called fixation threads. These fixation threads differ from ordinary infection threads by a significant thinner cell wall, enabling a more efficient exchange of nutrient across the membrane interfase.
Mycorrhizae and Rhizobium Common Signaling Pathway
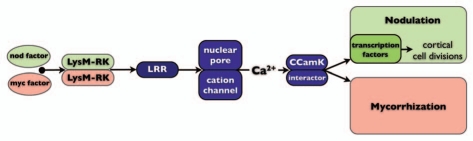
It has been known for a long time that mycorrhizae and rhizobium symbioses in legumes share some common key signaling genes.17 This so-called common symbiotic signaling pathway has been unravelled in pea (Pisum sativum) and the legume model species; Medicago truncatula (medicago) and Lotus japonicus (lotus), respectively. In both model species the common symbiotic signaling pathway comprises a rather conserved set of genes encoding a plasma membrane receptor kinase (MtDMI2 and LjSYMRK), several components in the nuclear envelope including a cation ion channel (MtDMI1, LjCASTOR and LjPOLLUX), a nuclear localized Calcium Calmodulin dependent Kinase (CCaMK; MtDMI3 and LjCCaMK) and a CCaMK interacting protein (MtIPD3 and LjCYCLOPS) (Fig. 1).18 Mycorrhizae and rhizobium induced signaling bifurcates downstream of CCaMK, possibly due to a different nature of the calcium signal.19
Figure 1.
Schematic representation of the genetically dissected symbiosis signaling pathway. In legumes rhizobium Nod factors and mycorrhizal Myc factors are perceived by distinct receptor complexes. In case of Nod factors these are the LysM-RK type receptors MtLYK3/LjNFR1 and MtNFP/LjNFR5, whereas Myc factors remain to be elucidated. In Parasponia PaNFP fulfils a dual function and acts in both symbioses. The subsequent common signaling pathway consists of several components including a plasma membrane localized LRR-type receptor (MtDMI2/LjSymRK), a cation channel in the nuclear envelope (MtDMI1/LjCASTOR/LjPOLLUX) and subunits of the nuclear pore (NUP85, NUP133), and a nuclear localized complex of calcium calmodulin dependent kinase (CCaMK) and interactor protein MtIPD3/LjCYCLOPS. Downstream of CCaMK the rhizobium and mycorrhiza induced responses bifurcate.
In legumes the common signaling pathway is activated by LysM-type Nod factor receptors. Nod factors are perceived by two distinct LysM-type receptor kinases (LysM-RKs) that form a heterodimeric complex to achieve symbiotic signaling.20,21 In medicago and lotus these receptors, named MtLYK3/LjNFR1 and MtNFP/LjNFR5, are not essential for mycorrhizal symbiosis, suggesting that in legumes Nod factor receptors have evolved specifically to support rhizobium Nod factor signaling. This raises immediate questions concerning the evolutionary origin of rhizobium Nod factor perception and how mycorrhizae achieve activation of the common signaling pathway. Research on Parasponia and the mycorrhizal fungus G. intraradices revealed first answers to these questions.
Nod Factor Signaling in Parasponia
The genus Parasponia comprises about six species and is part of the Celtidaceae.22–26 Recent molecular phylogenetic studies combine this family with the Cannabaceae,27 and according to nomenclature rules this family should be named Urticaceae.28 All Parasponia species can establish a nitrogen fixing endosymbiosis with rhizobium species that also can nodulate legumes.29–33 Interestingly, the Parasponia-rhizobium symbiosis is also Nod factor driven.5,34 Because the Celtidaceae/Cannabaceae and Fabaceae are only remotely related, it is most probable that both lineages have gained the symbiotic capacity independently. Therefore, a comparison of Parasponia and legumes will provide insights in genetic constrains underlying rhizobium symbiosis.
The Parasponia-rhizobium symbiosis is most likely relatively young as Parasponia is very closely related to its non-symbiotic sister genus Trema.27 This hypothesis is further supported by the rather primitive nature of Parasponia root nodules. First, the nodule ontology differs with that the ontology of legume nodules. Parasponia nodules are modified lateral roots with a central vascular bundle and infected cells in the peripheral zone. In contrast, legume nodules have a peripheral vasculature with a central zone of infected cells.34 Furthermore, there is a distinct difference in the infection mode. Rhizobium enters the Parasponia root intercellularly by crack entry and only when a bacteria reaches a nodule primoridium, intracellular infection occurs. Once inside a nodule cell, fixation threads are formed, similar as found for some basal legumes.33
As the Parasponia-rhizobium symbiosis is relatively young when compared with legumes, it can provide additional insights in the evolutionary origin of symbiotic genes. Such comparative evolutionary studies have been conducted on LysM-type Nod factor receptors. Plant LysM-RK genes can be divided into three major clades, two of which contain a legume Nod factor receptor, MtNFP/LjNFR5 and MtLYK3/LjNFR1, respectively.35–39 The latter clade includes also the chitin innate immune receptor, AtCERK1 of Arabidopsis thaliana (arabidopsis).40–43 Legume gene duplication events of different nature have occurred in all three classes resulting in a relative large number of 15–17 genes in for example lotus and medicago, whereas in arabidopsis only 5 such genes are found.38,39 Especially the MtLYK3/LjNFR1-class expanded significantly in both legumes. As the Nod factor receptor MtLYK3/LjNFR1 is a close homolog of arabidopsis AtCERK1, it suggests that both genes share a recent common ancestral gene. Comparative studies revealed that in evolution the kinase domain of both proteins underwent specific amino acid substitutions enabling it to trigger specific responses.21
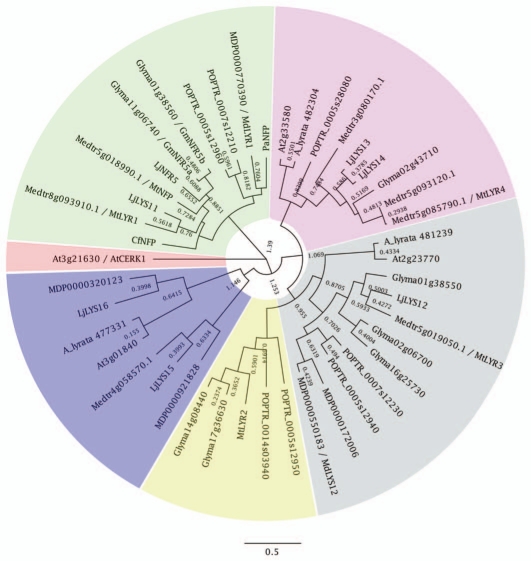
Interestingly, the MtNFP/LjNFR5-class of LysM-type receptors is less expanded in legumes, and MtNFP/LjNFR5 underwent only a single legume specific duplication event in the subfamiliy of medicago, lotus and soybean (Glycine max).5,37 MtNFP/LjNFR5-homologs can be found in many non-legume species, including Parasponia (green orthology group Fig. 2). Functional analysis of this gene in Parasponia andersonii revealed a dual symbiotic function. PaNFP controls intracellular infection of rhizobium and mycorrhizal fungi.5 PaNFP knockdown lines are blocked specifically in the formation of symbiotic interfaces. In case of rhizobium this is the switch from initial intercellular infection to the formation of fixation threads. In case of mycorrhizae, root cortical cells get infected by cell wall bound fungal hyphae (known as trunks), but arbuscules are not formed. Arbuscles represent the symbiotic interface that supports nutrient exchange in this ancient symbiosis. As Parasponia has only a single MtNFP/LjNFR5-homolog, as is the case in other non-legume species, it suggests that in Parasponia the mycorrhizal signaling perception mechanism has been co-opted to achieve rhizobium Nod factor signaling (green orthology group Fig. 2).5 Interestingly, arabidopsis lacks a clear MtNFP/LjNFR5-homologous gene (green orthology group Fig. 2),38 which is in line with the inability of this species to establish a mycorrhizal symbiosis.44
Figure 2.
Bayesian phylogenic tree of MtNFP/LjNFR5-class of LysM-RKs using Arabidopsis thaliana AtCERK1 as outgroup. Five orthology groups can be recognized indicated in different colors. Parasponia andersonii (Pa), Malus domesticus (Md), Populus trichocarpa (POPTR), Lotus japonicus (Lj), Glycine max (Glyma), Medicago truncatula (Mt/Medtr), Chamaecrista fasciculata (Cf), Arabidopsis thaliana (At) and Arabidopsis lyrata (A_lyrata). Branch lengths are proportional to the number of amino acid substitutions per site. The analysis was run for 290,000 generations, sampling every 200 generations using Geneious software with default settings.
Mycorrhizae Secrete Nod Factor-Like Signal Molecules
The finding that in Parasponia a single LysM-type Nod factor receptor is essential for mycorrhizal symbiosis implies that mycorrhizae produce Nod factor-like LCOs. This is indeed the case.4 G. intraradices secretes symbiotic LCOs that are a mixture of Nod factor-like molecules. These LCOs stimulate the mycorrhizal symbiosis in legumes as well as non-legumes. Like Nod factors, such mycorrhizal signaling factors, or Myc factors, also contain a tetrameric or pentameric N-acetyl glucosamine backbone that is acylated at the non-reducing end with either a C16:0 or C18:1 acyl chain. Such lipids are common in microbes and also found on Nod factors of several rhizobium species.6 Furthermore, a sulfate group can be present at the reducing end of the Myc factor, similar as can be found on Nod factors of certain rhizobium species; e.g., Sinorhizobium meliloti.1,4 Based on these structural similarities we can conclude that Myc factors and Nod factors are very related, with the notion that in case of Nod factors more structural variation in side-groups and/or acyl chains are known. This finding, together with the knowledge that a MtNFP/LjNFR5-type Nod factor receptor can control two symbioses, discloses the evolutionary origin of rhizobium Nod factor signaling. In evolution, an ancestral free-living nitrogen fixing rhizobium species has gained a biosynthetic pathway of LCOs. This could be the result of horizontal gene transfer of fungal genes, a hypothesis that is supported by the finding of rhizobial endosymbionts in Glomeromycota species.45 Alternatively, such ancestral rhizobium species has phenocopied the Myc factor by synthesizing LCOs along an different pathway, an hypothesis that find support by the unique nature of the rhizobial N-acetylglucosaminyl transferase NodC that is most homologous to animal hyaluronan synthases.46 Either way, the net result is that an LCO producing rhizobium can activate the mycorrhizal signaling cascade of higher plants. In some plant species, like ancestral legumes and Parasponia, it has gained intracellular access, which would have lead to the formation of nodular organs to host the microsymbionts. Ultimately, this resulted in the nitrogen fixing symbioses as we see today.
Perspectives for the Near Future
The finding that in Parasponia an orthologous gene of a legume Nod factor receptor has been recruited to support rhizobium symbiosis strongly suggests that there are genetic constraints underlying evolution of this symbiotic association. A further comparison of the rhizobium symbioses in legumes and Parasponia can reveal an in depth characterization of these genetic constraints.
Interestingly, the PaNFP receptor also controls mycorrhization in Parasponia, which provides leads for future research on the evolution of Nod factor signaling. First of all, this finding, together with the identification of Nod factor-like molecules secreted by the fungus G. intraradices, points to the evolutionary origin of Rhizobium Nod factor signaling; namely the ancient mycorrhizal symbiosis. Simultaneously, these findings underline that the shared common signaling cascade extends from initial signal perception down to CCaMK controlled gene expression. Possibly the shared elements extend even further as the intracellular infection structures of mycorrhiza and Rhizobium display quite some similarities in Parasponia. Therefore it is tempting to speculate that both symbioses also share cellular processes.
The fact that mycorrhizal symbiosis can occur with the vast majority of land plants underlines that LCO signaling is wide spread. Identification of the underlying genes in non-legumes will provide insight in the evolutionary events that have occurred in legumes to exploit the Nod factor-based rhizobium symbiosis. The fact that some species have lost the ability to establish a mycorrhizal symbiosis, e.g., species of the genus Arabidopsis, can be exploited to identify the LCO signaling cascade in non-legumes. For example, Arabidopsis species have lost most genes of the common signaling pathway as well as MtNFP/LjNFR5 (green orthology group Fig. 2).3 Following this argumentation, one could postulate that also root expressed genes of the orthology group of MtLYR2 have a symbiotic function as Arabidopsis species have lost this gene (yellow orthology group Fig. 2).
In legumes MtNFP/LjNFR5 interacts with MtLYK3/LjNFR1, which is essential for symbiotic signaling. Such heterodimerization is essential as MtNFP/LjNFR5 does not contain a functional kinase domain, and therefore is dependent on a interacting partner for downstream signaling.35 Identification of the interacting counterpart of MtNFP/LjNFR5-type receptors in non-legumes is therefore important to understand the functioning of the LCO perception mechanism in non-legumes; including Parasponia. Ultimately such comparative studies on Parasponia and legumes as well as Parasponia and it non-nodulating sister Trema could provide a blueprint for a future transfer of the rhizobium symbiosis to the major non-legume crops.
Acknowledgments
R.G. is funded by the Dutch Science Organisation NWO (VIDI 864.06.007 and ALW 864.06.2007).
References
- 1.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome JC, et al. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 2.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang B, Yeun LH, Xue JY, Liu Y, Ane JM, Qiu YL. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 2010;186:514–525. doi: 10.1111/j.1469-8137.2009.03137.x. [DOI] [PubMed] [Google Scholar]
- 4.Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 5.Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 6.D'Haeze W, Holsters M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology. 2002;12:79R–105R. doi: 10.1093/glycob/12.6.79R. [DOI] [PubMed] [Google Scholar]
- 7.Mergaert P, van Montagu M, Holsters M. Molecular mechanisms of Nod factor diversity. Mol Microbiol. 1997;25:811–817. doi: 10.1111/j.1365-2958.1997.mmi526.x. [DOI] [PubMed] [Google Scholar]
- 8.Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- 9.Pueppke SG, Broughton WJ. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact. 1999;12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 10.Broughton WJ, Jabbouri S, Perret X. Keys to symbiotic harmony. J Bacteriol. 2000;182:5641–5652. doi: 10.1128/JB.182.20.5641-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison NA, Hau CY, Trinick MJ, Shine J, Rolfe BG. Heat curing of a sym plasmid in a fast-growing Rhizobium sp. that is able to nodulate legumes and the nonlegume Parasponia sp. J Bacteriol. 1983;153:527–531. doi: 10.1128/jb.153.1.527-531.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathesius U, Schlaman HR, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- 13.Truchet G, Roche P, Lerouge P, Vasse J, Camut S, de Billy F, et al. Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Cell. 1991;67:1131–1143. [Google Scholar]
- 14.Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, et al. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Faria SM, Sutherland JM, Sprent JI. A new type of infected cells in root nodules of Andria spp. (Leguminosae) Plant Sci. 1986;45:143–147. doi: 10.1016/0168-9452(86)90050-6. [DOI] [Google Scholar]
- 16.Naisbitt T, James EK, Sprent JI. The evolutionary significance of the legume genus Chamaecrista, as determined by nodule structure. New Phytol. 1992;122:487–492. doi: 10.1111/j.1469-8137.1992.tb00077.x. [DOI] [PubMed] [Google Scholar]
- 17.Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S. First report of non-mycorrhizal plant mutants (Myc-) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.) Plant Sci. 1989;60:215–222. doi: 10.1016/0168-9452(89)90169-6. [DOI] [Google Scholar]
- 18.Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, et al. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, et al. Differential and chaotic calcium signatures in the symbiosis signalling pathway of legumes. Proc Natl Acad Sci USA. 2008;105:9823–9828. doi: 10.1073/pnas.0803499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011;65:404–417. doi: 10.1111/j.1365-313X.2010.04431.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanash K, et al. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 2011;65:169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 22.Akkermans ADL, Abdulkadir S, Trinick MJ. Nitrogen-fixing root nodules in Ulmaceae. Nature. 1978;274:190. doi: 10.1038/274190c0. [DOI] [Google Scholar]
- 23.Trinick MJ. Structure of nitrogen-fixing nodules formed by Rhizobium on roots of Parasponia andersonii Planch. Can J Microbiol. 1979;25:565–578. doi: 10.1139/m79-082. [DOI] [PubMed] [Google Scholar]
- 24.Tjepkema JD, Cartica RJ. Diffusion Limitation of Oxygen Uptake and Nitrogenase Activity in the Root Nodules of Parasponia rigida Merr. and Perry. Plant Physiol. 1982;69:728–733. doi: 10.1104/pp.69.3.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sujatha Lilly S, Kannaiyan S. Nitrogen fixation in Parasponia. In: Kannaiyan S, editor. Biotechnology of biofertilizers. New Delhi, India: Narosa Publishing House; 2002. pp. 107–121. [Google Scholar]
- 26.Yesson C, Russel SJ, Parrish T, Dalling JW, Garwood NC. Phylogenetic Framework for Trema (Celtidaceae) Plant Syst Evol. 2004;248:85–109. doi: 10.1007/s00606-004-0186-3. [DOI] [Google Scholar]
- 27.Sytsma KJ, Morawetz J, Pires JC, Nepokroeff M, Conti E, Zjhra M, et al. Urticalean rosids: circumscription, rosid ancestry, and phylogenetics based on rbcL, trnL-F, and ndhF sequences. Am J Bot. 2002;89:1531–1546. doi: 10.3732/ajb.89.9.1531. [DOI] [PubMed] [Google Scholar]
- 28.APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linn Soc. 2003;141:399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x. [DOI] [Google Scholar]
- 29.Becking JH. The Parasponia-Parviflora - Rhizobium Symbiosis - Host Specificity, Growth and Nitrogen-Fixation under Various Conditions. Plant Soil. 1983;75:309–342. doi: 10.1007/BF02369969. [DOI] [Google Scholar]
- 30.Becking JH. The Rhizobium Symbiosis of the nonlegume Parasponia. In: Stacey G, editor. Biological Nitrogen Fixation. New York: Chapman and Hall; 1992. pp. 497–559. [Google Scholar]
- 31.Trinick MJ. Symbiosis between Rrizobium and the non-legume, Trema aspera. Nature. 1973;244:459–460. doi: 10.1038/244459a0. [DOI] [Google Scholar]
- 32.Trinick MJ, Galbraith J. Structure of Root Nodules Formed by Rhizobiurn on the Non-Legume Trema cannabina var. scabra. Arch Microbiol. 1976;108:159–166. doi: 10.1007/BF00428946. [DOI] [Google Scholar]
- 33.Webster G, Poulton PR, Cocking E, Davey MR. The nodulation of micro-propagated plants of Parasponia andersonii by tropical legume rhizobia. J Exp Bot. 1995;46:1131–1137. doi: 10.1093/jxb/46.9.1131. [DOI] [Google Scholar]
- 34.Pawlowski K, Sprent JI. Comparison between actinorhizal and legume symbioses. In: Pawlowski K, Newton WE, editors. Actinorhizal Symbioses. Dordrecht: Kluwer Academic Publishers; 2007. pp. 261–288. [Google Scholar]
- 35.Arrighi JF, Barre A, Ben Amor B, Bersoult A, Campos Soriano L, Mirabella R, et al. The Medicago truncatula Lysine Motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H, Riely BK, Burns NJ, Ane JM. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, et al. Molecular evolution of lysin motiftype receptor-like kinases in plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XC, Cannon SB, Stacey GC. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann GV, Shimodam Y, Nielsen MW, Jørgensenm FG, Grossmann C, Sandal N, et al. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact. 2010;23:510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- 40.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signalling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, et al. A LysM receptor-like kinase plays a critical role in chitin signalling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith SE, Read DJ. Arbuscular mycorrhizas. In: Smith SE, Read DJ, editors. Mycorrhizal symbiosis. 3rd edition. New York: Elsevier; 2008. pp. 11–145. [Google Scholar]
- 45.Bonfante P, Anca IA. Plants, Mycorrhizal Fungi, and Bacteria: A Network of Interactions. Annu Rev Microbiol. 2009;63:363–383. doi: 10.1146/annurev.micro.091208.073504. [DOI] [PubMed] [Google Scholar]
- 46.Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]