Abstract
Plant natriuretic peptides (PNPs) are signaling molecules that are secreted into the apoplast particularly under conditions of biotic and abiotic stress. At the local level, PNPs modulate their own expression via feed forward and feedback loops to enable tuning of the response at the transcript and protein level and to prevent overexpression. PNPs also employ a systemic signal, possibly electrical, to rapidly alter photosynthesis and respiration not only in treated leaves but also in upper and lower leaves thereby modulating and integrating physiological responses at the level of the whole plant.
Key words: abiotic stress, photorespiration, photosynthesis, plant homeostasis, plant natriuretic peptides plant stress responses
Plant natriuretic peptides (PNPs) are immunological analogs of mammalian atrial natriuretic peptides that have homeostatic roles in plants. We have previously demonstrated that PNPs and PNP analogs have an emerging role in the regulation of ion fluxes, stomatal movement and fluid circulation in plants.1,2 In addition, PNP affects plant biological activities such as photosynthesis and respiration both locally and at long distances.3–5 Moreover, we recently demonstrated that PNPs were secreted into the apoplast and their expression was upregulated under several stress conditions and importantly PNP was able to modulate its own expression.6 Therefore, here we attempt to further address the control mechanisms of PNP production in planta and the relationship between PNP and plant homeostasis.
Feedback-Regulation of PNP Production
Using a “promoter-reporter” assay, where the AtPNP-A promoter controls luciferase expression (Fig. 1), we show that AtPNP-A can modulate its own gene expression. At the lower concentration AtPNP-A promoted its expression, while at higher concentrations, AtPNP-A repressed its expression. Apparently, the control system of AtPNP-A promoter operates via a feedback loop to prevent excessive AtPNP-A production. A reasonable explanation to this control system may be that in the initial stage AtPNP-A induces itself via positive feedback. Thereby AtPNP-A can be amplified in a very short time. Such a biosynthetic pathway which is promoted by end-products reflects the importance of rapid hormone accumulation for signaling on demand. However, when AtPNP-A production reaches the threshold level, the self-regulation turns to negative feedback to suppress continued expression of AtPNP-A thereby avoiding any harmful effects from overexpression. Consistently, there are similar reports on the positive and negative feedback of ethylene,7 abscisic acid8 and brassinosteroids9 on their own expression. Most plant hormones are biologically active within a certain concentration range. Hormone-expressed genes are often involved in a quantitative control to initiate a qualitative change.10 For effective coordination, hormone levels need to be modulated in a rather complicated network of operations.11 This means that turning on transcription regulators and turning off negative controllers can happen at the same time, and vice versa. Perhaps, this is a universal rule in the plant hormone regulation.
Figure 1.
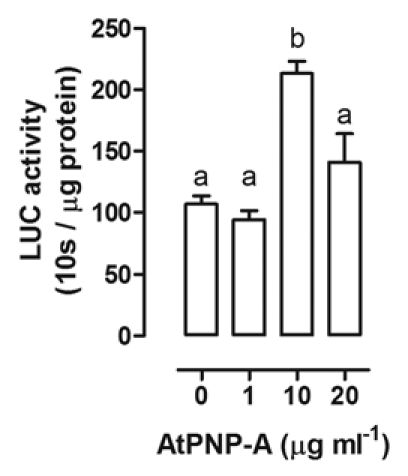
Effect of AtPNP-A treatment on its own expression. LUC activity was measured in protoplasts transfected with ProPNP-A:LUC fusion constructs after 18 h incubation with different concentrations of purified recombinant AtPNP-A as described previously in reference 6. Columns with different letters were significantly different from the others (p < 0.05; one way ANOVA followed by Tukey-Kramer post test).
Our previous report in reference 6 demonstrated that the wave of gene expression of PNP-A did not always correspond to the wave of PNP-A produced at the protein level. Such unmatched production between RNA and protein is probably related to distinctive control mechanisms for different expression layers in biological organisms. Plants may adopt a similar receptor-mediated system as animals12 to maintain the appropriate amount of PNP. Certain amount of PNP can bind to a receptor conceivably with a cytosolic guanylate cyclase domain13 that could allow signaling via the second messenger cGMP.14 The downstream actions of cGMP consequently lead to a series of biological changes.15,16 Meanwhile, the concentration and distribution of PNP may be regulated by a clearance receptor. Once the clearance receptor binds to PNP, the “ligand-receptor” complex will be internalized into the cell and disassociated. Then PNP is degraded while the receptor recycles back to plasma membrane. Such a clearance receptor may be an important player in controlling the dynamic equilibrium of the total PNP pool in circulation. Protein degradation is an essential mechanism for cells to manipulate component homeostasis and to maintain cellular behaviour. Excessive proteins are usually rapidly degraded in cells. Thus protein processing has the possibility to simultaneously include synthesis and degradation. In terms of PNP, both the protein synthesis and degradation need to be considered equally so that the activity of PNP in cells can be defined properly.
The ubiquitin proteasome system is a common biological process for degradation of major proteins in eukaryotic cells.17 A broad array of cellular activities including cell cycle, DNA repair, cell growth and differentiation, protein quality control and regulation of receptors (membrane and intracellular) and ion channels are regulated by the ubiquitin proteasome system.11,18 The system works via enzymes that attach ubiquitin proteins to target proteins, and subsequently subject the target proteins for degradation.17,19 The ubiquitin proteasome system is probably involved in PNP degradation, although other subcellular sites for protein degradation include lysosomes and vacuoles in plants.20 Receptor degradation is an important control mechanism in plant signaling and ligand degradation is also likely to be important. For instance, the auxin receptor, TIR1 is a member of the Skp1/Cullin/F-box ubiquitin ligase (E3) family and so regulates degradation of auxin regulated transcription factors.21
PNP as a Systemic Signal for Plant Homeostasis
AtPNP-A also is involved in signaling long distances within the plant. AtPNP-A application to leaves rapidly increases the rate of dark respiration after ≥5 min. Interestingly, increases in respiration also occur in lower leaves followed with a lag time of ≥10 min in the upper and subsequently (after ≥15 min) the opposite leaves. This response pattern is indicative of a signal with phloem mobility that rapidly influences plant homeostasis in distal parts.5
The exact nature of the long distance signal is still under investigation, but given the velocity, we have hypothesized that it could be electrical.5 Long distance electrical signaling can invoke action potentials or variation potentials.22 Action potentials are phloem transmitted rapid self-propagating waves of membrane depolarization with constant velocity and amplitude. The membrane depolarization is likely to be caused by an initial Ca2+ influx that is strengthened by Cl− efflux followed K+ efflux required for re-polarization mediated by voltage gated channels. Variation potentials are due to local changes in membrane potential possibly caused by ligand activated channels or loss of hydraulic pressure from the xylem. Variation potentials do not involve voltage gated channels and their amplitude and velocity alters with distance from the stimulus and they have longer and more delayed repolarization periods than action potentials. The “electrical signal” hypothesis is supported by the previous findings that PNPs can cause rapid and significant membrane depolarization,23 induce rapid ion fluxes24,25 as well as promote the movement of water out of the xylem.26
In conclusion, since local increases of PNP can have marked influences on plant function distantly,5 it therefore becomes evident that for the plant it is highly important to maintain levels of PNP within a defined range at the local level where it is produced. Hence rapid responses in the leaves to abiotic or biotic stresses will increase transcription and production of PNP at the local level.6 This in turn will result in “electrical” signals altering distant plant homeostasis events such as dark respiration and photosynthetic rates5 so that plant homeostasis is modulated to respond the stress.
Acknowledgments
This work was supported by the Australian Research Council's Discovery project funding scheme (DP0557561, DP0878194) and the South African National Research Foundation.
Abbreviations
- PNP
plant natriuretic peptide
Addendum to: Wang YH, Gehring C, Irving HR. Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 2011;52:837–850. doi: 10.1093/pcp/pcr036. and Ruzvidzo O, Donaldson L, Valentine A, Gehring C. The Arabidopsis thaliana natriuretic peptide AtPNP-A is a systemic regulator of leaf dark respiration and signals via the phloem. J Plant Physiol. 2011;168:1710–1714. doi: 10.1016/j.jplph.2011.03.011.
References
- 1.Gehring CA, Irving HR. Natriuretic peptides—a class of heterologous molecules in plants. Int J Biochem Cell Biol. 2003;35:1318–1322. doi: 10.1016/s1357-2725(03)00032-3. [DOI] [PubMed] [Google Scholar]
- 2.Meier S, Irving HR, Gehring C. Plant natriuretic peptides—emerging roles in fluid and salt balance. In: Vesely DL, editor. Cardiac Hormones. Kerala, India: Transworld Research Network; 2008. pp. 1–17. [Google Scholar]
- 3.Garavaglia BS, Thomas L, Zimaro T, Gottig N, Daurelio LD, Ndimba B, et al. A plant natriuretic peptide-like molecule of the pathogen Xanthomonas axonopodis pv. citri causes rapid changes in the proteome of its citrus host. BMC Plant Biol. 2010;10:51. doi: 10.1186/1471-2229-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottig N, Garavaglia BS, Daurelio LD, Valentine A, Gehring C, Orellano EG, et al. Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide-like protein to modify host homeostasis. Proc Natl Acad Sci USA. 2008;105:18631–18636. doi: 10.1073/pnas.0810107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruzvidzo O, Donaldson L, Valentine A, Gehring C. The Arabidopsis thaliana natriuretic peptide AtPNP-A is a systemic regulator of leaf dark respiration and signals via the phloem. J Plant Physiol. 2011;168:1710–1714. doi: 10.1016/j.jplph.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Wang YH, Gehring C, Irving HR. Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 2011;52:837–850. doi: 10.1093/pcp/pcr036. [DOI] [PubMed] [Google Scholar]
- 7.Fellner M, Franklin JA, Reid DM, Sawhney VK. Increased sensitivity to, and reduced production of, ethylene in an ABA-overproduction tomato mutant. Acta Biol Cracoviensia: Series Botanica. 2005;47:205–212. [Google Scholar]
- 8.Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 9.Szekeres M. Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends Plant Sci. 2003;8:102–104. doi: 10.1016/S1360-1385(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 10.Teale WD, Ditengou FA, Dovzhenko AD, Li X, Molendijk AM. Auxin as a model for the integration of hormonal signal processing and transduction. Mol Plant. 2008;1:229–237. doi: 10.1093/mp/ssn006. [DOI] [PubMed] [Google Scholar]
- 11.Wang YH, Irving HR. Developing a model of plant hormone interactions. Plant Signal Behav. 2011;6:494–500. doi: 10.4161/psb.6.4.14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewicki JA, Protter AA. Molecular determinants of natriuretic peptide clearance receptor function. In: Samson WK, Levin ER, editors. Natriuretic Peptides in Health and Disease. Totowa, NJ: Humana Press; 1997. pp. 51–69. [Google Scholar]
- 13.Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C. The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One. 2010;5:8904. doi: 10.1371/journal.pone.0008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwezi L, Ruzvidzo O, Wheeler JI, Govender K, Iacuone S, Thompson PE, et al. The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependant signaling in plants. J Biol Chem. 2011;286:22580–22588. doi: 10.1074/jbc.M110.168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YH, Gehring C, Cahill DM, Irving HR. Plant natriuretic peptide active site determination and effects on cGMP and cell volume regulation. Funct Plant Biol. 2007;34:645–653. doi: 10.1071/FP06316. [DOI] [PubMed] [Google Scholar]
- 16.Pharmawati M, Maryani MM, Nikolakopoulos T, Gehring CA, Irving HR. Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiol Biochem. 2001;39:385–394. [Google Scholar]
- 17.Viestra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nature Reviews Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 18.Reinstein E, Clechanover A. Narrative review: Protein degradation and human diseases: The ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 19.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muentz K. Protein dynamics and proteolysis in plant vacuoles. J Exp Bot. 2007;58:2391–2407. doi: 10.1093/jxb/erm089. [DOI] [PubMed] [Google Scholar]
- 21.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 22.Fromm J, Lautner S. Electrical signals and their physiological significance in plants. Plant Cell Environ. 2007;30:249–257. doi: 10.1111/j.1365-3040.2006.01614.x. [DOI] [PubMed] [Google Scholar]
- 23.Maryani MM, Shabala SN, Gehring CA. Plant natriuretic peptide immunoreactants modulate plasma-membrane H+ gradients in Solanum tuberosum L. leaf tissue vesicles. Arch Biochem Biophys. 2000;376:456–458. doi: 10.1006/abbi.2000.1736. [DOI] [PubMed] [Google Scholar]
- 24.Ludidi N, Morse M, Sayed M, Wherrett T, Shabala S, Gehring C. A recombinant plant natriuretic peptide causes rapid and spatially differentiated K+, Na+ and H+ flux changes in Aradidopsis thaliana roots. Plant Cell Physiol. 2004;45:1093–1098. doi: 10.1093/pcp/pch113. [DOI] [PubMed] [Google Scholar]
- 25.Pharmawati M, Shabala SN, Newman IA, Gehring CA. Natriuretic peptides and cGMP modulate K+, Na+ and H+ fluxes in Zea mays roots. Mol Cell Biol Res Commun. 1999;2:53–57. doi: 10.1006/mcbr.1999.0151. [DOI] [PubMed] [Google Scholar]
- 26.Suwastika IN, Gehring CA. Natriuretic peptide hormones promote radial water movements from the xylem of Tradescantia shoots. Cell Mol Life Sci. 1998;54:1161–1167. [Google Scholar]


