Abstract
Plant diversity in nature is to a large extent reflected by morphological diversity of their leaves. Both simple and dissected (with multiple blades or leaflets) leaves are initiated from shoot apical meristem (SAM) in a highly ordered fashion. Similarly, development of leaflets from leaf marginal meristem (marginal blastozone) is also highly ordered. How morphological diversity of plant leaves is regulated remains an important topic of studies on plant form evolution. Here, we describe isolation and characterization of loss-of-function mutants of auxin efflux transporter MtPIN10 of a legume species, Medicago truncatula. Mtpin10 mutants exhibit defects in diverse developmental processes including leaf and leaflet development. Cross species genetic complementation demonstrates that MtPIN10 and Arabidopsis PIN1 are functional orthologs. Double mutant analyses reveal complex genetic interactions between MtPIN10 and Medicago SINGLE LEAFLET1 (SGL1) and CUP-SHAPED COTYLEDON2 (MtCUC2), three regulatory genes involved in developmental processes including dissected leaf and flower development.
Key words: auxin, auxin transport, compound leaf development, MtPIN10, SGL1, MtCUC2, Medicago truncatula
Introduction
In nature, plant leaves exhibit remarkable morphological diversity and can be classified as either simple with a single blade or compound with multiple blades known as leaflets. How divergent leaf morphology is determined remains an important topic of studies on plant form evolution. Both simple and compound leaves are initiated from the periphery of shoot apical meristem (SAM) that consists of pluripotent stem cells capable of self renewal. Development of leaf primordia from the SAM requires downregulation of KNOTTED1-like homeobox transcription factors (KNOXIs) and convergence of activity maxima of the plant hormone auxin at incipient sites of organ initiation.1–4 In simple leafed species such as Arabidopsis thaliana, the KNOXI genes are permanently downregulated in leaves. In tomato and several other species with compound leaves, the KNOXI genes are re-activated in developing leaves and are required for compound leaf development. In some compound leafed species of the legume family (Fabacaea), including pea (Pisum sativum) and Medicago truncatula, FLORICAULA (FLO)/LEAFY(LFY) transcription factor, UNIFOLIATA(UNI)/SINGLE LEAFLET1(SGL1), plays a key role in place of KNOXI in compound leaf development,5–7 consistent with independent origins of compound leafed species during evolution. Recent studies have shown, however, that differential deployment of conserved molecular mechanisms contributes to diversification of leaf forms during evolution.8–10
In Cardamine hirsuta and tomato (Solanum lycopersicum), both with compound leaves, auxin activity maxima converge at incipient sites of leaf primordia in the SAM and of leaflet primordia in leaf margins (marginal blastozones) and condition the outgrowth of leaf and leaflet primordia.11,12 In C. hirsuta, loss-of-function mutations in auxin efflux transporter PIN-FORMED1 (PIN1) gene impair polarized outgrowth and result in simplified leaves, demonstrating involvement of a fundamental mechanism in compound leaf development.12 In A. thaliana, auxin is required for initiation of lateral organs in the SAM, development of serrations in leaf margins and differentiation of vascular tissues.13–17 Polarized outgrowth during organ initiation requires auxin efflux transporter PIN1-mediated auxin activity maxima.13,14,17,18 Consistent with this, Arabidopsis pin1 mutants exhibit pin-like inflorescence stems devoid of reproductive lateral organs, proliferation of vascular tissues and reduced serrations in leaf margins.19–22
Recent studies demonstrate that plant-specific NAC transcription factors play a regulatory role in compound leaf development in diverse species.8,9 The NAC family of transcription factors, including CUP-SHAPED COTYLEDON1, 2, 3 (CUC1, 2, 3) in A. thaliana, NO APICAL MERISTEM (NAM) in petunia, GOBLET (GOB) in tomato and CUPULIFORMIS (CUP) in Antirrhinum majus, are expressed in boundary cells and function to suppress boundary cell growth and promote boundary formation.23–26 In addition, NAC transcription factors activate KNOXI gene expression and are involved in meristem maintenance. Loss-of-function mutants of NAC transcription factors are impaired in shoot meristem development and boundary formation, leading to fusion of leaflets and simplified leaves in several compound leafed species and smooth leaf margins in both compound and simple leafed species.8,9,27
It has been shown that the lineage in the legume family, the inverted repeat-lacking clade (IRLC), including pea and M. truncatula, utilizes the FLO/LFY pathway in place of KNOXI genes in compound leaf development.5–7,28 To investigate the role of auxin efflux transporter PIN1-mediated polar auxin transport in compound leaf development and interactions with the FLO/LFY ortholog, SGL1, and the NAM/CUC ortholog, MtCUC2, we isolated two pin10 mutant alleles in M. truncatula and characterized mutant phenotypes in detail. Double mutants were constructed between Mtpin10 and sgl1, and Mtcuc2 to examine their genetic interactions. Cross-species genetic complementation was performed to examine the degree of functional conservation of MtPIN10 and the Arabidopsis PIN1.
Results and Discussions
Isolation of Medicago truncatula pin10 mutants.
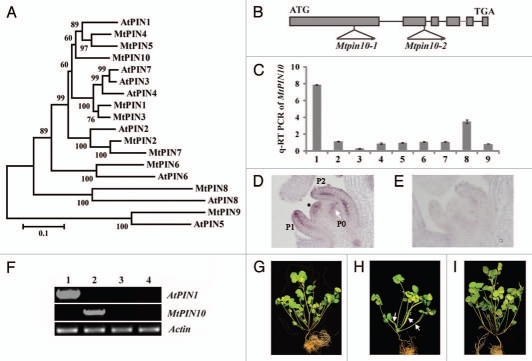
Arabidopsis pin1 mutants exhibit pronounced defects in shoot apical meristem, inflorescence stem, lateral organ and vascular tissue development. To study the role of M. truncatula PIN1 homolog in compound leaf development, we compared sequences of Medicago PIN proteins (MtPIN1–MtPIN10) 29 with the Arabidopsis counterparts (PIN1–PIN8). Phylogenetic analysis indicated that MtPIN4, MtPIN5 and MtPIN10 are clustered with the Arabidopsis PIN1 (Fig. 1A). In addition, MtPIN4, MtPIN5 and MtPIN10 genes share a similar intron-exon structure as the Arabidopsis PIN1 (Fig. 1B) and the encoded proteins share 71%, 65% and 65% amino acid sequence identities with PIN1, respectively. In silico gene expression analysis indicated that MtPIN4 and MtPIN10 are expressed in similar types of tissues in M. truncatula, albeit MtPIN4 being much highly expressed; whereas MtPIN5 expression was not detectable in almost all tissues except that a low level of expression was detected in seed coat (Fig. S1).
Figure 1.
Phylogenetic relationships of Medicago and Arabidopsis PINs and genetic complementation of Mtpin10. (A) Phylogenetic relationships of Medicago and Arabidopsis PINs. MtPIN4, MtPIN5 and MtPIN10 are clustered with AtPIN1. (B) MtPIN10 gene structure and Tnt1 insertion sites in Mtpin10-1 and Mtpin10-2 mutants. (C) Quantitative RT-PCR analysis of MtPIN10 expression pattern. MtPIN10 expression was normalized with an internal control, MtActin. 1, shoot bud; 2, young leaf; 3, mature leaf; 4, rachis; 5, petiole; 6, stem; 7, root; 8, flower and 9, immature pod. (D) RNA in situ hybridization. MtPIN10 transcripts were detected in the shoot apical meristem (asterisk), and P0, P1 and P2 compound leaf primordia. In P1 and P2 leaf primordia, MtPIN10 transcripts were detected in both epidermal and vascular cells. (E) No signal was detected in an adjacent tissue section hybridized with a sense probe. (F) RT-PCR analysis of AtPIN1 and MtPIN10 gene expression. Lanes 1–4, Mtpin10-1 mutant transformed with an Arabidopsis PIN1::PIN1:GFP construct, wild-type (R108), Mtpin10-1 and Mtpin10-2 mutants, respectively. MtActin was used as a loading control. (G–I) Growth defects and genetic complementation of Mtpin10 mutants. Shown were six-week-old wild type (G), Mtpin10-1 (H) and Mtpin10-1 transformed with an Arabidopsis PIN1::PIN1:GFP construct (I). In Mtpin10 mutants, leaves were often fused (H, arrows).
Using a reverse genetics approach, we isolated two independent alleles of Mtpin10 mutant, Mtpin10-1 and Mtpin10-2, from a M. truncatula Tnt1 retrotransposon insertion mutant population.30 However, no mutants of MtPIN4 were uncovered from the screen. Flanking sequence analysis showed that Mtpin10-1 has the Tnt1 inserted in the first exon and Mtpin10-2 in the second exon of MtPIN10 (Fig. 1B). RT-PCR using primers flanking the full-length coding sequence did not detect any MtPIN10 transcripts in both alleles (Fig. 1E), indicating that both are knockout mutants. Both mutants were backcrossed with the wild-type parent. In F2 populations, one quarter of F2 plants (135/536 and 43/180, respectively) were homozygous for Tnt1 insertion in MtPIN10 and exhibited mutant phenotypes, including abnormal phyllotaxy, and leaf and flower development (Fig. 1G and H; Fig. S2). Because Mtpin10 mutants are sterile, heterozygous plants are maintained. Functional conservation between MtPIN10 and the Arabidopsis PIN1 is confirmed by cross-species genetic complementation of Mtpin10-1 with an Arabidopsis PIN1::PIN1:GFP contruct (Fig. 1I). Mtpin10 mutant phenotypes are discussed below.
Abnormal cotyledon development in Mtpin10 mutants.
In contrast to wild-type plants that had two separate cotyledons, Mtpin10 mutants frequently exhibited three or four cotyledons and fusion between them (Fig. 2A and B). This is similar to Arabidopsis pin1 mutants that frequently exhibit three partially fused cotyledons. To investigate early developmental stages in which cotyledon abnormality occurred, embryos from different developmental stages were dissected. Microscopic analysis indicated that abnormal embryos with an increased number of cotyledons were frequently observed in heart-stage embryos in Mtpin10 heterozygous plants (Fig. 2F). By contrast, all heartstage embryos in wild-type plants had two distinct cotyledons (Fig. 2E). No apparent abnormalities were observed in embryos at earlier stages in Mtpin10 mutants (data not shown). These results suggest that loss-of-function mutations in MtPIN10 results in an early embryonic defect, leading to development of an increased number of partially fused cotyledons.
Figure 2.
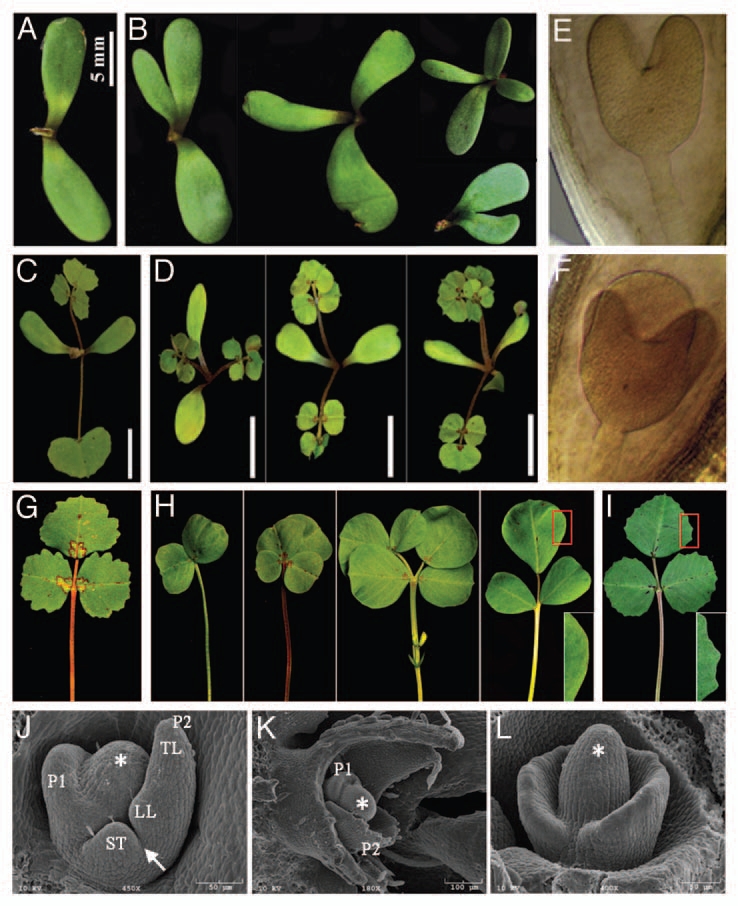
Mtpin10 phenotypes. (A and B) Mtpin10 mutants frequently exhibited three to four cotyledons and partial fusion of cotyledons (B). By contrast, all wild-type plants exhibited two separate cotyledons (A). (C and D) The juvenile leaf was always compound in Mtpin10 mutants (D), but simple in wild-type plants (C). (E and F) Heart-stage embryos always had two cotyledons in wild-type plants (E). However, in Mtpin10 (+/−) plants, three cotyledons were frequently observed (F). (G and H) Compound leaves of wild-type (G) and Mtpin10-1 mutant (H). (G) A representative compound leaf of wild-type plants (R108). (H) Representative compound leaves of Mtpin10-1 mutant. Observed were a wide range of abnormal leaves, including leaves lacking a terminal leaflet, leaves with multiple leaflets with fusion of terminal leaflets, leaves with multiple leaflets presumably derived from fusion of two leaves and trifoliate leaves with altered symmetry of lateral leaflets. In all cases, leaf margins were always smooth (H, inset), unlike leaf margin serrations in wild-type plants (G). (I) Genetic complementation of Mtpin10-1 mutant with an Arabidopsis PIN1::PIN1:GFP construct. Shown was a representative compound leaf of Mtpin10-1 transformed with the Arabidopsis PIN1 gene. Wild-type compound leaves and leaf margin serrations (inset) were restored. (J–L) SEM analysis of leaf development. Compound leaf primordia were initiated from the periphery of SAM (asterisk). (J) In wild-type P2 leaf primordia, terminal (TL) and lateral leaflet (LL) and stipule (ST) primordia were clearly recognizable and a boundary between lateral leaflet and stipule primordia (arrow) was established. (K) In Mtpin10 mutants, compound leaf primordia were initiated from the periphery of SAM but appeared to be much broader and without recognizable leaflet primordia at P2. (L) In some cases, almost the entire periphery of SAM gave rise to compound leaf primordia, which may explain fusion of leaves in mutants.
Compound juvenile leaf in Mtpin10 mutants.
In wild-type Medicago plants, the first leaf, or the so-called juvenile leaf, is always simple and leaves developed later are trifoliate, consisting of a pair of lateral leaflets, a terminal leaflet, a petiole and a pair of stipules. By contrast, the juvenile leaf in Mtpin10 mutants was always compound (Fig. 2C and D). In fact, the first few leaves including the juvenile leaf frequently had four leaflets with an even pinnate configuration in Mtpin10 mutants (Fig. 2D). Occasionally, the two distal leaflets fused, leaving two visible mid-veins.
Mtpin10 mutants developed abnormal compound leaves with smooth margins.
Mtpin10 mutants exhibited pronounced defects in compound leaf development, including an increase in leaflet number, fusion of leaves and leaflets and an altered placement of lateral leaflets (Figs. 1G, H, 2G and H). SEM analysis indicated that compound leaf primordia were initiated from the periphery of the SAM in a sequential order in wild-type plants (Fig. 2J). In a P2 stage leaf primordium, terminal and lateral leaflet, and stipule primordia were clearly visible and a boundary between stipule and lateral leaflet primordia was established (Fig. 2J). By contrast, compound leaf primordia appeared to be much broader with less recognizable leaflet primordia at the P2 stage in Mtpin10 mutants (Fig. 2K). In some cases, almost the entire periphery of the SAM developed into compound leaf primordia (Fig. 2L), which may explain fusion of leaves and altered phyllotaxy seen in Mtpin10 mutants (Figs. 1H and 2H).
Plant leaves, both simple and compound, exhibit characteristic leaf margin morphologies such as smooth, serrated or lobed margins due to secondary morphogenesis. Auxin activity maxima mediated by PIN1 are required not only for the initiation and differentiation of compound leaf primordia but also for the elaboration of margins of both simple and compound leaves.14,27,31,32 Wild-type Medicago plants had serrated leaf margins (Fig. 2G). However, all leaves in Mtpin10 mutants had smooth margins (Fig. 2H and inset). In Mtpin10-1 plants stably transformed with an Arabidopsis PIN1::PIN1:GFP construct, which is sufficient to complement A. thaliana pin1 mutants, all mutant phenotypes including smooth leaf margins were rescued (Figs. 1I and 2I). These results indicate that MtPIN10 and the Arabidopsis PIN1 are functional orthologs and MtPIN10 plays a key role in diverse developmental processes ramified in Mtpin10 mutants.
Abnormal floral development.
In contrast to Arabidopsis pin1 mutants, which exhibit a naked inflorescence meristem devoid of lateral organs, Mtpin10 mutants developed flowers. However, inflorescence and flower were abnormal and plants were sterile. In wild-type Medicago plants, one to three flowers developed on a single stalk (Fig. 3A). In Mtpin10 mutants, however, up to 10 flowers were formed and clustered on a stalk (Fig. 3B and C). And, unlike wild-type flowers that have a pea flower-like closed structure, all flowers were precociously opened in Mtpin10 mutants (Fig. 3A–C). Medicago flowers have pentamerous organs in the outermost four whorls (sepals, petals, and outer and inner stamens) and a central carpel (Fig. 3D).7,33 Floral organs were reduced in Mtpin10 mutants, although the degree of reduction was variable among flowers (Fig. 3E–G). In the mutants, different floral organs were frequently fused. Unlike the central carpel enclosed by a stamen tube in wild-type plants, Mtpin10 mutants had an exposed central carpel (Fig. 3D–G). In some flowers (5/60), two carpels were formed (Fig. 3F). It is noticeable that petals were much less serrated and petal teeth were fatter and shorter in Mtpin10 mutants than wild-type counterparts (Fig. 3H–K).
Figure 3.
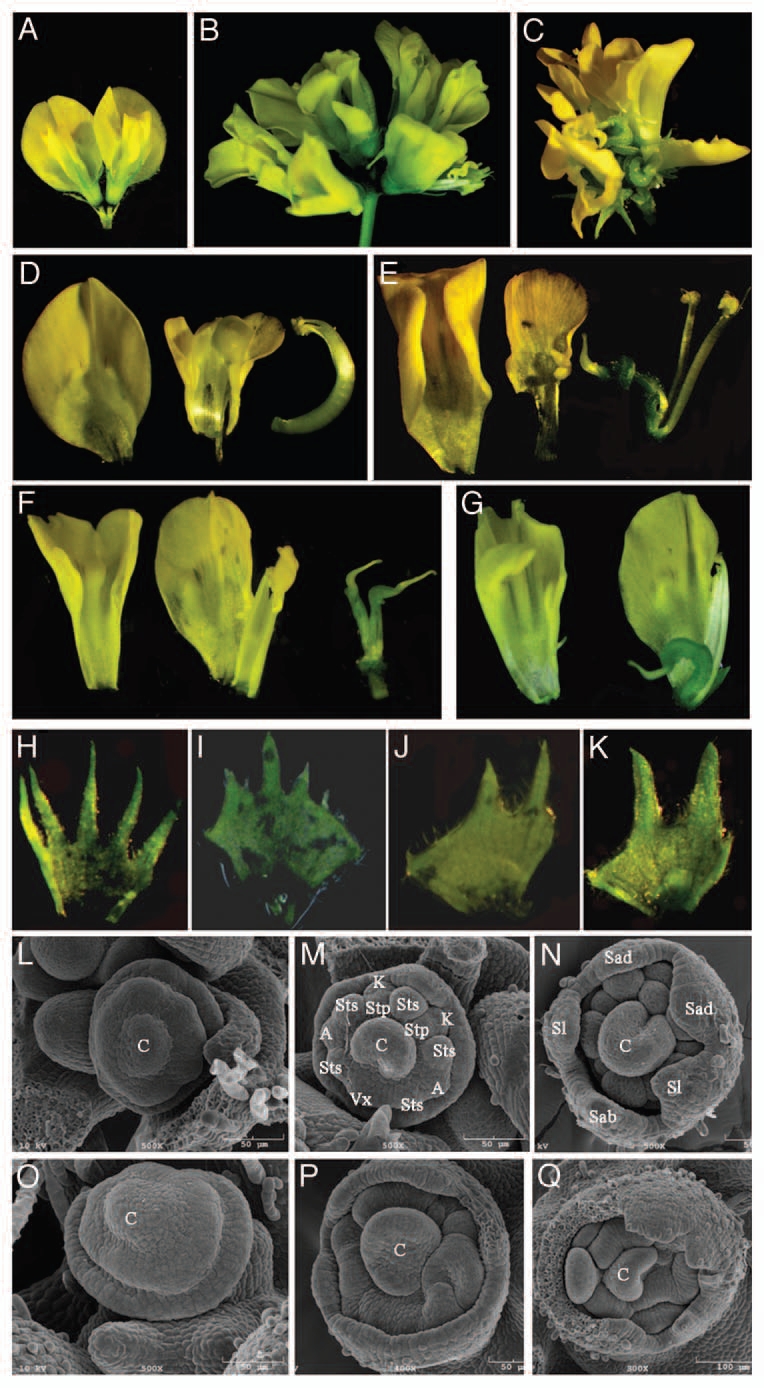
Floral defects in Mtpin10 mutants. (A–C) Flowers developed on inflorescence stalks. Wild-type plants developed 1–3 flowers on each stalk (A), whereas Mtpin10 mutants developed up to 10 flowers on a stalk (B). In contrast to a closed pea flower-like structure of wild-type flowers (A), flowers in Mtpin10 mutants were always precociously opened (B and C). (D–K) Dissected floral organs. A wild-type flower had a standard petal (or vexillum), two keel petals, two fused alae petals, a central carpel and 10 fused stamens (D), and a circular sepal with five teeth (H). In Mtpin10 flowers, floral organs were frequently missing and had irregular shapes (E–G); the central carpel was always exposed (E–G); stamens were not fused (E); two carpels were occasionally formed (5/60); and the circular sepal had a reduced number of teeth that were shorter and fatter than the wild-type counter parts (H–K). (L–Q) SEM analysis of flower development. Early stages of floral meristem development appeared to be similar in wild-type (L) and Mtpin10-1 mutant (O). Between S4 and S5 stages, common primordia in the second whorl gave rise to petal and stamen primordia along the abaxial-adaxial direction in wild type (M). In Mtpin10-1, the developmental polarity was less pronounced and common primordia in the second whorl failed to properly differentiate, resulting in misshaped floral organ primordia (P). At a late S5 stage, all floral organ primordia were clearly developed in wild-type (N), but not in Mtpin10-1 mutant (Q). C, carpel; Vx, vexillum; K, keel petal; A, alae petal; Sts, antesepal stamen; Stp, antepetal stamen; Sab, abaxial sepal; Sad, adaxial sepal; Sl, lateral sepal.
SEM analysis indicated that early flower development appeared to be normal in Mtpin10 mutants (Fig. 3L and O). In wild-type plants, development of floral organ primordia occurred along the abaxial-adaxial axis (Fig. 3M).7,33 In Mtpin10 mutants, the developmental polarity along the abaxial-adaxial axis was less pronounced (Fig. 3P). In wild-type plants, all floral organ primordia were clearly differentiated in a late S5 stage (Fig. 3N). By contrast, floral organ primordia were misshaped and differentiation of common primordia in the second whorl, which would normally give rise to petals and stamens, was impaired in Mtpin10 mutants (Fig. 3P and Q).
MtPIN10 was expressed in the shoot apical meristem and leaf primordial.
Quantitative RT-PCR was first performed to investigate tissue specific expression pattern of MtPIN10. The results showed that MtPIN10 was expressed in vegetative shoot apices and flowers (Fig. 1C). In mature leaves, however, MtPIN10 expression was relatively low (Fig. 1C). These results are consistent with in silico expression analysis of MtPIN10 (Fig. S1).
Next, RNA in situ hybridization was performed to identify cell types in which MtPIN10 was expressed. Using an MtPIN10-specific probe, MtPIN10 transcripts were detected in the SAM and P0 leaf primordia (Fig. 1D). In P2 and P3 leaf primordia, a high level of MtPIN10 expression was confined to vascular and epidermal cells, as well as in tips of leaf and leaflet primordia (Fig. 1D; data not shown). As a negative control, a sense probe did not detect any signals (Fig. 1E). MtPIN10 expression in the SAM suggests that it may play a role in SAM maintenance, whereas its expression in incipient sites of leaf initiation, and vascular and epidermal cells in developing leaf primordia is consistent with its role in mediating auxin transport in leaf and leaflet initiation and differentiation.
Cross-species genetic complementation of Mtpin10.
To evaluate functional conservation of MtPIN10 and the Arabidopsis PIN1, we introduced an Arabidopsis PIN1::PIN1-GFP construct that is sufficient to rescue A. thaliana pin1 mutants into Mtpin10-1 mutant via Agrobacterium tumefaciense-mediated transformation. Independent transgenic lines that were obtained exhibited wild-type morphologies, including two separate cotyledons, simple juvenile leaf, trifoliate adult leaves, serrated leaf margins, normal phyllotaxy and fertile flowers (Figs. 1I and 2I). These results indicate that the Arabidopsis PIN1 completely rescued Mtpin10 mutant phenotypes and MtPIN10 is the Medicago PIN1 ortholog.
Genetic interactions between sgl1 and Mtpin10.
Previous studies have shown that M. truncatula SGL1 is required for compound leaf and flower development.7 To examine genetic interactions between Mtpin10 and sgl1, we made crosses between sgl1-1 and Mtpin10-1 and generated sgl1-1 Mtpin10-1 double mutants. Phenotypic analysis indicated that all leaves in the double mutants were simple, resembling the sgl1 single mutant (Fig. 4). However, leaf margins of the double mutants were smooth, resembling the Mtpin10 single mutant. On the other hand, flowers of the sgl1 Mtpin10 double mutants were cauliflower-like, resembling sgl1. Taking together, these results indicate that sgl1 is genetically epistatic to Mtpin10 in leaflet development and inflorescence differentiation, and Mtpin10 is epistatic to sgl1 in control of leaf margin serration.
Figure 4.
Genetic Interactions of Mtpin10 and single leaflet1 (sgl1), and cup-shaped cotyledon2 (Mtcuc2). Shown were mature leaves of wild-type, sgl1-1, sgl1-1 Mtpin10-1, Mtcuc2-2 and Mtcuc2-1 Mtpin10-1 plants. Leaves of sgl1 Mtpin10 double mutants are simple with smooth margins. Leaves of Mtcuc2-2 Mtpin10-1 double mutants are partially fused with smooth margins.
Genetic interactions between Mtcuc2 and Mtpin10.
It has been recently shown that MtCUC2 encoding a NAC domain transcription factor is required for maintenance of the SAM and development of organ boundaries in M. truncatula (Cheng et al., unpublished results). Mtcuc2-1, a strong allele, does not form any shoot structures; whereas Mtcuc2-2, a weak allele, exhibits partial fusion of cotyledons and leaflets but with wild type-like leaf margin serrations (Fig. 4). To evaluate genetic interactions between Mtcuc2 and Mtpin10, we constructed Mtcuc2-2 Mtpin10-1 double mutants. The double mutants exhibited fusion of cotyledons and leaflets that resembles the Mtcuc2-2 single mutant and smooth leaf margins that resembles the Mtpin10-1 single mutant. These results indicate that Mtcuc2 is genetically epistatic to Mtpin10 in boundary separation and leaf patterning; whereas Mtpin10 is epistatic to Mtcuc2 in leaf margin serration. Similar to tomato GOB, MtCUC2 is the only CUC-like gene identified in M. truncatula genome. Yet, Mtcuc2 mutants exhibited pronounced defects in shoot apical meristem maintenance and boundary separation. Shoots can be developed in regenerated Mtcuc2-1 mutant through tissue culture. However, both Mtcuc2-1 and Mtcuc2-2 mutant alleles had wild type-like serrated leaf margins (Fig. 4). These results suggest that MtCUC2 may not play a prominent role in leaf margin serration, like NAM/CUC genes from other species do. Alternately, some unidentified MtCUC2 homologs that may exist in the M. truncatula genome mask the role of MtCUC2 in leaf margin formation.
Our mutant studies identified a key role for MtPIN10 encoding an auxin efflux transporter in diverse developmental processes in M. truncatula. The role of MtPIN10 in embryonic cotyledon development is similar to the Arabidopsis PIN1. However, the role of MtPIN10 in compound leaf development appears to be different from the PIN1 ortholog in C. hirsuta, a compound leafed close relative of A. thaliana. The role of MtPIN10 in inflorescence and floral meristem development also appears to be different from the Arabidopsis PIN1. In addition, our double mutant analysis revealed complex genetic interactions of MtPIN10 with SGL1 and MtCUC2 in both leaf and flower development. Our studies revealed that MtPIN10 but not MtCUC2 plays a prominent role in leaf margin serration. Cross-species genetic complementation studies not only support functional conservation of MtPIN10 and the Arabidopsis PIN1 but also raise some interesting questions how fundamental mechanisms are differentially deployed to regulate developmental processes in independent lineages of plants during evolution.
Materials and Methods
Plant growth.
M. truncatula seeds were treated and germinated as previously described in reference 7.
Phylogenetic analysis.
Cluster X was used for multiple sequence alignments as previously described in reference 34. Phylogenetic trees were constructed using neighbor-joining, maximum parsimony and UPGMA algorithms implemented in MEGA software suite (www.megasoftware.net)35 with 1,000 bootstrap replicates.
Reverse genetics screening of Mtpin10 mutants.
To isolate Mtpin10 mutants, PCR-based reverse genetics screening of the Medicago Tnt1 mutant population was performed as previously described in reference 30. Briefly, nested PCRs using the following nested forward and reverse primers, MtPIN10-N1-F, MtPIN10-N2-F, MtPIN10-N1-R and MtPIN10-N2-R, Tnt1-F, Tnt1-F1, Tnt1-R and Tnt1-R1 were performed and a total of 18 super-pools of DNA samples prepared from 9,000 Tnt1 insertion mutant lines were screened. Positive PCR products were subsequently confirmed in lower level DNA pools and individual lines. The PCR products were purified, cloned and sequenced using Tnt1-F2 or Tnt1-R2 primer. Resulting flanking sequences were used to determine Tnt1 insertion sites in MtPIN10. MtPIN10-F and MtPIN10-R primers were used to amplify MtPIN10 transcripts by reverse transcription (RT)-PCR, and MtPIN10-q-F and MtPIN10-q-R primers were used in quantitative RT-PCR analysis of MtPIN10 gene expression.
RNA in situ hybridization.
RNA in situ hybridization was performed as previously described in references 7, 36 and 37. Briefly, MtPIN10 probes were generated from a non-conserved region of MtPIN10. Ten micrometer sections prepared from shoot apices of 2-week-old wild type plants were hybridized with digoxigenin-labeled sense or antisense probes. Primer sequences are listed in Table 1.
Table 1.
Primers used in this study
| Name | Forward primer | Reverse primer |
| MtPIN10-RT | ATG ATA AGT GCT TTA GAC TTA TAC | TCA AAG TCC CAA TAA AAT GTA GTA AAC |
| MtPIN10-N1 | AAA GTC CTC CCC TTT TTA CCA TAT CC | AAA TAT GCA AAA GTT GGG TTC ACA GT |
| MtPIN10-N2 | ACC ATA TCC CTG TCT TCT TCC A | ACT ACC TAA CTA GCT ATT GCC CT |
| AtPIN1-RT | CTA TGA TCC TCG CTT ACG GCTC | GTT TAG CAG GAC CAC CGT CTT C |
| MtPIN10-q | CCA TGT CTT TAG AGG TGG TGG TG | CAC TGG TCC TTC TTT GTC CAC AC |
| MtActinB-RT | TCT TAC TCT CAA GTA CCC CAT TGA GC | GTG GGA GTG CAT AAC CTT CAT AGA TT |
| MtActinB-q | TCA ATG TGC CTG CCA TGT ATG T | ACT CAC ACC GTC ACC AGA ATC C |
Tissue clearing.
Flowers and seeds dissected from young pods 1∼4 d after pollination were cleared in Hoyer's solution (7.5 g gum Arabic, 100 g chloral hydrate, 5 ml glycerol and 60 ml ddH2O). Ovules and embryos were further dissected and observed under microscope (specify the type of microscope used).
Scanning electron microscopy.
Shoot apices of 2- to 4-week-old seedlings were subjected to vacuum infiltration in a fixative solution (3% glutaraldehyde in 25 mM phosphate buffer, pH 7.0) for 1 h and then incubated at 4°C overnight. Plant tissues were further fixed with 1.0% osmium tetroxide in the same fixative buffer overnight and dehydrated in a graded ethanol series. Scanning electron microscopy (SEM) was performed as described previously in reference 7.
RNA Extraction, RT-PCR, qRT-PCR.
Roots, stems, leaves, vegetative shoots, inflorescence shoots, flower buds, young pods and young seeds were collected from M. truncatula wild-type (R108) plants. Vegetative shoot buds were collected from wild-type and homozygous Mtpin10-1 and Mtpin10-2 mutants. Total RNA was extracted using Trizol (Invitrogen) and treated with Turbo DNase I (Ambion). Three micrograms of total RNA were used for reverse transcription using SuperScript III Reverse Transcriptase (Invitrogen) with olig(dT)20 primer. Two microliters of 1:20 diluted cDNA were used as templates. Gene-specific primers used are listed in Table 1. Quantitative RT-PCR was performed as previously described in reference 38. The expression level was normalized with M. truncatula ACTIN (tentative consensus no. 107326).
Stable plant transformation.
An Arabidopsis PIN1::PIN1:GFP construct was introduced into Agrobacterium tumefaciens EHA105 strain by electroporation. Mtpin10 heterozygous plants were transformed following the protocol previously reported in reference 39.
Acknowledgments
The authors wish to thank members of the Chen laboratory for helpful comments on the manuscript, Xiaofei Cheng, Junying Ma, Yingqing Guo and Guangming Li for expert technical support. This work was supported by the National Science Foundation (DBI 0703285) and the Samuel Roberts Noble Foundation.
Supplementary Material
References
- 1.Moon J, Hake S. How a leaf gets its shape. Curr Opin Plant Biol. 2011;14:24–30. doi: 10.1016/j.pbi.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Efroni I, Eshed Y, Lifschitz E. Morphogenesis of simple and compound leaves: a critical review. Plant Cell. 2010;22:1019–1032. doi: 10.1105/tpc.109.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakayama N, Kuhlemeier C. Leaf development: untangling the spirals. Curr Biol. 2009;19:71–74. doi: 10.1016/j.cub.2008.11.036. [DOI] [PubMed] [Google Scholar]
- 4.Hasson A, Blein T, Laufs P. Leaving the meristem behind: the genetic and molecular control of leaf patterning and morphogenesis. C R Biol. 2010;333:350–360. doi: 10.1016/j.crvi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Hofer J, Gourlay C, Michael A, Ellis TH. Expression of a class 1 knotted1-like homeobox gene is downregulated in pea compound leaf primordia. Plant Mol Biol. 2001;45:387–398. doi: 10.1023/A:1010739812836. [DOI] [PubMed] [Google Scholar]
- 6.Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol. 1997;7:581–587. doi: 10.1016/S0960-9822(06)00257-0. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Chen J, Wen J, Tadege M, Li G, Liu Y, et al. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 2008;146:1759–1772. doi: 10.1104/pp.108.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, et al. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136:823–832. doi: 10.1242/dev.031625. [DOI] [PubMed] [Google Scholar]
- 9.Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, et al. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 10.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 11.Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N. Auxin patterns Solanum lycopersicum leaf morphogenesis. Development. 2009;136:2997–3006. doi: 10.1242/dev.033811. [DOI] [PubMed] [Google Scholar]
- 12.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 13.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 14.Hay A, Barkoulas M, Tsiantis M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–3961. doi: 10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]
- 15.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 17.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, Prusinkiewicz P, et al. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009;23:373–384. doi: 10.1101/gad.497009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gälweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 20.Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, et al. VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development. 2005;132:1699–1711. doi: 10.1242/dev.01716. [DOI] [PubMed] [Google Scholar]
- 21.Mattsson J, Sung ZR, Berleth T. Responses of plant vascular systems to auxin transport inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- 22.Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development. 2004;131:915–922. doi: 10.1242/dev.00993. [DOI] [PubMed] [Google Scholar]
- 24.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 25.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 27.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 29.Schnabel EL, Frugoli J. The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Genet Genomics. 2004;272:420–432. doi: 10.1007/s00438-004-1057-x. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X, Wen J, Tadege M, Ratet P, Mysore KS. Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol. 2011;678:179–190. doi: 10.1007/978-1-60761-682-5_13. [DOI] [PubMed] [Google Scholar]
- 31.Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, et al. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA. 2011;108:3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, et al. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23:54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benlloch R, Navarro C, Beltran JP, Canas LA. Floral development of the model legume Mediago truncatula: ontogeny studies as a tool to better characterize homeotic mutations. Sex Plant Reprod. 2003;15:231–241. [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-F. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Yu J, Ge L, Wang H, Berbel A, Liu Y, et al. Control of dissected leaf morphology by a Cys(2) His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc Natl Acad Sci USA. 2010;107:10754–10759. doi: 10.1073/pnas.1003954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, et al. A gene expression atlas of the model legume. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 39.Cosson V, Durand P, d'Erfurth I, Kondorosi A, Ratet P. Medicago truncatula transformation using leaf explants. Methods Mol Biol. 2006;343:115–127. doi: 10.1385/1-59745-130-4:115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.