Abstract
The mammalian genome harbors a large number of long non-coding RNAs (lncRNAs) that do not code for proteins, but rather exert their function directly as RNA molecules. LncRNAs are involved in executing several vital cellular functions. They facilitate the recruitment of proteins to specific chromatin sites, ultimately regulating processes like dosage compensation and genome imprinting. LncRNAs are also known to regulate nucleocytoplasmic transport of macromolecules. A large number of the regulatory lncRNAs are retained within the cell nucleus and constitute a subclass termed nuclear-retained RNAs (nrRNAs). NrRNAs are speculated to be part of crucial gene regulatory networks, and act as structural scaffolds of subnuclear domains. NrRNAs modulate gene expression by influencing chromatin modification, transcription and post-transcriptional gene processing. The cancer-associated Metastasis-associated lung adenocarcinoma transcript1 (MALAT1) is one such long nrRNA that regulates pre-mRNA processing in mammalian cells. Thus far, our understanding about the roles played by nrRNAs and their relevance in disease pathways is only ‘a tip of an iceberg’. It will therefore be crucial to unravel the functions for the vast number of long nrRNAs, buried within the complex mine of the human genome.
Key words: MALAT1, MALAT-1, non-coding RNA, nuclear RNA, nuclear domains, pre-mRNA splicing, alternative splicing
Introduction
The human transcriptome analyses have revealed that ∼50–70% of the human genome is transcribed, out of which only 2% of the transcriptional output represents protein coding sequences (International Human Genome Sequencing Consortium 2004).1,2 Thus, the large fraction of the genome transcribes RNAs that do not code for proteins, and such transcripts are classified as non-coding RNAs (ncRNAs).3–5 Although many housekeeping non-coding transcripts, like tRNAs, rRNAs, snRNAs, snoRNAs, RNaseP RNAs, are known to play crucial roles in key cellular processes, there remains an ongoing debate regarding the biological relevance of the vast majority of the uncharacterized ncRNAs.6,7 The evolutionary conservation,6,8,9 specific subcellular localization,10–18 spatial and temporal expression pattern,19–25 and association with disease5,26–30 of a significant number of these transcripts favor the argument that these ncRNAs are not merely by products of promiscuous transcription. Rather, they are more likely an integral part of an extensive RNA control network that co-exist along with proteins.31 Moreover, genomic analyses have shown that the protein-coding portion of genomes scales down relative to increased developmental and physiological complexity of organisms, suggesting the existence of a parallel regulatory network governed by the non-coding RNA counterparts.3,31,32
Among these mysterious non-coding RNAs, transcripts larger than 200 nucleotides have been classified as long noncoding RNAs (lncRNAs).5,31,33–35 Number of lncRNA genes is conservatively estimated as ∼17,000 in human and ∼10,000 in the mouse genome (www. Invitrogen.com).36 Also, based on their genomic position with respect to protein-coding genes, lncRNAs are further classified as overlapping, cis-antisense, bidirectional, promoter-associated, 3′UTR-associated, enhancer-associated, intronic or inter-genic lncRNAs.33,37 This shifts our linear idea of genome organization to a more complex view in which sense and antisense and/or coding and non-coding transcripts can arise from the same genomic segment. Most lncRNAs are transcribed by RNA polymerase II (RNA pol II), but several of them are also transcribed by RNA polymerase III.5,38 Recent studies have indicated that lncRNAs may utilize transcriptional regulation strategies similar to that of protein coding genes including conventional chromatin signatures such as histone 3 lysine 4 [H3K4] trimethylation at the promoter and H3K36 trimethylation within the transcribed region of RNA Pol II transcription units.8,39,40 The 5′CAP site has been found in ∼1,600 of the abundant and conserved mammalian lncRNAs,8 and several of the well-known RNA pol II transcription factors also regulate lncRNA transcription units.20,41
Sub-cellular localization of lncRNAs is highly regulated and context dependent. Specific lncRNAs are sequestered either within the nucleus or the cytoplasm and execute their functions within specific subcellular compartments.11,42 Several of the nuclear-retained ncRNAs (nrRNAs) localize to specific subnuclear compartments and are suggested to play structural or gene regulatory roles.11,42 In this 'point of view', we will focus our attention on the function and dynamics of some recently identified long nrRNAs and their roles in organizing chromatin and regulating gene expression. RNA sequence and structure offer nrRNAs two inherent functional properties: (1) sequence-mediated interactions with genomic DNA or other RNA and (2) secondary/tertiary structure-mediated interactions with RNA-binding proteins. NrRNAs can modulate gene expression by their potential capability to function as cofactors for their protein partners and even as scaffold molecules for combinatorial complex assembly at a specific DNA/RNA locus. In support of this concept, increasing numbers of nuclear RNAs have been shown to participate in regulating gene expression at the epigenetic, transcriptional and post-transcriptional level.
Chromatin Modifications
Recent genome-wide studies have suggested that a large proportion of nuclear lncRNAs are involved in regulating gene expression through chromatin modification in an allele or cell-type specific manner by targeting chromatin modifying complexes in cis or trans to specific genomic loci.43 Examples of nuclear noncoding RNAs involved in dosage compensation include Xist, Tsix, RepA, Jpx and roXs. Similarly, nuclear RNAs implicated in genome imprinting are Air and Kcnq1ot1. Multiple lncRNAs participate in the inactivation of one of the two X-chromosomes in female mammals.44–46 XIST/Xist (X-inactive specific transcript) (∼17 kb) is the best-studied long nuclear RNA that is primarily transcribed from the female inactive X-chromosome in mammals and is involved in X-chromosome inactivation (XCI). Xist and the recently identified RepA transcripts facilitate the recruitment of chromatin-remodeling complexes to silence the future inactive X-chromosome (Xi).47 RepA, which shares sequence similarity with the 5′end of Xist, directly interacts with the polycomb repressive complex 2 (PRC2) and recruits it to the future Xi. Interestingly, Tsix, which is the antisense transcript of Xist and is known to function as the negative regulator of Xist, competes with RepA for PRC2 binding. However, during the initial stages of Xi, Tsix is downregulated on the future Xi, which in turn facilitates Xist transcription as well as enhances interaction of RepA with the PRC2 complex. The upregulated Xist in Xi then interacts with PRC2 and promotes the spreading of the polycomb complex and the trimethylated H3 lysine 27 throughout the Xi. Jpx is another X-chromosome-encoded lncRNA that is developmentally regulated and accumulates during XCI. A recent study from Lee and coworkers demonstrated that Jpx acts as a positive regulator of Xist RNA.48 These results indicate that Xist expression is regulated by multiple lncRNAs; Tsix on Xa and Jpx on Xi. In Drosophila, the dosage compensation of X-chromosome is achieved by the hyperactivation of the single X-chromosome in male flies. Nuclear-retained, roX lncRNAs (roX-1 and 2) are specifically transcribed from the male X-chromosome and are part of the chromatin remodeling complex that facilitates the hyperactivation of genes encoded on the male X-chromosome.49 Similarly, allele-specific gene silencing is also brought about by imprinted nuclear lncRNAs such as Air and Kcnq1ot1 that recruit polycomb proteins and histone methyltransferases to specific gene loci.34
In a recent genome-wide analysis to elucidate the polycomb-associated transcriptome of mouse embryonic stem cells, more than 9,000 PRC2-interacting RNAs were identified, the majority being antisense, intergenic or promoter-associated ncRNAs.50 In a separate study, more than 20% of long intergenic ncRNAs (lincRNAs) were found to physically associate with the repressive chromatin modifying complexes including PRC2, CoREST and JARIDIC/SMCX and the functional significance of such interaction has been supported by the finding that siRNAmediated depletion of several of the PRC2-associated lincRNAs resulted in derepression of PRC2 target genes at distant loci.39 HOTAIR is one such example of a lncRNA that is involved in regulating polycomb-mediated gene repression.25 HOTAIR lncRNA (∼2.2 kb) is transcribed from the boundary of the two diametric chromatin domains in the HOX C gene locus, which is one of the four HOX gene clusters located on different human chromosomes.25,51,52 The noncoding regions in HOX clusters are ultraconserved among mammalian genomes and give rise to ∼230 novel ncRNAs. HOTAIR recruits the PRC2 complex in trans and represses the HOX D locus.25,51,52 Interestingly, HOTAIR is highly upregulated in primary and metastatic breast tumors, and its overexpression is correlated with both metastasis and poor survival rate.53 Overexpression of HOTAIR results in genome-wide retargeting of PRC2, an altered pattern of H3K27 methylation and increased invasiveness, whereas depletion of HOTAIR impedes metastasis of PRC2overactive cells.53 Furthermore, HOTAIR was found to bridge PRC2 and LSD1/COREST/REST complexes together via its 5′ and 3′ domains, respectively.52 It was therefore proposed that lncRNAs such as HOTAIR coordinate H3K27 methylation and H3K4 demethylation.
Antisense transcript-mediated silencing is also emerging as a widely applied strategy in epigenetic gene silencing. Examples include ANRIL lncRNA, which recruits PRC1 and 2 complexes to the INK4A/ARF/INK4b gene cluster and influences the silencing of the genes within this locus.54,55 Similarly, P15AS, Khps1 and rDNA-promoter antisense ncRNAs modulate region-specific DNA methylation by recruiting different combinations of DNA methylases and demethylases.56–58
Besides lncRNAs like HOTAIR and ANRIL that act as negative regulators of gene expression, there are several examples of lncRNAs that maintain active chromatin status, indicating the existence of an RNA network that controls transcription by influencing chromatin structure. Examples include the recently identified HOTTIP RNA, which targets WDR5/MLL complexes across the HOX A cluster via chromosome looping59 and Jpx RNA that acts as an activator of Xist expression by antagonizing Tsix in trans.48
In light of many examples of epigenetic control of gene regulation by lncRNAs, it is tempting to speculate that lncRNAs may provide modular scaffolds for chromatin modifying complexes. A small repertoire of chromatin modifying complexes that have limited DNA specificity could use specific RNA binding domains to interact with their lncRNA partner and modulate genome-wide chromatin modification events.34,52
Transcriptional Regulation
LncRNAs regulate transcription through diverse mechanisms.33,34,40,42 LncRNAs directly influence general transcription factor loading at specific chromatin sites. For example, a lncRNA transcribed from the upstream region of the DHFR gene locus impedes transcription from the DHFR major promoter region by forming a triplex structure with the DHFR promoter DNA.60 This lncRNA also interacts with the general transcription factor TFIIB and facilitates the disassociation of the pre-initiation complex from the DHFR major promoter. Similarly, human Alu RNA and mouse SINE B2 RNA are suggested to bind and block the DNA-binding channel of RNA pol II during heat shock and thus repress global RNA pol II-mediated transcription.61–64 Some lncRNAs can also serve as cofactors to recruit and modulate the activity of transcription factors. For example, the steroid receptor RNA activator (SRA) is a 700 nt lncRNA, operating as part of a ribonucleoprotein complex, which coactivates several steroid-hormone receptors.65–67 It was recently shown that lincRNA-p21, transcribed from the intergenic region upstream of the p21/cdkn1a locus, is induced by p53 and is required for p53-mediated repression of genes involved in apoptosis, partially via orienting hnRNP-K to the promoters of specific genes.68 LncRNAs transcribed from the 5′regulatory regions of the Cyclin D1 (CCND1) gene negatively regulate CCND1 transcription during genotoxic stress by recruiting chromatin modifying enzymes.40,69
The role of lncRNAs in enhancer activity in human cells has advanced significantly with the recent identification of a large set of lincRNAs with enhancer-like functions.37,70 Evf2 is a mouse brain specific lncRNA, transcribed from the inter-genic enhancer region of Dlx-5/6 loci. EvF2 recruits the homeodomain transcription factor Dlx-2 in cis and induces the Dlx-5/6 enhancer activity resulting in the elevated expression of Dlx-5 and -6.71 Earlier studies have reported that many of the well-known enhancer elements are actively transcribed, including those in the iconic globin locus control region,72 HS2 enhancer73 and neuronal activity-regulated enhancers.74 It is proposed that enhancer lncRNAs provide a platform at specific promoter sequences, to recruit and organize appropriate transcription factors and chromatin modification complexes. Thus, enhancer-encoded lncRNAs may be integral to enhancer function.
Besides the above examples in which the lncRNA molecules have a gene regulatory mode of action, there are also examples where the transcription process of lncRNAs itself is enough to promote or repress the transcription of adjacent genes. This is achieved either via progressive opening of chromatin structure to increase the accessibility of transcription factors, or conversely via transcription interference, as ongoing lncRNA transcription impedes the initiation and/or progression of another transcription from the overlapping region. Examples of transcription activation and interference are observed in case of Fbp1+ in S. pombe75 and SRG1 lncRNA in S.cerevisae respectively.76,77
Post-Transcriptional Regulation
It is now clear that ∼95% of the multiexoncontaining human pre-mRNAs undergo alternative splicing, implying that post-transcriptional processing of pre-mRNAs plays a much more important role in generating genetic diversity than previously anticipated.78,79 Alternative splicing represents one of the major mechanisms of regulation of gene expression, enabling a high degree of proteomic complexity.80–82 The precise removal of the introns during pre-mRNA processing is catalyzed by spliceosomes that consist of well-characterized small uridine-rich nuclear RNAs (snRNAs) and a large number of splicing factors, including hnRNPs and serine-arginine (SR) splicing factors.83 SR proteins are characterized by the presence of a RS dipeptide-rich domain as well as RNA recognition motif (RRM) that together mediate simultaneous interactions with RNA and other proteins.84,85 SR proteins play crucial roles in multiple steps of RNA metabolism, ranging from constitutive and alternative pre-mRNA splicing to mRNA export, nonsense mediated decay and translation.84,85
LncRNAs are also known to function as regulators of pre-mRNA splicing.86 Natural antisense transcripts (NATs) regulate splicing of overlapping sense transcripts,87–89 as has been observed in the case of Zeb2/Sip1 mRNA splicing.90 Zeb2/Sip1 is a transcriptional repressor of E-cadherin that is upregulated during epithelial-mesenchymal transition (EMT).90 The Zeb2/Sip1 intron contains an internal ribosomal entry site (IRES) that is necessary for the translation of Zeb2/Sip1. In normal epithelial cells, the Zeb2 is not expressed due to the removal of the intron containing the IRES. Upon EMT, a NAT is transcribed from a region that is complementary to the intron containing the IRES. This NAT prevents the splicing of the IRES-containing intron from the pre-mRNA, resulting in the efficient translation of Zeb2 mRNA.90
Stress-induced repeat-containing lncRNAs have also been suggested to regulate pre-mRNA splicing by sequestering pre-mRNA splicing factors.91–93 Examples include the hsrω-n RNA in fruit flies and the Sat III transcripts in mammalian cells (see, Structural Organization of Nuclear Bodies).91–93 Others and we have recently demonstrated that Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA also plays a pivotal role in alternative splicing regulation.18,94–96
MALAT1 is among the most abundant nuclear lncRNAs, and is highly conserved in mammals.18,94,97,98 The deregulation of MALAT1 has been correlated with a distinct pathological event, making it one of the few lncRNAs whose aberrant expression has a clear phenotypic manifestation. It was originally identified based on its overexpression in various cancers, especially cancers with aggressive metastatic tumors.97 Multiple studies have correlated the differential expression of MALAT1 with tumor development, progression or survival in several cancerous conditions including non-small cell lung carcinoma, uterine endometrial stromal sarcoma, hepatocellular carcinoma and breast carcinoma.28,97–102 Recent studies have demonstrated that inhibition of MALAT1 impairs proliferative and invasive properties of cancer cells,96,103 further supporting the involvement of MALAT1 in cellular transformation and/or tumorigenesis.
Understanding how the aberrant expression of a gene results in a specific disease phenotype necessitates uncovering the underlying molecular mechanism/s. MALAT1 exhibits tissue- and developmental-specific expression, showing particularly high expression in neurons.94 In mouse hippocampal neurons, depletion of MALAT1 decreases synaptic density, whereas its overexpression increases synaptic density.94 Further, it was demonstrated that MALAT1 modulates synaptogenesis by regulating the expression of protein-coding genes, including neuroligins and syncams that are involved in synapse formation and/or maintenance.94 Since MALAT1 is enriched in nuclear speckles (Fig. 1Aa), a sub-nuclear domain that is involved in pre-mRNA processing, it was proposed that MALAT1 executes its gene regulatory functions by modulating one or several steps of pre-mRNA processing.14,94 Nuclear speckles are enriched with various pre-mRNA splicing proteins including SR splicing factors and act as storage/modification/assembly centers of these factors.104 SR splicing factors rapidly exchange between sites of gene transcription and nuclear speckles, and this regulated exchange is crucial for efficient transcription and pre-mRNA splicing.104–106 However, the mechanism/s that modulate cycling of SR splicing factors are not yet understood. MALAT1 is a very stable nuclear transcript that associates with nuclear speckles in a RNA pol II transcription-dependent manner.94 Based on this, it was speculated that MALAT1 could either act as a structural RNA of nuclear speckles or regulate the intranuclear trafficking of key splicing factors between nuclear speckles and transcription sites. Biochemical analyses showed interaction of MALAT1 with a subset of SR proteins. Based on bioinformatic analyses that predict potential SR protein binding sites and CLIP-seq analyses using antibodies against SR proteins, it was estimated that each MALAT1 transcript contains a large number of potential SR protein binding sites (∼50 sites for SRSF1 on each MALAT1 RNA).18,107 However, SR proteins do not influence the localization of MALAT1 to nuclear speckles. On the contrary, MALAT1 is required for the localization of SR proteins and several other pre-mRNA splicing factors to nuclear speckles.18 Also, the recruitment of SR proteins to a transcriptionally active gene locus was compromised in cells lacking MALAT1.94 Interestingly, polyA+ RNA, speckle-associated RNA transport factors continue to localize at nuclear speckles in MALAT1-depleted cells. Based on these observations, it is inferred that MALAT1 is not a structural component of nuclear speckles, but it modulates the nuclear distribution of a subset of premRNA splicing factors.
Figure 1.
(A) Co-RNA-FISH using probes against MALAT1 (a, green) and Neat1 (a′, red) reveals distribution of MALAT1 and Neat1 RNA in nuclear speckles and paraspeckles respectively. (B) Depletion of MALAT1 (MALAT1-AS) in HeLa cells results in nuclear fragmentation. Note that the fragmented nuclei contain nuclear pores (green) and nuclear lamina (red). DNA is counterstained with DAPI. The bar represents 5 µm.
Interestingly, depletion of several of the pre-mRNA processing factors compromises the nuclear speckle distribution of MALAT1.18 Of particular interest is SON, an SR-related speckle-associated protein, which is suggested to act as the scaffold for the proper organization of speckle components and also to function as a coactivator for efficient RNA processing.108–110 Depletion of SON resulted in the redistribution of MALAT1 from nuclear speckles to a more homogenous nuclear distribution.18 On the other hand, SON continued to localize to speckles in MALAT1-depleted cells, indicating that only SON influences the MALAT1 association to speckles and not vice versa.18 Furthermore, the C-terminal domains of SON (including SR and G-patch domains) were necessary for the localization of MALAT1 to speckles and also for the splicing activator function of SON.18,108 Depletion of SON in cancerous cells resulted in mitotic arrest, with severe impairment of spindle pole segregation and genome integrity, due to aberrant splicing of pre-mRNAs whose protein products are involved in cell cycle progression and mitosis.108–110 Prolonged depletion of MALAT1 in human cancerous cells also revealed mitotic arrest with abnormal microtubule organization and chromosome segregation defects.18 MALAT1-depleted cells also showed abnormal nuclear structures, including micronuclei and nuclear lobes similar to the phenotypes observed upon SON depletion. The individual nucleus of the multinucleated structure formed in MALAT1-depleted cells contained nuclear pores and nuclear lamina, indicating that the formation of nuclear lobes upon MALAT1-depletion is a post-mitotic event (Fig. 1B).18 In general, MALAT1 or SON depleted human cells displayed similar mitotic abnormalities, indicating that both the genes could be part of a common pathway. It is possible that mislocalization of MALAT1 in SON-depleted cells could alter the distribution and/or splicing activity of specific splicing proteins, including SR proteins, which in turn affects the splicing of key cell cycle regulated genes. Future studies will test how MALAT1 and SON coordinate key splicing events.
Besides SR proteins, MALAT1 also interacts with other pre-mRNA splicing factors including PRP6 and TAR DNA binding protein-43 (TDP-43).18,111,112 Human PRP6 is a homolog of yeast PRP6p, an essential nuclear speckle-associated splicing factor that functions as a bridging molecule between U5 and U4/U6 snRNPs during tri-snRNP assembly.113 Interestingly, depletion of PRP6 also results in the homogenous distribution of MALAT1, indicating that PRP6 could influence the speckle association as well as splicing regulatory roles of MALAT1.18 TDP-43 is a highly conserved hnRNP, suggested to regulate several steps of RNA metabolism, including transcription, RNA splicing, mRNA transport and stability.114,115 Abnormal expression and mislocalization of TDP-43 have been implicated in several neurological disorders, including fronto-temporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS).115,116 By utilizing CLIP-seq assays in human and mouse tissue samples as well as cell lines, two independent groups have recently shown the interaction of TDP-43 with lncRNAs, including MALAT1 and NEAT1; another nrRNA that localize to paraspeckle domains (see, Fig. 1Aa′).111,112 Further, TDP-43 is required for the stability of MALAT1, as observed by the 2-fold decrease in MALAT1 transcript levels in TDP-43 depleted cells.111 Further, TDP-43 regulates the expression and premRNA processing of several genes that are involved in neuronal functions including synaptic transmission, implicating TDP43 in brain function.111,112 Both MALAT1 and TDP-43-depleted neuronal cells show downregulation of cell adhesion molecules such as neurologin-1, components involved in neuronal functions including synaptogenesis.94,111 Interestingly, elevated expression of MALAT1 is observed in brain samples from patients with FTLDTDP-43 inclusions (FTLD-TDP).112 Also, FTLD-TDP samples show increased interaction between TDP-43 and MALAT1 in comparison to brain samples from healthy individuals, supporting the involvement of MALAT1 in neurological disorders.112 The interaction of MALAT1 with TDP-43 and the proposed role of TDP-43 in the alternative splicing of key genes involved in synapse functions indicate that MALAT-1 and TDP-43 could act cooperatively in regulating neuronal functions. Based on these observations, it is tempting to speculate that MALAT1 by interacting with specific splicing factors like TDP-43 could modulate tissue or cell specific splicing of pre-mRNAs.
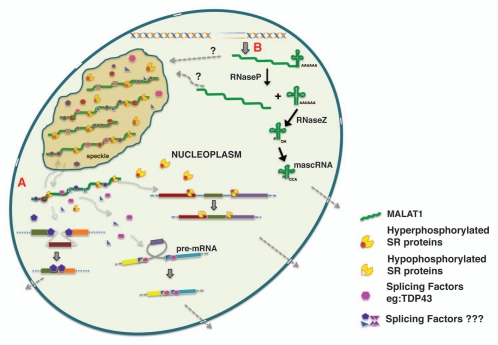
Several recent studies have demonstrated that MALAT1 modulates alternative splicing of endogenous pre mRNAs.18,64,96 MALAT1-depletion increased exon inclusion in a majority of pre-mRNAs, an event that is usually associated with differential SR protein activity. Further, the alternative splicing changes observed in MALAT1-depleted cells were sensitive to cellular levels of SR proteins.18 These results indicate that MALAT1 influences alternative splicing by modulating the cellular levels or activity of one or more SR splicing factors (Fig. 2). Pre-mRNA splicing is regulated by dynamic phosphorylation/dephosphorylation cycle of SR proteins, which is also an important regulator of alternative splicing patterns. The phosphorylation status of SR proteins also influences the intranuclear trafficking of SR proteins between speckles and transcription sites.105,117 MALAT1-depleted cells showed increased levels of SR proteins, and majority were dephosphorylated.18 Such changes alter the cellular ratio of phosphorylateddephosphorylated SR proteins and could thus influence the alternative splicing of pre-mRNAs. Based on these results, we hypothesize that MALAT1 regulates the cellular levels of phosphorylated SR proteins. It is possible that MALAT1 stabilizes the cellular pool of phosphorylated SR proteins. Alternatively MALAT1 could modulate the activity of kinases (SRPKs or Clk/Sty family) or phosphatases (PP1) that modify SR proteins.118 In any case, by controlling the levels of phosphorylated SR-proteins, MALAT1 may modulate not only alternative splicing but also many other post-transcriptional gene regulatory mechanisms (RNA export, NMD, translation) that are controlled by phosphorylated SR proteins.85,118
Figure 2.
Model depicting the biogenesis of MALAT1 and the involvement of MALAT1 in alternative splicing regulation. (A) By associating with splicing factors in nuclear speckles and the nucleoplasm, MALAT1 regulates the nuclear distribution and recruitment of splicing factors to pre-mRNA, thereby modulating alternative splicing. the color boxes indicate exons and the lines indicate introns. (B) the nascent MALAT1 transcript is cleaved at its 3′end to generate a mature long MALAT1 nuclear RNA and a small ∼61 nucleotide tRNA-like mascRNA, which is transported into the cytoplasm.147 ‘?’ indicates lack of clear evidence on whether nuclear speckles contain both the precursor and mature long MALAT1 transcripts or only the mature transcript.
In multicellular organisms, splicing regulatory proteins are generally constitutively expressed in all cell types and tissues. Despite this fact, alternative splicing is tightly regulated in a tissue-specific and context-dependent manner, during embryonic development and cellular differentiation, or during certain physiological conditions or in response to various types of cellular stress signals.80,85,119 This raises an exciting question of how a combination of ubiquitously expressed splicing regulators can lead to a cell type-specific alternative splicing. Variable expression levels of splicing factors having antagonistic effects in splice site usage (examples include SRSF1 and HnRNP A1) can result in cell-type specific alternative splicing.85 Besides changing the cellular levels of splicing proteins, cells also have developed various strategies to directly modify the activity of the existing pool of splicing factors, either by altering their intrinsic ability to act as splicing regulators or by changing their intracellular or sub-nuclear distribution. Based on our results, we propose that nuclear RNAs like MALAT1 act as a ‘molecular sponges’ by titrating and balancing the cellular pool of splicing factors and other RNA processing proteins (Fig. 2A). In this model, an interaction between MALAT1 and splicing factors will create a gradient of functionally competent splicing factors in the cell, which in turn regulates splicing events.
Recent transcriptome analyses comparing tumor cells and normal cells have indicated differential expression of several lncRNAs in various cancers including breast, prostate, ovary, liver as well as in leukemias and lymphomas.27,28,31,33,53,97,98,120,121 At the molecular level, cancer is a disease that is manifested by alterations in gene expression. Since many of the ncRNAs regulate gene expression, their abnormal expression in cells can alter the levels of critical gene products that are involved in cell proliferation and survival, leading to tumorigenesis. Though the deregulation of MALAT1 has been observed during tumorigenesis, it is not clear whether its altered expression observed during tumorigenesis is a cause or effect of tumorigenesis. Several pieces of evidence indicate that MALAT1 contributes to tumorigenesis and cellular transformation. Overexpression of MALAT1 RNA fragments in human cells promotes tumorigenicity, while MALAT1 down-regulation suppresses tumorigenicity.122 Secondly, several mutations across the entire MALAT1 gene have been detected in a large number of colorectal cancer samples.123 Further, overexpression of a fragment from the 3′ region of MALAT1 contributes to proliferation, migration and invasion in colorectal cancer cells. Depletion of MALAT1 in lung cancer cells impairs in vitro cellular motility and alters the pre-mRNA processing of several genes that are involved in cell motility and migration.96 Finally, MALAT1 depleted cancerous cells show changes in alternative splicing of several pre-mRNAs, including B-Myb and Mgea6, whose protein products are involved in tumorigenesis.18 There is strong evidence that establishes the connection among the aberrant expression of pre-mRNA splicing factors, the changes in alternative splicing patterns of key genes and cancer.85,124–129 For example, SRSF1, which interacts with MALAT1, is over-expressed in various human tumors and slight overexpression of SRSF1 in mouse has been shown to cause sarcomas.125,127,128 Based on the proposed role of MALAT1 as a splicing regulator, we speculate that abnormal expression of MALAT1 in cells results in altered splicing factor distribution and pre-mRNA splicing activity. Such aberrant alternative splicing leading to misexpression of several genes involved in cell cycle, proliferation and/or cell death, could ultimately result in complex diseases like cancer.
Structural Organization of Nuclear Bodies
Besides the proposed role of nrRNAs in chromatin modifications, transcriptional and post-transcriptional gene regulation, they also actively participate in the establishment of specific subnuclear structures or domains.11,130 Examples include MEN ε/β (or NEAT1), and satellite III (sat III) transcripts in mammalian cells and hsrω-n in flies. NEAT1 locus codes for two nuclear lncRNAs (>3 and 17 kb) that localize to paraspeckle subnuclear domain (Fig. 1Aa″).10,12,15,17 Paraspeckles constitute a distinct sub-nuclear domains that depend on RNA for its structural integrity.131–135 Though the exact role of para-speckles remain to be elucidated, several studies have indicated that these domains represent nuclear retention sites for a population of RNAs that have undergone adenosine-to-inosine editing.11,131,134–136 Paraspeckles contain numerous RNA binding proteins, including the members of the DBHS (Drosophila Behavior and Human Splicing) family of RNA binding proteins: PSP1α, P54nrb and PSF.134 NEAT1 interacts with several of the para-speckle proteins including PSP1α and P54nrb. Depletion of NEAT1 disrupts paraspeckles concomitant with enhanced nucleocytoplasmic export of hyperedited mRNAs.10,12,15,17,131,134 On the other hand, overexpression of NEAT1 in several cell lines increases the number of paraspeckles.12 Recently, by utilizing a live-cell imaging system Spector and co-workers have demonstrated that NEAT1 ncRNA is sufficient to nucleate de novo formation of functional paraspeckles.137 These results indicate that NEAT1 may act as a scaffold for the assembly of macromolecular complexes in paraspeckles.
NEAT1 is not expressed in undifferentiated human embryonic stem cells (ESCs) but shows elevated levels of expression when the cells are differentiated into trophoblasts.10 Interestingly, paraspeckles are formed only in differentiated ESCs, when NEAT1 is expressed.10 This observation further supports the argument that NEAT1 is required for the formation of paraspeckles. Similar to ESCs, NEAT1 is upregulated upon differentiation of myoblasts into myotubes17 suggesting a potential role of NEAT1 in developmental regulation. However, a very recent study demonstrated that in mouse, paraspeckles are formed only in specific tissues.138 Furthermore, NEAT1 knockout mice are viable with no apparent phenotypes despite the loss of paraspeckles.138 Based on this, it is proposed that paraspeckles constitute a subpopulation of specific nuclear bodies that are formed as a result of particular environmental triggers.138
In Drosophila melanogaster, the developmentally regulated and stress-inducible 93D locus encodes hsrω (heat shock RNA omega) ncRNA gene that transcribes multiple lncRNAs through alternative polyadenylation and splicing.93,139,140 The nuclear-retained long (∼15 kb) hsrω-n ncRNA is transcribed from the 93D locus and localizes to nuclear omega speckles along with several other pre-mRNA processing factors.141 Further, hsrω-n transcript is essential for omega speckle integrity, as cells devoid of hsrω-n fail to form omega speckles.141 Similar to omega speckles, a specific sub-nuclear domain called nuclear stress bodies (nSBs) in mammalian cells also contain several pre-mRNA processing factors including SR splicing factors and an lncRNA transcribed from the Sat III repeats from chromosome 9q12 region.91,142–145 The stress-induced nSBs are formed at the Sat III lncRNA transcription sites, and the accumulation and relocalization of nuclear proteins to nSBs is strictly dependent on the presence of Sat III transcripts, indicating the crucial role played by Sat III transcripts in the formation of nSBs.91 It is hypothesised that both omega speckles and nSBs are dynamic sites where several of the pre-mRNA processing factors are sequestered during cellular stress. The long nrRNAs localized within play vital roles in the formation and/or maintenance of these domains and are also involved in modulating pre-mRNA processing.92,146
Recent studies have demonstrated that nuclear RNAs can also initiate the assembly of the nuclear bodies preferentially by acting as nucleation sites for the recruitment and accumulation of specific nuclear proteins.130,137 Together, these observations reveal a crucial role for RNA in the dynamic assembly, organization and maintenance of nuclear bodies.
Future Directions
MALAT1 is a nuclear regulatory lncRNA that is involved in modulating key cellular regulatory networks and proper cell functions. Several studies have indicated the role of MALAT1 in pre-mRNA processing regulation, yet a large number of exciting questions need to be answered in order to understand the exact mechanism that is utilized by MALAT1 to regulate premRNA splicing. First of all, the basis for nuclear retention and speckle distribution of MALAT1 remains to be understood. Characterization of the protein components that interact with MALAT1 may provide clues to the mechanism/s responsible for the nuclear-restricted distribution of MALAT1. Additionally, the way in which MALAT1 controls the distribution or activity of splicing factors remains to be determined. RNA expression analyses revealed that mammalian tissues display differential expression of MALAT1.14,94,98 MALAT1 also shows elevated expression in several cancer conditions and altered expression of MALAT1 leads to defects in cell division, proliferation, migration and invasion.18,95–98,103,122 These results indicate that cellular levels of MALAT1 are tightly regulated. MALAT1 RNA has been shown to undergo an unusual 3′end cleavage to produce a mature, long, nuclear-retained MALAT1 transcript and a small (61 nt) cytoplasmic Masc RNA (MALAT1-associated small cytoplasmic RNA) (Fig. 2B).147 The function of Masc RNA remains to be determined. It is possible that such 3′end cleavage could be part of a post-transcriptional mechanism that regulates cellular levels of functional MALAT1 RNA. Further, a recent study showed the presence of a natural antisense transcript (NAT) against MALAT1 that interacts with PRC2 complex.50 Future studies will determine the involvement of 3′end cleavage of MALAT1 and MALAT1-NAT in the expression and activity of MALAT1.
Protein-coding genes have always been assumed to play the central role in regulating eukaryotic gene expression. However, the recent identification of the nuclear-retained lncRNAs in the genome clearly indicates that nrRNAs play equally important roles in gene expression control. NrRNAs are involved in diverse and complex biological processes that range from cell proliferation to cell death. We are also beginning to appreciate the roles played by nuclear lncRNAs in regulating gene expression as well in controlling the cellular distribution and activities of specific RNA binding proteins. Out of the large number of lncRNAs that are expressed in mammalian cells, only less than 1% of them have been well characterized, suggesting that there is quite a long way to go before we unravel the functional relevance of all these regulatory lncRNAs.
Acknowledgments
We thank the members of Prasanth lab for discussion. We also would like to thank Drs. Paula Bubulya and Supriya Prasanth for critical reading of the manuscript and for providing valuable suggestions. This work was supported by the University of Illinois start-up funds and American Cancer Society, Research Scholar grant (RSG-11-174-01-RMC) to K.V.P.
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS. The genetic signatures of noncoding RNAs. PLoS genetics. 2009;5:1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309:1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV. Non-coding RNA. Human Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 6.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature Structural Molecular Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen LL, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen HR, Panning B. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1. Chromosoma. 2007;116:373–383. doi: 10.1007/s00412-007-0100-1. [DOI] [PubMed] [Google Scholar]
- 14.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Nat Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 17.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN varepsilon/beta nuclear-retained non-coding RNAs are upregulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic analysis of mouse retinal development. PLoS biology. 2004;2:247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 21.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome research. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:122–126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Nat Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome research. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Human molecular genetics. 2010 doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Human Mol Genetics. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 29.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochimica et biophysica acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Rev. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 32.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 33.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory and biomedical significance of mammalian long nonprotein-coding RNA. Biochimica et biophysica acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Nagano T, Fraser P. No-Nonsense Functions for Long Noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:146–151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Current Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet: TIG. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large inter-genic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Nat Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harbor Perspect Biol. 2011;3:3756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, et al. Distribution of NFkappaB-binding sites across human chromosome 22. Proc Nat Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Seminars Cell Develop Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Nagano T, Fraser P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm Genome. 2009;20:557–562. doi: 10.1007/s00335-009-9218-1. [DOI] [PubMed] [Google Scholar]
- 44.Lee JT. The X as model for RNA's niche in epigenomic regulation. Cold Spring Harbor perspectives in biology. 2010;2:003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes & development. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science (New York, NY) 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA biology. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 50.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 57.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Develop. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 61.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nature Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 62.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nature Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 63.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proceed Nat Acad Sci USA. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 66.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 67.Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through post-transcriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 68.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 76.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 77.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes & development. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 80.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 81.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 82.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hocine S, Singer RH, Grunwald D. RNA processing and export. Cold Spring Harbor Perspect Biol. 2010;2:000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Advances Experimental Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 85.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochemical J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 86.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J Biol Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- 89.Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. The J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 90.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Devel. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harbor perspectives in biology. 2010;2:695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochemical Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 93.Jolly C, Lakhotia SC. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34:5508–5514. doi: 10.1093/nar/gkl711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, et al. Control of RNA processing by a large non-coding RNA overexpressed in carcinomas. FEBS letters. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS letters. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 97.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 98.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006 doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 99.Fellenberg J, Bernd L, Delling G, Witte D, Zahlten-Hinguranage A. Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod Pathol. 2007;20:1085–1094. doi: 10.1038/modpathol.3800937. [DOI] [PubMed] [Google Scholar]
- 100.Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97:106–112. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta biochimica et biophysica Sinica. 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 104.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harbor Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Misteli T. RNA splicing: What has phosphorylation got to do with it? Curr Biol. 1999;9:198–200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- 106.Misteli T, Spector DL. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- 107.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome research. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahn EY, Dekelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, et al. SON Controls Cell-Cycle Progression by Coordinated Regulation of RNA Splicing. Molecular cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huen MS, Sy SM, Leung KM, Ching YP, Tipoe GL, Man C, et al. SON is a spliceosome-associated factor required for mitotic progression. Cell Cycle. 2010;9:2679–2685. doi: 10.4161/cc.9.13.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long premRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Makarov EM, Makarova OV, Achsel T, Luhrmann R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J Mol Biol. 2000;298:567–575. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- 114.Buratti E, Baralle FE. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- 115.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human Mol Genet. 2010;19:46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 119.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annual Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 120.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 121.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proceed Nat Acad Sci USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: A long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011 doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 124.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 126.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Current Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proceed Nat Acad Sci USA. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, et al. Cancer-associated regulation of alternative splicing. Nature structural & molecular biology. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 130.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 131.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. The Journal of cell biology. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Molecular biology of the cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, et al. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 134.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harbor Perspectives Biol. 2010;2:687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 136.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lakhotia SC. The non-coding, developmentally active and stress inducible hsrw gene of Drosophila melanogaster integrates post-transcriptional processing of other nuclear transcripts. Georgetown, TX: Kluwer Acdemic/Plenum Publishers; 2003. [Google Scholar]
- 140.Mutsuddi M, Lakhotia SC. Spatial expression of the hsr-omega (93D) gene in different tissues of Drosophila melanogaster and identification of promoter elements controlling its developmental expression. Dev Genet. 1995;17:303–311. doi: 10.1002/dvg.1020170403. [DOI] [PubMed] [Google Scholar]
- 141.Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles—a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;113:3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 142.Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Denegri M, Moralli D, Rocchi M, Biggiogera M, Raimondi E, Cobianchi F, et al. Human chromosomes 9, 12 and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol Biol Cell. 2002;13:2069–2079. doi: 10.1091/mbc.01-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, et al. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- 145.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, et al. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jolly C, Lakhotia SC. Human sat III and Drosophila hsr{omega} transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34:5508–5514. doi: 10.1093/nar/gkl711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]