Abstract
Plant microRNAs (miRNAs) have an impact on the regulation of several biological processes such as development, growth and metabolism by negatively controlling gene expression at the post-transcriptional level. However, the role of these small molecules in the symbiotic interaction of plant roots and arbuscular mycorrhizal (AM) fungi remained elusive. To elucidate the role of miRNAs during AM symbiosis we used a deep sequencing approach to analyze the small RNA and degradome sequence tags of Medicago truncatula non-mycorrhizal and mycorrhizal roots. We identified 243 novel Medicago microRNAs and 118 mRNA cleavage targets of miRNA mature and star sequences. Several AM symbiosis-relevant genes were identified as miRNA targets. The transcript of MtNsp2, encoding a GRAS transcription factor involved in the nodule and mycorrhizal signaling pathway, is cleaved by a novel member of the miR171 gene family, namely miR171 h. Here, we carried out a detailed analysis of the genomic structure of the MIR171 h gene comprising our deep sequencing data. The results suggest a feedback circuit between mature miR171 h and its own primary transcript showing the ability of this miRNA to regulate itself.
Key words: deep sequencing, degradome, small RNAs, self-regulation, miRNA cleavage, miR171, MtNsp2, AM symbiosis, DCL1-processing
Small regulatory RNAs such as the microRNAs play a crucial role in the post-transcriptional and translational gene regulation in both plants and animals. MiRNAs in plants are mostly transcribed from intergenic located MIR genes through RNA polymerase II activity. The resulting 5′ capped and 3′ poly(A)-tailed primary transcript1,2 is processed in the nucleus by the ribonuclease III enzyme DICER-LIKE1 (DCL1) yielding in a miRNA/miRNA* duplex.3–5 The duplex is subsequently methylated by HUA ENHANCER1 (HEN1),6,7 and exported to the cytosol presumably by HASTY.8 The mature strand then binds to the AGO1 protein and directs the miRNA mediated cleavage or translational inhibition of target transcripts.9
Some miRNAs, like miR162 and miR168, can auto-regulate their biogenesis through the targeting of DCL1 and AGO1, respectively.10,11 Moreover, it has been shown that, in Arabidopsis thaliana, miR167 regulates the auxin response factor (ARF) 6 transcription factor by a negative feedback circuit.12 Several ARF-binding motifs were found in the miR167 promoter region, indicating that miR167 itself is regulated by this transcription factor.12 Also the recent analysis of small RNA and degradome tag sequencing indicated miRNA self-regulation phenomena, including the miR399*-mediated cleavage of miR399 primary transcripts.13
Deep sequencing coupled miRNA prediction and expression profiling resulted in the identification of 181 known and 243 novel miRNAs in Medicago truncatula non-mycorrhizal (nm) and mycorrhizal (myc) roots. Several of these miRNAs showed an increased expression in mycorrhizal roots compared to non-mycorrhizal roots, like miR5229a/b, miR160f*, miR160c, miR5204, miR169d/d*, miR167 and miR171 h. This indicates that miRNAs are an important part of the regulatory network leading to AM symbiosis development. Interestingly, the degradome analysis revealed that miR171 h cleaves MtNsp2 transcripts, which encode a GRAS transcription factor essential for nodule symbiosis signaling.14 It is worth mentioning, that mtnsp2 mutants also showed a reduced AM colonization, suggesting that MtNsp2 is involved in AM symbiosis development.15 Remarkably, miR171 h seems to be the only member of the M. truncatula miR171 family targeting MtNsp2, whereas miR171a-g mediate the cleavage of other GRAS transcription factors.13 However, repression of MtNSP2 translation by other members of the M. truncatula miR171 family cannot be ruled out at the moment.
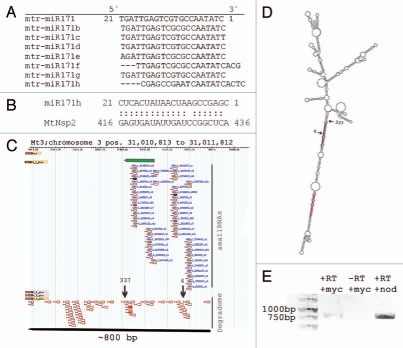
The sequence alignment of all known members of the M. truncatula miR171 family (Fig. 1A) revealed a 4 nucleotide shift towards the 3′ end of the mature miR171 h sequence compared to the mature sequences of miR171a/b/c/d/e/g. Based on the fact that the perfect complementary binding between the 10th and 11th nucleotide of the miRNA/target duplex is essential for transcript cleavage,16 this nucleotide shift leads to a cleavage target switch (Fig. 1B). To analyze the gene structure of MIR171 h, we mapped the predicted miR171 h sequence to the M. truncatula genome. The miR171 h derives from an independent locus on chromosome 3 and is processed by DCL1 resulting in specific mature miRNA and miRNA* sequences. Hence, the observed shift in the mature miR171 h sequence is not the result of alternative DCL1-cleavage of another miR171 primary transcript and was therefore annotated as a separate member of the miR171 family. The phenomenon of a nucleotide shift in the miR171 family is also observed in other plant species (miRBase version 17; reviewed in ref. 17). In rice, 9 members of this family are known so far. Interestingly, the 3 nt-shifted osa-miR171 h is the only member that targets the transcript of the rice Nsp2 orthologue (pmiRKG database, bis.zju.edu.cn/pmirkb), whereas the others mediate the cleavage of other GRAS TF transcripts.
Figure 1.
Analysis of the mtr-miR171 h genomic locus. (A) Sequence alignment of all known Medicago truncatula miR171 family members. (B) Alignment of miR171 h to its cleavage target MtNsp2. (C) Distribution pattern of small RNA and degradome tags within 1 kb containing the genomic locus of the miR171 h predicted precursor. The upper short arrow indicates the predicted 109 nt long precursor. Gradient shading of the small RNA and degradome marks indicates the absolute abundance of reads at this position. (D) Secondary structure of the miR171 h primary transcript depicting the miRNA/miRNA * duplexes (bold) and DCL 1 as well as RISC cleavage sites (arrows labeled with 337 and 4, respectively). (E) Agarose gel analysis of RT PCR products validating the presence of a long primary transcript in mycorrhizal (myc) and nodulated (nod) roots. -RT terms the reverse transcription control without reverse transcriptase.
The mtr-miR171 h is transcribed as a long primary transcript of at least 800 bp (Fig. 1C). This transcript is comprised of two loci encoding identical mature miR171 h sequences and two loci encoding slightly different miR171 h* sequences. Remarkably, the information about MIR171 gene structure were obtained after integrated analysis of both small RNA and degradome sequence tags in the genomic region of the predicted miR171 h precursor (Fig. 1C). This long primary transcript was confirmed by RT-PCR (Fig. 1E). Zhang et al.18 also found evidence that several distinct miRNA duplexes can be generated from one long miRNA hairpin structures.
Our degradome analysis proved the base-to-loop DCL1-processing of miR171 h (Fig. 1D). During miRNA biogenesis two sequential cleavages of the long miRNA hairpin structure by DCL1-activity releases a miRNA/miRNA* duplex.19,20 The number of precise DCL1-cleavage sites within a miRNA primary transcript indicates whether the miRNA is processed from stem to loop or vice versa.21,22 We found that 337 of 399 degradome sequence tags are mapping precisely to one position in the miR171 h primary transcript indicating one DCL1-cleavage at this position and a stem-to-loop processing of this miRNA.
The observed cleavage of one of the miR171 h* between the 10th and 11th nucleotide, suggests a self-regulation of the miR171 h. This self-regulation of miRNA expression, where miRNAs are able to bind and cleave their primary transcripts, has been previously reported for A. thaliana.23 However, this mechanism does not seem to occur very frequently in rice.21,24 The miR171 family is one example of miRNA self-regulation in M. truncatula. Interestingly, the accumulation of both mature miR171 h and its target MtNsp2 is significantly increased in mycorrhizal roots compared to non-mycorrhizal roots.13 We showed that MtNsp2 is negatively regulated by miR171 h implying that MtNSP2 expression is tightly controlled in roots at least at two regulatory levels. Moreover, the miR171 h which controls MtNSP2 expression shows a negative self regulation. In conclusion we demonstrated the power of a combined degradome and small RNA sequencing data analysis for the investigation of MIR gene structure and regulation using the example of miR171 h.
References
- 1.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Han J, Yeom KH, Lee S, Beak SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 5.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucl Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 11.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng Y, Chen D, Ma X, Mao C, Cao J, Wu P, et al. Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant Signal Behav. 2010;5:252–254. doi: 10.4161/psb.5.3.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devers EA, Branscheid A, May P, Krajinski F. Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011;156:1990–2010. doi: 10.1104/pp.111.172627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 15.Maillet F, Poinsot V, Andre O, Puech-Pages V, Haouy A, Gueunier M, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 16.Mallory AC, Bouché N. MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci. 2008;13:359–367. doi: 10.1016/j.tplants.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang WX, Gao S, Zhou XF, Xia J, Chellappan P, Zhou XA, et al. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010;11:81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 20.Voinnet O. Origin, biogenesis and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 21.Meng Y, Gou L, Chen D, Wu P, Chen M. High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J Exp Bot. 2010;61:3833–3837. doi: 10.1093/jxb/erq209. [DOI] [PubMed] [Google Scholar]
- 22.Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28:3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 24.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, et al. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]