Abstract
Proteolysis-related genes have diverse functions across taxa and have long been considered as key players for intracellular protein turnover. Growing evidence indicates the biological significance of peptidases in degradation, maturation and modulation of bioactive peptides/proteins. By screening T-DNA tagged lines and functional analysis approaches we unraveled the Arabidopsis leucine aminopeptidase (AtLAP2) function in amino acid turnover. Transcriptomics and metabolomics profiling data suggested involvement of AtLAP2 in specific metabolic pathways. Loss-of-function of AtLAP2 resulted in early-leaf senescent and stress-sensitive phenotypes. Our work indicates an important in planta role for AtLAP2 contributing to a further understanding of the proteases having several implications in higher plants.
Key words: leucine aminopeptidase, Arabidopsis, stress response, plant growth, senescence
Leucine aminopeptidase (LAP) is ubiquitously found in all living organisms. LAPs are members of M1 or M17 families1–3 that cleave the leucine (Leu) residue from N-terminal of proteins or peptides. Substantial activities of LAPs may also be evident on other amino acids. Functional diversification of a LAP family across taxa has been reported. In mammals, LAPs contribute in generating antigenic peptides and processing of bioactive peptide hormones.4 In addition, LAP along with other peptidases is important in the degradation of crystalline protein in the eye after oxidative stress.5 Most recently, the role of ERAP1 in innate immunity by processing particular substrate(s) was also suggested.6 In prokaryotes, LAPs have a role in proteolysis, potential virulence,7 breakdown of hemoglobin in infected red blood cells,8 and replication.9
In higher plants, the LAP family has been extensively studied in tomato but to a lesser extent from other plant species. Given our interest in understanding biological roles of LAPs in higher plants, we recently performed functional and expression analyses of LAP2 in a model dicot plant genome model, Arabidopsis thaliana. We demonstrated Arabidopsis LAP2 (AtLAP2) is an enzymatically active aminopeptidase, which is responsible for the cleavage of Leu, Met (methionine), and Phe (phenylalanine) from N-terminal peptides, and plays important roles in various cellular processes in planta.10 In this addendum, we elaborate our discussion on a distinct biological function of AtLAP2, expression pattern, compared with well-characterized plant LAPs.
AtLAP2 is Expressed in all Organs but Unresponsive to Mechanical Wounding
To date, the genome sequences of several model plants have been completed. Information of numerous Expression Sequenced Tag (EST) libraries is now available. Analysis of the LAPs among the sequenced genomes using KEGG GENES database (www.genome.jp/kegg/genes.html) has shown a multiplicity of LAPs (e.g., Lycopersicum esculentum, A. thaliana, A. lyrata, Oryza sativa and Sorghum bicolor). The redundancy of LAPs might reflect various requirements, overlapping functions and/or different localization in plants. Extensive studies of the tomato LAP (LeLAP) revealed two classes (LeLAP-A, acidic pI and LeLAP-N, neutral pI) exhibiting distinct biochemical characteristics.11 Immunoblotting analysis revealed the presence of two additional LeLAP-like proteins.12 LeLAP-A modulates plant defense against pathogen and is expressed in response to various stimuli.13,14 Suppression of LeLAP-A impaired the wound response.14 By contrast, LeLAP-N is not known to respond to environmental stresses.11 In the Arabidopsis genome, three putative LAP genes have been identified. We recently presented a new functional study of the AtLAP2 and showed that AtLAP2 loss-of-function leads to early leaf senescence and rendered plant more sensitive to applied stresses.10
AtLAP2 was expressed in all organs (R, root; RO, rosette; CA, cauline; ST, stem; FL, flower) examined. Expression pattern of AtLAP1 and AtLAP2 was very similar whereas the AtLAP3 was expressed at very low levels (Fig. 1). Spatial expression of promotor-AtLAP2 showed high levels of expression in apices, vascular tissue and quiescent center.10 Unlike the LeLAP-A, AtLAP2 was not induced by mechanical wounding (Fig. 2) and applied environmental stimuli (data not shown). Isoelectric point analysis of the predicted AtLAP proteins revealed that AtLAP1, 2, 3 have theoretical pIs of 5.56, 6.62 and 6.26, respectively. Thus, the putative AtLAP2 and AtLAP3 are likely neutral LAPs while AtLAP1 is likely to be an acidic LAP.
Figure 1.
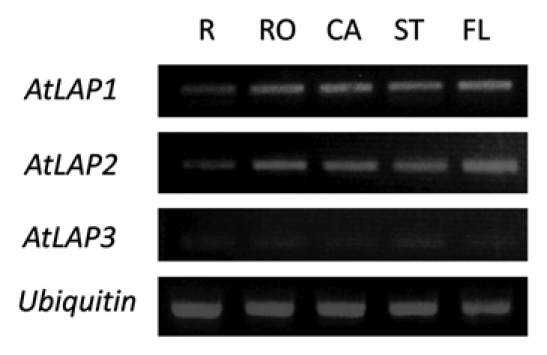
Expression of the AtLAP genes in Arabidopsis organs. The expression level of a ubiquitin gene was used as an internal control. Two-week-old roots (R) were used. Mature (rosette, RO and cauline, CA ) leaves including stems (ST) were obtained from 6-week-old plants. Flowers (FL) were from 8-week-old plants.
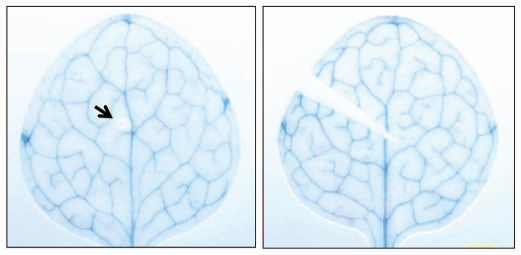
Figure 2.
AtLA P2 promoter activity as determined by histochemical GUS staining in respective reporter gene lines was not induced upon mechanical wounding by piercing with a needle (left) and cutting off the leaf with scissors (right).
Exploring Natural Substrate(s) for Plant LAPs: Tougher than Expected!
LAPs are aminopeptidases that usually cleave Leu most preferentially among synthetic substrates. Indeed in vitro analysis demonstrated that all plant LAPs exhibit a broad specificity toward Leu-, Met-, Phe-, Arg (arginine)-, Pro (proline)-, Ile (isoleucine)-, Val (valine)-, Ala (alanine)-MCAs.10,15–17 Although LAP can cleave many peptides in vitro, the biological relevant target should be determined not only by substrate specificity but also by the natural substrates. In mammals, substrate specificity of LAPs toward natural substrates is usually rather broad. For instance, Laeverin, a novel bestatin-sensitive LAP, was able to cleave the N-terminal amino acid of several natural peptides such as angiotensin III, kisspeptin-10 and endokinin C.18 Another LAP, P-LAP/IRAP cleaves oxytocin, vasopressin and angiotensin III efficiently.19 However, identification of physiological substrate in vivo is still rare.20 Information of plant natural substrate peptide is also scarce. To date, very few examples show possible natural peptides for plant aminopeptidases. Cortes et al. (2006) reported aminopeptidase activity from pollen Parietaria judaica was able to degrade two neuropeptides, namely substrate P and VIP angiotensin.21 In 2011, it was reported that CLE-3, a well known plant active peptide, was possibly degraded by serine protease and carboxypeptidase.22 Thus, identification of the natural substrates for LAP and/or other aminopeptidases would be a very interesting topic for future research.
Acknowledgments
R.W.S. appreciates support of the Ratchadapisek Somphot Endowment Fund. This work was also in part funded by Research Funds from the Faculty of Science, Chulalongkorn University.
References
- 1.Herrera-Camacho I, Rosas-Murrieta NH, Rojo-Domínguez A, Millán L, Reyes-Leyva J, Santos-López G, et al. Biochemical characterization and structural prediction of a novel cytosolic leucyl aminopeptidase of the M17 family from Schizosaccharomyces pombe. FEBS J. 2007;274:6228–6240. doi: 10.1111/j.1742-4658.2007.06142.x. [DOI] [PubMed] [Google Scholar]
- 2.Walling LL. Recycling or regulation? The role of amino-terminal modifying enzymes. Curr Opin Plant Biol. 2006;9:227–233. doi: 10.1016/j.pbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Tsujimoto M, Goto Y, Maruyama M, Hattori A. Biochemical and enzymatic properties of the M1 family of aminopeptidases involved in the regulation of blood pressure. Heart Fail Rev. 2008;13:285–291. doi: 10.1007/s10741-007-9064-8. [DOI] [PubMed] [Google Scholar]
- 4.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Sharma KK, Kester K. Peptide hydrolysis in lens: role of leucine aminopeptidase, aminopeptidase III, prolyloligopeptidase and acylpeptidehydrolase. Curr Eye Res. 1996;15:363–369. doi: 10.3109/02713689608995826. [DOI] [PubMed] [Google Scholar]
- 6.Goto Y, Ogawa K, Hattori A, Tsujimoto M. Secretion of endoplasmic reticulum aminopeptidase 1 is involved in the activation of macrophages induced by lipopolysaccharide and interferon-{gamma} J Biol Chem. 2011;286:21906–21914. doi: 10.1074/jbc.M111.239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jobin MC, Grenier D. Identification and characterization of four proteases produced by Streptococcus suis. FEMS Microbiol Lett. 2003;220:113–119. doi: 10.1016/S0378-1097(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 8.Allary M, Schrevel J, Florent I. Properties, stage-dependent expression and localization of Plasmodium falciparum M1 family zinc-aminopeptidase. Parasitology. 2002;125:1–10. doi: 10.1017/s0031182002001828. [DOI] [PubMed] [Google Scholar]
- 9.Devroede N, Huysveld N, Charlier D. Mutational analysis of intervening sequences connecting the binding sites for integration host factor, PepA, PurR and RNA polymerase in the control region of the Escherichia coli carAB operon, encoding carbamoylphosphate synthase. J Bacteriol. 2006;188:236–245. doi: 10.1128/JB.188.9.3236-3245.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waditee-Sirisattha R, Shibato J, Rakwal R, Sirisattha S, Hattori A, Nakano T, et al. The Arabidopsis aminopeptidase LAP2 regulates plant growth, leaf longevity and stress response. New Phytol. 2011;191:958–969. doi: 10.1111/j.1469-8137.2011.03758.x. [DOI] [PubMed] [Google Scholar]
- 11.Tu CJ, Park SY, Walling LL. Isolation and characterization of the neutral leucine aminopeptidase (LapN) of tomato. Plant Physiol. 2003;132:243–255. doi: 10.1104/pp.102.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu YQ, Pautot V, Holzer FM, Walling LL. A Complex Array of Proteins Related to the Multimeric Leucine Aminopeptidase of Tomato. Plant Physiol. 1996;110:1257–1266. doi: 10.1104/pp.110.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu YQ, Walling LL. Identification of residues critical for activity of the wound-induced leucine aminopeptidase (LAP-A) of tomato. Eur J Biochem. 2002;269:1630–1640. doi: 10.1046/j.1432-1327.2002.02795.x. [DOI] [PubMed] [Google Scholar]
- 14.Fowler JH, Narváez-Vásquez J, Aromdee DN, Pautot V, Holzer FM, Walling LL. Leucine aminopeptidase regulates defense and wound signaling in tomato downstream of jasmonic acid. Plant Cell. 2009;21:1239–1251. doi: 10.1105/tpc.108.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marinova M, Dolashki A, Altenberend F, Stevanovic S, Voelter W, Tchorbanov B. Purification and characterization of L-phenylalanine aminopeptidase from chick-pea cotyledons (Cicer arietinum L.) Protein Pept Lett. 2009;16:207–212. doi: 10.2174/092986609787316333. [DOI] [PubMed] [Google Scholar]
- 16.Lomate PR, Hivrale VK. Induction of leucine aminopeptidase (LAP) like activity with wounding and methyl jasmonate in pigeonpea (Cajanas cajan) suggests the role of these enzymes in plant defense in leguminosae. Plant Physiol Biochem. 2011;49:609–616. doi: 10.1016/j.plaphy.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 17.Herbers K, Prat S, Willmitzer L. Functional analysis of a leucine aminopeptidase from Solanum tuberosum L. Planta. 1994;194:230–240. [PubMed] [Google Scholar]
- 18.Maruyama M, Hattori A, Goto Y, Ueda M, Maeda M, Fujiwara H, et al. Laeverin/aminopeptidase Q, a novel bestatin-sensitive leucine aminopeptidase belonging to the M1 family of aminopeptidases. J Biol Chem. 2007;282:20088–20096. doi: 10.1074/jbc.M702650200. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto H, Rogi T, Yamashiro K, Kodama S, Tsuruoka N, Hattori A, et al. Characterization of a recombinant soluble form of human placental leucine aminopeptidase/oxytocinase expressed in Chinese hamster ovary cells. Eur J Biochem. 2000;267:46–52. doi: 10.1046/j.1432-1327.2000.00949.x. [DOI] [PubMed] [Google Scholar]
- 20.Wallis MG, Lankford MF, Keller SR. Vasopressin is a physiological substrate for the insulin-regulated aminopeptidase IRAP. Am J Physiol Endocrinol Metab. 2007;293:1092–1102. doi: 10.1152/ajpendo.00440.2007. [DOI] [PubMed] [Google Scholar]
- 21.Cortes L, Carvalho AL, Todo-Bom A, Faro C, Pires E, Veríssimo P. Purification of a novel aminopeptidase from the pollen of Parietaria judaica that alters epithelial integrity and degrades neuropeptides. J Allergy Clin Immunol. 2006;118:878–884. doi: 10.1016/j.jaci.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 22.Ni J, Guo Y, Jin H, Hartsell J, Clark SE. Characterization of a CLE processing activity. Plant Mol Biol. 2011;75:67–75. doi: 10.1007/s11103-010-9708-2. [DOI] [PubMed] [Google Scholar]