Abstract
The rapid and sensitive detection and characterization of human viruses and bacteriophage is extremely important in a variety of fields, such as medical diagnostics, immunology and vaccine research, and environmental contamination and quality control. We introduce an optical detection scheme for real-time and label-free detection of human viruses and bacteriophage as small as ~ 24 nm in radius. Combining the advantages of heterodyne interferometry and dark-field microscopy, this label-free method enables us to detect and characterize various biological nanoparticles with unsurpassed sensitivity and selectivity. We demonstrate the high sensitivity and precision of the method by analyzing a mixture containing HIV virus and bacteriophage. The method also resolves the distribution of small nano-impurities (~ 20 − 30 nm) in clinically relevant virus samples.
Keywords: Heterodyne Interferometry, Dark-field microscopy, Viruses, Nanofluidics, Phage, Nanoparticles, Biosensing, Biodetection, Label-free flow cytometry
1. Introduction
Nanoparticles play a significant role in various fields such as biomedical imaging and diagnostics (Choi et al. (2007); Huang et al. (2007)), process control in semiconductor manufacturing (Wali et al. (2009)), environmental monitoring and climate change (Ramanathan and Carmichael (2008); Morawska (2010)). Inhalation of ultrafine particulates in air has been shown to have adverse effects, such as inflammation of lungs or pulmonary and cardiovascular diseases (Oberdörster (2000); Somers et al. (2004)). Nano-sized biological agents and pathogens such as viruses are known to be responsible for a wide variety of human diseases such as flu, AIDS and herpes, and have been used as biowarfare agents (Krug (2003); Anderson et al. (2006)).
It has become increasingly important to rapidly and accurately quantify viruses. Accurate quantification of the presence of human viruses such as HIV, herpes or influenza in blood samples is essential for clinical diagnosis and also for vaccine development. It is also highly important to be able to distinguish between different kinds of viruses present in a sample. For example, a single patient may be infected with multiple viral pathogens such as HIV and HCV, and it is important to identify and also quantify both kinds of viruses in order to treat the patient.
Water contamination control is another application, where detecting and quantifying nanoscale contaminants such as bacteriophages is important (Salter et al. (2010); Santiago-Rodrguez et al. (2010)).
Most of the existing virus particle quantification techniques either suffer from significant technical glitches or are extremely time and cost consuming. For example, the Quantitative Electron Microscopy (QEM) technique (Tsai et al. (1996); Chuan et al. (2007)), which counts polystyrene beads constructed to presumably contain a certain number of HIV-1 particles, assumes that the number of beads per virus particle is constant, a fact that cannot be experimentally confirmed given the low-resolution of electron microscopy for small particles such as viruses. The Image Enhanced Microscopy (IEM) technique counts virus particles labeled with fluorescent dyes (Dimitrov et al. (1993); Hübner et al. (2009)), but the dye-labeling efficiency could not be experimentally confirmed, and hence quantification is unreliable. The quantitative-PCR method for counting viral RNA genome copy numbers is also popular, but it only indirectly determines the number of the viral particles, and does not actually count them (Hockett et al. (1999); Engelmann et al. (2008)). The plaque titer method (Dulbecco and Vogt (1954); Cromeans et al. (2008)), on the other hand, can only be used to quantify viral particles that cause visible cell-damage. At present there does not exist any virus quantification method available to biologists which can quickly and reliably detect, quantify and characterize virus particles with single particle sensitivity.
Recently there have been several studies focused on developing sensitive optical or electrical techniques for label-free viral biosensing. Electrical sensors have been demonstrated to be able to detect single viruses in solution (Patolsky et al. (2004); Fraikin et al. (2011)), but they suffer from the drawback that they are extremely sensitive to changes in ionic strengths of the media (Stern et al. (2007)). Optical techniques based on sensing discrete resonance shifts in whispering gallery mode (WGM) microcavities due to binding of single virus particles have been developed (Vollmer and Arnold (2008); Vollmer et al. (2008); Zhu et al. (2009)), but they cannot be used to distinguish between viruses of different sizes present in a heterogeneous mixture. Other optical sensing platforms such as those based on nanoplasmonics (Yanik et al. (2010)) or interferometry (Ymeti et al. (2007); Daaboul et al. (2010)) have been developed ; but while some of them are time-consuming and unconducive to real-time sample characterization, others rely on extensive surface preparation steps or availability of specific antibodies for the target viruses in a sample. A single method which can quickly and accurately quantify levels of different viruses present in clinically relevant samples without additional sample preparation steps, has remained elusive for practical implementation.
Optical detection of nanoscale biological agents (such as viruses) using light scattering is difficult due to their low scattering cross-section and low index contrast to the surrounding medium. Light scattering from a homogeneous sphere has a rigorous solution, as derived by Mie (Mie (1908)). Particles much smaller than the wavelength of the excitation light can be described by a dipolar polarizability α. The polarizability is given by
| (1) |
where R is the particle radius, and εp and εm are the dielectric permittivities of the particle and the surrounding medium, respectively. An incident oscillating electric field Eexc induces a dipole p in the particle according to p = αEexc (Bohren and Huffmann (1983)). The induced dipole radiates (i.e. scatters) a secondary electric field Es ∝ αEexc. Evidently, α defines the scattering and absorption efficiencies and bears information on both particle size (R) and composition (εp), and hence provides an important fingerprint in nanoparticle characterization. In nanoparticle detection techniques such as dynamic light scattering (Berne and Pecora (2000)) or flow cytometry (Givan (2001)), which probe the intensity of the scattered light I ∝ |Es|2, the detector signal scales with |α|2 ∝ R6. The strong size dependence makes it extremely difficult to detect small particles such as viruses based on standard light scattering. Virus detection approaches based on flow cytometry rely upon fluorescent labeling of segments of the viral genome(Brussaard et al. (2000); Ferris et al. (2002); Stoffel et al. (2005)), and hence are not label-free. In addition, no quantitative information can be obtained about the size of the virus particles(Porter et al. (1997)). On the other hand, interferometric detection (Lindfors et al. (2004); Batchelder and Taubenblatt (1991); Batchelder et al. (1991); Plakhotnik and Palm (2001)) exhibits a weaker size dependence and therefore provides significantly better signal-to-noise for small particles. For interferometric detection, the detector signal is proportional to the amplitude of the scattered light |Es|, and hence scales with α ∝ R3. Interferometric detection can provide single particle sensitivity and has the potential for real-time detection (Ignatovich and Novotny (2006); Mitra et al. (2010); Person et al. (2011); Deutsch et al. (2010)).
In real-time interferometric nanoparticle monitoring, particles typically are made to traverse a stationary laser focus, and the scattered field from a single particle is combined with a reference field and recorded interferometrically with a photodetector. In this article we introduce a new technique, which combines heterodyne interferometry with dark-field microscopy (Braslavsky et al. (2001)). The dark-field approach prevents any background light from reaching the detector in the absence of a particle at the laser focus, and hence improve detection sensitivity by reducing the background noise. Using heterodyne interferometry it is possible to effectively decouple the amplitude and phase of the detector signal and hence improve detection accuracy. Using this combined approach, we demonstrate a sensitivity superior to other interferometric techniques, and can clearly differentiate between single biological nanoparticles (phage and viruses) in a mixture. Such high sensitivity and resolution enables us to detect even impurities in virus samples.
2. Materials and Methods
2.1. Dark-field Heterodyne Interferometric Detection
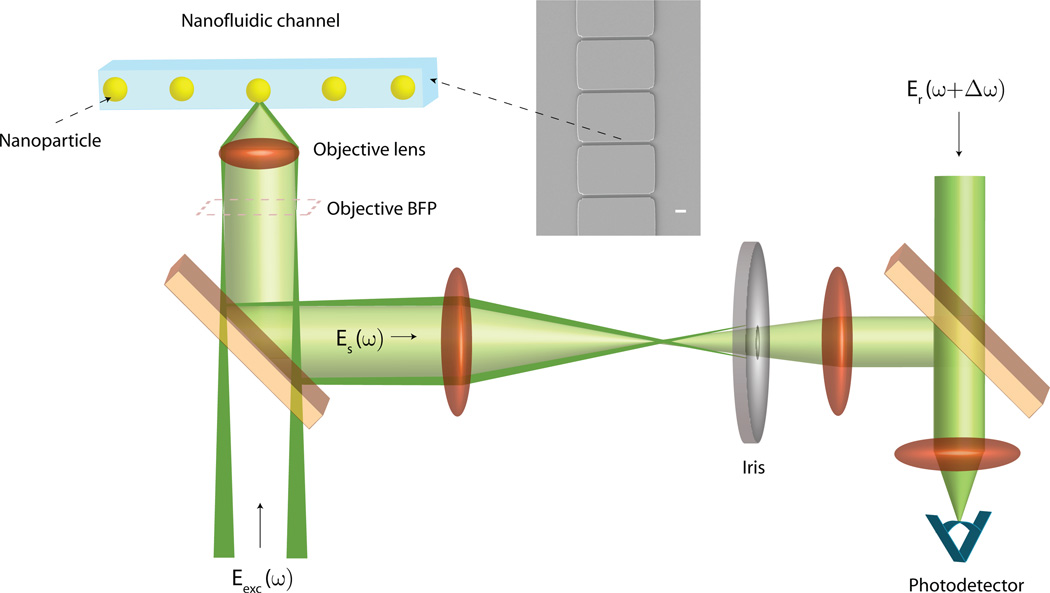
Fig. 1 illustrates the basic concept of the detection scheme (Refer to Supplementary Section 1 for a detailed description). Structured illumination is used to create converging annular illumination at the focal plane of high-NA objective, where particles such as viruses traverse the illumination spot inside a glass nanofluidic channel (see Section 2.2). Such configuration allows to separate the light back-scattered by the nanoparticle from the portion of the incident light specularly reflected by the channel interfaces, i.e. the background light. Eliminating the background lowers the noise floor in the detector signal and therefore results in high detection sensitivity.
Figure 1.
Conceptual schematic of the dark-field heterodyne interferometric nanoparticle detection technique. (Inset) SEM micrograph of a series of nanofluidic channels in our flow-cell. Scale Bar = 2 µm.
Without dark-field detection, such as for interferometric detection strategies which employ a ‘bright-field’ scheme where a collimated gaussian beam is tightly focused to illuminate a particle, the signal S recorded by the photodetector shown in Fig. 1 would be
| (2) |
where Δϕrb is the phase difference between Eb and the frequency-shifted reference field Er. Δϕsr is the phase difference between the field scattered Es and the reference field Er, and Δω is the heterodyne detuning frequency, that is, the frequency difference between Er and Es. Note that since the last term in Eq. 2 does not depend on the scattered field Es, it does not contain any information about the particle, but only increases the noise floor and thus reduces the method’s sensitivity. Because lock-in detection uses Δω as a reference frequency, the third term cannot be eliminated, unlike the second term. Usually a differential detection strategy is employed where a split detector or a balanced detector is used to eliminate the third term (Mitra et al. (2010)). However, such a strategy relies on ideal detector alignment and a perfectly stable system. In practice, an interferometric system is not sufficiently stable to completely eliminate the contribution of Eb in reduction of detection sensitivity, unless Eb is eliminated as good as possible. In the present approach, Eb is suppressed by means of dark-field detection, which eliminates the need for differential detection, ideal detector alignment, and beam stability to eliminate background.
With dark-field detection, the terms containing Eb in Eq. (2) are eliminated and only the first term survives. The detector signal S hence becomes
| (3) |
After lock-in demodulation of the detector signal S we obtain the two orthogonal signals
| (4) |
where we have used the fact that Es ∝ αEexc, where Eexc is the excitation electric field on a particle. The modulus reflects the particle’s polarizability α and hence is a measure of the particle’s radius R. Note that A is independent of phase, and hence variations in phase due to differences in trajectories of individual particles traversing the laser focus do not affect particle characterization. The elimination of background also means that the noise floor in the absence of a particle at the focus is mainly shot noise from the reference field, which greatly enhances the sensitivity over that achievable without dark-field detection (see Supplementary Section 3 for an experimental demonstration of the reduction in noise floor).
In addition to significantly influencing the phase of the scattered field, differences in flow trajectories of nanoparticles through the laser focus can also affect the resolution of a detection scheme due to intensity variations across the focus. In the present dark-field approach, the illumination spot at the focal plane is considerably larger (~ 18 microns) than the nanochannel width (500 nm)(see Supplementary Section 2 for details), ensuring that all particle trajectories experience practically the same excitation field amplitude. Thus, different particle trajectories do not cause any meaningful signal variations, which results in close to real particle size distributions. The same cannot be said for other methods which rely on a tightly focused laser beam with a diffraction-limited spot-size (Mitra et al. (2010)), since the intensity of the excitation field varies significantly across the width of a nanochannel.
2.2. Nanofluidic channels and flow mechanism
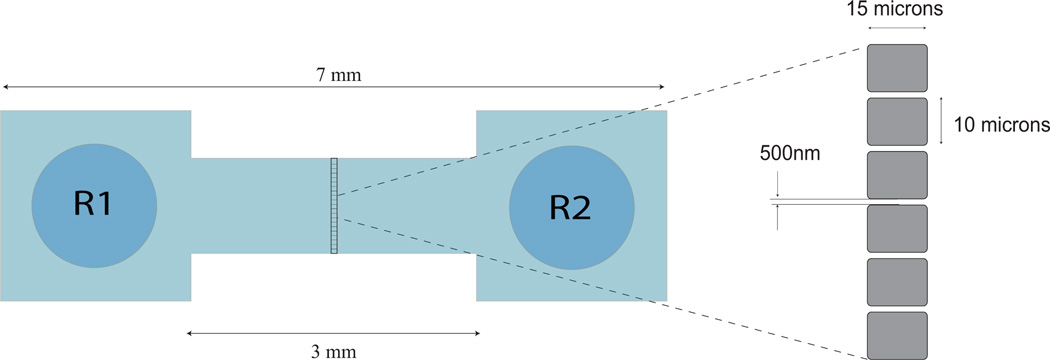
In order to detect nanoparticles such as viruses and phages in a solution in real-time, we make them flow through flow-cells fabricated on fused silica wafers using UV lithography (Ignatovich and Novotny (2003)). Each wafer-assembly consists of two wafers bonded to each other and it contains 82 isolated from each other flow-cells. As shown in Fig. 1, each flow-cell consists of two reservoirs separated from each other by 3 mm, and a 1 mm wide and 400 nm deep channel that connects the reservoirs. Halfway along the length of a flow-cell, there is a 15 µm long and 400 nm high ridge, on which an array of 500 nm wide nanochannels are fabricated .
A pressure driven flow-mechanism is employed to establish flow of nanoparticles such as viruses inside a flow-cell (Mitra et al. (2010)). Briefly, the end of a luer-lock barbed coupler is cut off so that its internal diameter is slightly larger than the diameter of a flow-cell reservoir. It is then attached on top of one of the reservoirs with glue and made air-tight with fast-cure epoxy. A PVC tubing is then fitted on to the coupler and an inflation device is attached to the tubing. The inflation device is used to create vacuum inside the flow-cell, which drives the sample introduced into the opposite reservoir. By adjusting the applied vacuum level, the flow-speed is made such that a single particle traverses the laser focus in ≤ 1 ms.
2.3. Determination of Sample concentration
The described detection scheme allows us to accurately measure particle concentrations. When a nanoparticle traverses the laser focus it yields a time-dependent signal of duration dt, which corresponds to the transit time through the focus. The particle velocity, and hence the fluid velocity, is defined by dl/dt, where dl is the size of the collection spot defined by the is defined by the NA of the objective. Using the cross-sectional dimensions of the nanochannel S we can then calculate the volume flow rate of the sample dV/dt = Sdl/dt. The concentration is equal to the ratio of the total particle counts N (in a defined size range) to the total volume V = τ dV/dt of the analyzed sample during the time of the experiment τ.
3. Results and Discussion
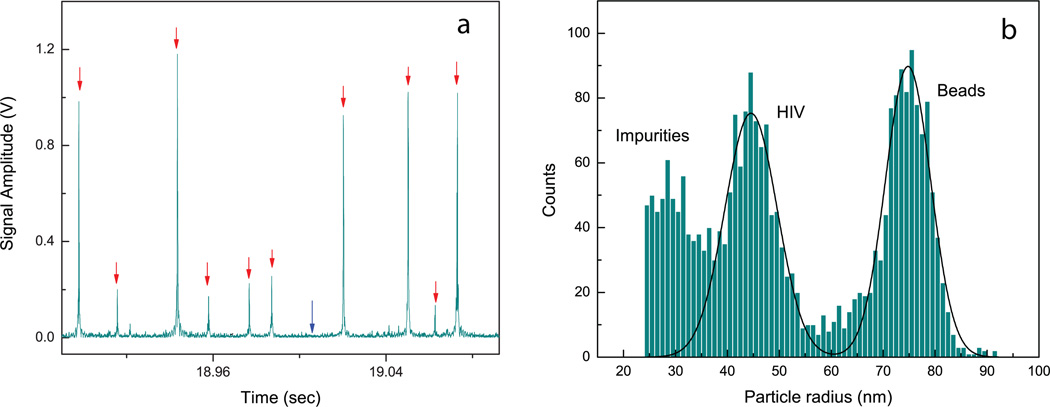
Fig. 3 shows the analysis of a sample containing HIV AT2WT wild-type virus in cell culture media, intermixed with 75 nm polystyrene beads for calibration purposes. Since the sample is directly extracted from a cell-line where HEK epithelial cells are transfected with the HIV AT2WT molecular clones, the media is expected to have a composition similar to clinically relevant samples. In Fig. 3a, we show a real-time trace for the detector signal, where individual particle detection events (shown with red arrows) are seen above the detector noise floor (shown with a blue arrow). The amplitudes of these single detection events are calibrated against particle size to obtain the size distribution of particles in the sample, shown in Fig. 3b. Two peaks for the virus particles and the beads respectively are clearly seen in the size distribution. Interestingly, we see some particles smaller than HIV virus in the sample, which are associated with impurities present in the sample. These are likely partially formed viral capsids, or exosomes secreted from the HIV transfected cells into the media from which the sample is extracted. The sensitivity and size resolution obtained for the sample is significantly better than that obtainable with ‘bright-field’ techniques where a diffraction-limited focal spot illuminates the sample (c.f. Supplementary Fig.4).
Figure 3.
Characterization of a sample containing HIV AT2WT wild-type virus in cell-culture media, mixed with 75 nm polystyrene beads. (a) Real-time time trace of the photodetector signal. Red arrows: Individual particle detection events, and Blue arrow: Detector noise floor. (b) Size distribution or nanoparticles in the sample. Notice the presence of particles smaller than HIV virus in the sample (labeled as impurities). These are likely free viral capsids or exosomes present the sample (see main text).
The concentration of HIV particles in our sample is measured to be 2.3 × 1011 particles/ml and the mean measured radius of HIV particles from Fig. 3a is 44.5 nm. This value is based on calibrating the data with 75 nm radius polystyrene particles. Difference in the refractive indices of polystyrene (1.59) and the HIV virus (assumed to be similar to that of Tabacco Mosaic Virus, 1.5 (Oster (1950))) is taken into account when calculating the true sizes of the virus. The mean radius for the HIV particles distribution is then 50.7 nm. This closely matches previously reported TEM measurements (Takasaki et al. (1997)) and is also consistent with TEM images obtained from our samples (c.f. Fig. 4b).
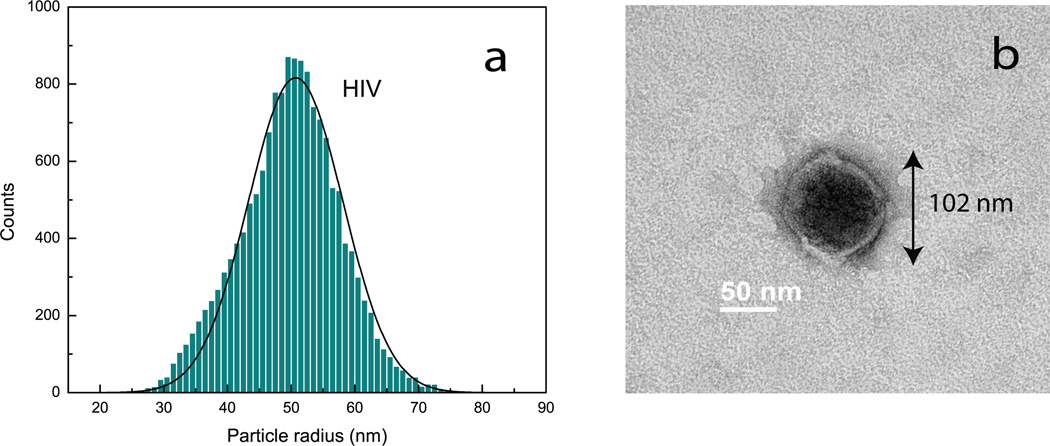
Figure 4.
(a) Size distribution obtained using dark-field interferometric detection for a purified sample of HIV virus (ADA strain).(b) TEM micrograph of a single HIV virus from the sample. The radius of the virus in the image is 51 nm, in close agreement with the mean size of the distribution.
In contrast to the commonly used p24 capsid ELISA HIV quantification method (Tehe et al. (2006)), our approach differentiates between intact virus particles and free capsids (which are smaller in size than the virus particles, as in Fig. 3b), and therefore the true virus concentration can be obtained. The ELISA method quantifies all p24 capsid proteins present in a sample, and hence any free and partial viral capsids in the sample are counted in addition to the intact virus.
Fig. 4a shows the size distribution obtained for a purified sample of HIV (ADA strain), free of the impurities. The viruses are suspended in TNE buffer (0.01 M Tris pH 7.2, 0.1 M NaCl, 1 mM EDTA), and to avoid sticking of virus particles to the surfaces of the nanochannels, the sample is diluted with a 2% solution of Polyvinylpyrrolidone (PVP, Sigma-Aldrich) in TNE buffer, which forms a protein-repellent layer in the nanochannel. For calibration, we used the mean size of HIV virus particles as 50.7 nm, determined from Fig. 3a. The standard deviation (SD) of the distribution is σ = 7.4 nm and the sample concentration is measured at 2.9 × 1012 particles/ml. As expected, Fig. 4a shows no impurities.
Fig. 4b shows a TEM micrograph of a HIV particle obtained for the sample characterized in Fig. 4a. The radius of the virus particle is 51 nm, which closely matches the mean radius determined from our optical measurements. Additional TEM images obtained for the HIV virus sample are shown in Supplementary Fig.5, to illustrate the size variation between individual particles.
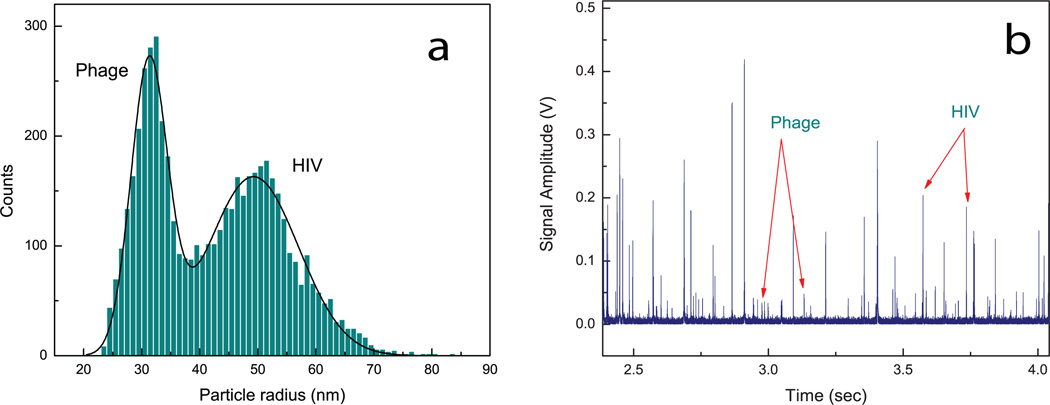
Fig. 5a shows the size distribution recorded for a mixture of bacteriophage lambda (or phage) and HIV particles. Phage is suspended in a dialysis buffer (0.05 M Tris-Cl pH 7.5, 0.01 M NaCl, 0.01 M MgCl2) and then mixed with the HIV ADA sample mentioned above. Both the phage and the HIV samples are diluted with a 2% solution of PVP in TNE buffer to prevent sticking to the nanochannels. The size distribution is calibrated using the mean size of HIV obtained from the previous experiments, assuming that the refractive index difference between phage and HIV is negligible. Two well-resolved peaks for phage and HIV can be easily seen in the mixture distribution. The mean size (radius) of phage from the mixture distribution is 31.6 nm. The standard deviations of the individual size distributions for phage and HIV are σ = 3.1 nm and σ = 7.8 nm respectively. Note that the standard deviation of the HIV size distribution closely matches that from Fig. 4a (σ = 7.4 nm). One can also see that the HIV particles have a wider size distribution than the phage particles. Refer to Supplementary Material (Supplementary Figs. 5 and 6), where TEM images obtained for the phage and HIV samples confirm this observation.
Figure 5.
Analysis of a mixture of bacteriophage lambda and HIV virus (ADA strain).(a) Size distribution obtained using dark-field interferometric detection.(b) Real-time trace (~ 1.5 sec) of the detector signal obtained for the virus/phage mixture. Signal peaks for single phage and HIV particles are shown with arrows, indicating a clear distinction based on the magnitude of the signal peaks.
Fig. 5b shows a time-trace of the detector signal obtained for the mixture of phage and HIV. Distinct peaks are clearly seen above the noise level, and these peaks represent single particles (phage or HIV) traversing the laser focus. The signal-to-noise obtained for single phage particles is ~ 3 − 4 and for single HIV particles ~ 15 − 30.
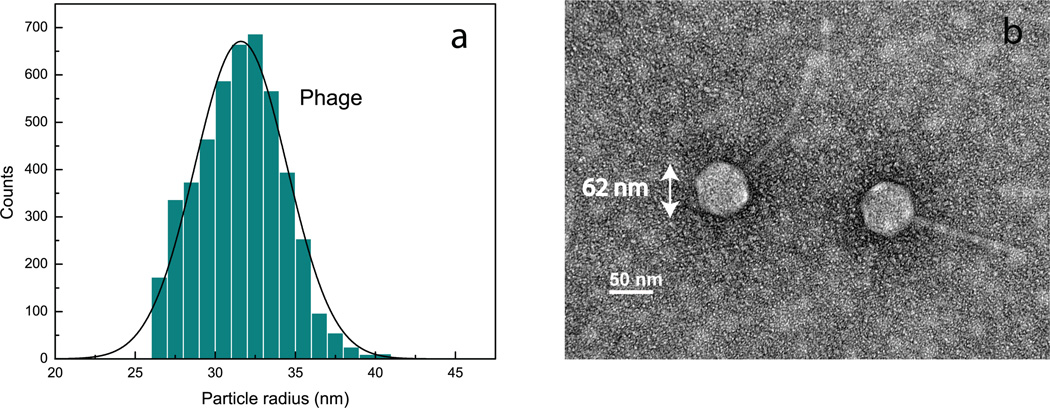
Fig. 6a shows the size distribution obtained for the phage sample to verify that mixing it with the HIV sample does not lead to aggregation. The standard deviation of the size distribution is 2.9 nm, similar to that (σ = 3.1 nm) determined from the mixture data (Fig. 5a). The concentration of phage particles in the sample using our method is 2 × 1012 particles/ml, which is in good agreement with the concentration obtained with the plaque titer method (1.5 × 1012 Plaque Forming Units (PFU) per ml). Our method required only 30 seconds to determine the phage concentration, compared to days when using the plaque titer method. Notice that the titer method quantifies the infective phage particles in the sample, whereas our method measures the total particle concentration. The difference of the measured concentration values indicates that the fraction of infective phage in the sample is ~ 76%.
Figure 6.
Analysis of a bacteriophage lambda sample.(a) Size distribution obtained using dark-field interferometric detection.(b)TEM micrograph from the sample used in (a). The hexagonal head along with the tail makes a phage particle easily recognizable in the image. Radii of the heads of the phage particles (half the distances between opposite corners of the hexagons) are determined from the image to be 31 nm, in close agreement with the mean size determined from our optical measurements.
Fig. 6b shows a TEM micrograph of two single phage particles. The mean radius of the heads in the image is 31 nm, which validates the mean size measured with our optical technique (the head of the phage is responsible for most of the scattered field). See Supplementary material for additional TEM images obtained for the phage sample.
4. Conclusions
We have developed and demonstrated a new single nanoparticle detection technique, which combines advantages of phase-sensitive heterodyne interferometry and dark-field detection. This approach allowed us to achieve high single-nanoparticle sensitivity, leading to detection and accurate particle-by-particle characterization of various microbiological samples, containing human and bacterial viruses. Importantly, this method is label-free and operates in real-time, without the need of surface immobilization or other invasive sample modifications. It makes it possible to detect impurities in clinically relevant media, and opens up new diagnostic and research applications, relevant for vaccine development, environmental monitoring and contamination control. When compared to methods such as flow-cytometry, our method does not require fluorescence labeling, and also provides better sensitivity and size resolution in heterogeneous samples. The detection sensitivity can be further improved by increasing the incident laser power on the test sample. Since the laser is not tightly focused into a spot, there is no danger of perturbing the particles with high light intensity. Higher incident laser power increases the signal from the virus, but not the background noise. This would enable detection of smaller viruses and even some proteins, and hence further broaden the range of applications of the technique. One can foresee that with the aid of a fast spectrometer with high sensitivity, it might be possible to also obtain specific chemical information about the virus particles being characterized, so that the virus types can be identified in unknown samples.
Highlights.
We developed a label-free optical technique for detection of single nanoparticles.
Combines advantages of heterodyne interferometry and dark-field microscopy.
Characterizes single human virus and bacteriophage particles down to ~24 nm size.
Can distinguish populations in heterogeneous mixtures of biological nanoparticles.
Resolves distribution of nano-impurities (~20–30 nm) in clinically relevant media.
Supplementary Material
Figure 2.
Schematic of a nanofluidic flow-cell fabricated on fused silica wafers, showing reservoirs R1 and R2 at its ends. Virus sample is introduced into reservoir R1 and are made to flow through the flow-cell by pressure-driven flow. Nanochannels are present in the zoomed in area of the flow-cell. Nanochannels (cross section 500 nm × 400 nm, length 15 µm) form an array across the 15 µm ridge along the center of the flow-cell, as shown in the figure. The laser focus is placed at the center a nanochannel to detect the passage of individual viruses or nanoparticles.
Acknowledgments
We thank Prof. Carrie Dykes for providing us the HIV virus samples, Prof. Steve Dewhurst and Dr. Jonelle Mattiacio for providing us the bacterio-phage lambda sample, and Zack Lapin for help with SEM imaging. We also thank Steve Person and Dr. John Lesoine for help in fabricating the nanofluidic channels. This work is supported by NIH (grant 1R21AI085543-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson B, Friedman H, Bendinelli M, editors. Microorganisms and bioterrorism. first edition. New York: Springer; 2006. [Google Scholar]
- Batchelder JS, DeCain DM, Taubenblatt MA, Wickramasinghe HK, Williams CC. 5,061,070. US Patent. 1991
- Batchelder JS, Taubenblatt MA. 5,037,202. US Patent. 1991
- Berne BJ, Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics. first edition. New York: Dover Publications; 2000. [Google Scholar]
- Bohren CF, Huffmann DR. Absorption and Scattering of Light by Small Particles. New York: Wiley; 1983. [Google Scholar]
- Braslavsky I, Amit R, Jaffar Ali BM, Gileadi O, Oppenheim A, Stavans J. Appl. Opt. 2001;40:5650–5657. doi: 10.1364/ao.40.005650. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD, Marie D, Bratbak G. J. Virol. Meth. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- Choi M, Stanton-Maxey K, Stanley J, Levin C, Bardhan R, Akin D, Badve S, Sturgis J, Robinson J, Bashir R, Halas N, Clare S. Nano Lett. 2007;7:3759–3765. doi: 10.1021/nl072209h. [DOI] [PubMed] [Google Scholar]
- Chuan YP, Fan YY, Lua L, Middelberg A. Biotechnology and Bioengineering. 2007;99:1425–1433. doi: 10.1002/bit.21710. [DOI] [PubMed] [Google Scholar]
- Cromeans TL, Lub X, Erdman DD, Humphrey CD, Hill VR. Journal of Virological Methods. 2008;151:140–145. doi: 10.1016/j.jviromet.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Daaboul GG, Yurt A, Zhang X, Hwang GM, Goldberg BB, nl MS. Nano Lett. 2010;10:4727–4731. doi: 10.1021/nl103210p. [DOI] [PubMed] [Google Scholar]
- Deutsch B, Beams R, Novotny L. Appl. Opt. 2010;49:4921–4925. doi: 10.1364/AO.49.004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D, Willey R, Sato H, Chang LJ, Blumenthal R, Martin M. Journal of Virology. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R, Vogt M. The Journal of Experimental Medicine. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I, Petzold D, Kosinska A, Hepkema B, Schulz T, Heim A. Journal of Medical Virology. 2008;80:467–477. doi: 10.1002/jmv.21095. [DOI] [PubMed] [Google Scholar]
- Ferris MM, McCabe MO, Doan LG, Rowlen KL. Analytical Chemistry. 2002;74:198. doi: 10.1021/ac011183q. [DOI] [PubMed] [Google Scholar]
- Fraikin JL, Teesalu T, McKenney CM, Ruoslahti E, Cleland AN. Nature Nanotech. 2011 doi: 10.1038/nnano.2011.24. [DOI] [PubMed] [Google Scholar]
- Givan AL. Flow cytometry: first principles. 2nd edition. New York: Wiley-Liss; 2001. [Google Scholar]
- Hockett RD, Kilby JM, Derdeyn CA, Saag MS, Sillers M, Squires K, Chiz S, Nowak MA, Shaw GM, Bucy RM. J Exp Med. 1999;189:1545–1554. doi: 10.1084/jem.189.10.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Nanomedicine. 2007;2:681–693. doi: 10.2217/17435889.2.5.681. [DOI] [PubMed] [Google Scholar]
- Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FYS, Li XD, Asmuth DM, Huser T, Chen BK. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatovich FV, Novotny L. Rev. Sci. Instr. 2003;74:5231–5235. [Google Scholar]
- Ignatovich FV, Novotny L. Phys. Rev. Lett. 2006;96 doi: 10.1103/PhysRevLett.96.013901. 013901. [DOI] [PubMed] [Google Scholar]
- Krug RM. Antiviral Research. 2003;57:147–150. doi: 10.1016/s0166-3542(02)00207-3. [DOI] [PubMed] [Google Scholar]
- Lindfors K, Kalkbrenner T, Stoller P, Sandoghdar V. Phys. Rev. Lett. 2004;93 doi: 10.1103/PhysRevLett.93.037401. 037401. [DOI] [PubMed] [Google Scholar]
- Mie G. Ann. d. Physik. 1908;330:376–445. [Google Scholar]
- Mitra A, Deutsch B, Ignatovich F, Dykes C, Novotny L. ACS Nano. 2010;4:1305–1312. doi: 10.1021/nn901889v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L. Air Quality and Climate Change. 2010;44:18–20. [Google Scholar]
- Oberdörster G. International Archives of Occupational and Environmental Health. 2000;74:1–8. doi: 10.1007/s004200000185. [DOI] [PubMed] [Google Scholar]
- Oster G. The Journal of General Physiology. 1950;33:445–473. doi: 10.1085/jgp.33.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patolsky F, Zheng G, Hayden O, Lakadamyali M, Zhuang X, Lieber CM. Proc. Natl. Acad. Sci. USA. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person S, Deutsch B, Mitra A, Novotny L. Nano Lett. 2011;11:257–261. doi: 10.1021/nl103656y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plakhotnik T, Palm V. Phys. Rev. Lett. 2001;87 doi: 10.1103/PhysRevLett.87.015504. 183602. [DOI] [PubMed] [Google Scholar]
- Porter J, Deere D, Hardman M, Edwards C, Pickup R. FEMS Microbiol. Ecol. 1997;24:93–101. [Google Scholar]
- Ramanathan V, Carmichael G. Nature Geosc. 2008;1:221–227. [Google Scholar]
- Salter R, Durbin G, Conklin E, Rosen J, Clancy J. Applied and Environmental Microbiology. 2010;76:7803–7810. doi: 10.1128/AEM.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rodrguez T, Dvila C, Gonzlez J, Bonilla N, Marcos P, Urdaneta M, Cadete M, Monteiro S, Santos R, Domingo J, Toranzos G. Water Research. 2010;44:4716–4725. doi: 10.1016/j.watres.2010.07.078. [DOI] [PubMed] [Google Scholar]
- Somers CM, McCarry BE, Malek F, Quinn JS. Science. 2004;304:1008–1010. doi: 10.1126/science.1095815. [DOI] [PubMed] [Google Scholar]
- Stern E, Wagner R, Sigworth FJ, Breaker R, Fahmy TM, Reed MA. Nano Lett. 2007;7 doi: 10.1021/nl071792z. 34053409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel C, Finch R, Christensen K, Edwards D, Rowlen KL. American Biotechnology Laboratory. 2005;37:24–25. [Google Scholar]
- Takasaki T, Kurane I, Aihara H, Ohkawa N, Yamaguchi J. Archives of Virology. 1997;142:375–382. doi: 10.1007/s007050050083. [DOI] [PubMed] [Google Scholar]
- Tehe A, Maurice C, Hanson DL, Borgeta MY, Abiolaa N, Marana M, Yavoa D, Tomasikc Z, Bnic J, Schpbachc J, Nkengasong JN. Journal of Clinical Virology. 2006;37:199–205. doi: 10.1016/j.jcv.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Tsai W, Conley S, Kung H, Garrity R, Nara P. Virology. 1996;226:205–216. doi: 10.1006/viro.1996.0648. [DOI] [PubMed] [Google Scholar]
- Vollmer F, Arnold S. Nature Methods. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- Vollmer F, Arnold S, Keng D. Proc. Natl. Acad. Sci. USA. 2008;105:20701–20704. doi: 10.1073/pnas.0808988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wali F, Knotter DM, Mud A, Kuper FG. Microelectronic Engineering. 2009;86:140–144. [Google Scholar]
- Yanik AA, Huang M, Kamohara O, Artar A, Geisbert TW, Connor JH, Altug H. Nano Lett. 2010;10 doi: 10.1021/nl103025u. 49624969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ymeti A, Greve J, Lambeck PV, Wink T, van Hvell SWFM, Beumer TAM, Wijn RR, Heideman RG, Subramaniam V, Kanger JS. Nano Lett. 2007;7:394–397. doi: 10.1021/nl062595n. [DOI] [PubMed] [Google Scholar]
- Zhu J, Ozdemir1 SK, Xiao YF, Li L, He L, Chen DR, Yang L. Nature Photon. 2009;4:46–49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.