Abstract
Multiple new prokaryotic C-terminal protein-sorting signals were found that reprise the tripartite architecture shared by LPXTG and PEP-CTERM: motif, TM helix, basic cluster. Defining hidden Markov models were constructed for all. PGF-CTERM occurs in 29 archaeal species, some of which have more than 50 proteins that share the domain. PGF-CTERM proteins include the major cell surface protein in Halobacterium, a glycoprotein with a partially characterized diphytanylglyceryl phosphate linkage near its C terminus. Comparative genomics identifies a distant exosortase homolog, designated archaeosortase A (ArtA), as the likely protein-processing enzyme for PGF-CTERM. Proteomics suggests that the PGF-CTERM region is removed. Additional systems include VPXXXP-CTERM/archeaosortase B in two of the same archaea and PEF-CTERM/archaeosortase C in four others. Bacterial exosortases often fall into subfamilies that partner with very different cohorts of extracellular polymeric substance biosynthesis proteins; several species have multiple systems. Variant systems include the VPDSG-CTERM/exosortase C system unique to certain members of the phylum Verrucomicrobia, VPLPA-CTERM/exosortase D in several alpha- and deltaproteobacterial species, and a dedicated (single-target) VPEID-CTERM/exosortase E system in alphaproteobacteria. Exosortase-related families XrtF in the class Flavobacteria and XrtG in Gram-positive bacteria mark distinctive conserved gene neighborhoods. A picture emerges of an ancient and now well-differentiated superfamily of deeply membrane-embedded protein-processing enzymes. Their target proteins are destined to transit cellular membranes during their biosynthesis, during which most undergo additional posttranslational modifications such as glycosylation.
INTRODUCTION
Most Gram-positive bacteria have cohorts of surface protein precursors that share a determinant at their carboxyl termini for sorting and covalent attachment to the cell wall. This sorting signal consists of the signature motif LPXTG, a hydrophobic transmembrane (TM) alpha helix, and a cluster of basic amino acids, primarily arginines (29). Sortase A (EC 3.4.22.70) cleaves these precursors after the Thr residue, transferring the protein first to its own active-site Cys and then to the Gram-positive cell wall (34). Homologous but distinct subfamilies of sortase enzymes cross-link pilin subunits (8), recognize a sorting signal with a signature motif that departs from LPXTG (4, 7), or occur only in members of the class Proteobacteria. Several of these alternate systems appear to be dedicated systems, meaning that a single protein substrate occurs per organism for sortases of the corresponding subfamily.
The LPXTG-sortase relationship is closely paralleled by the PEP-CTERM/exosortase system, which has been studied so far through bioinformatic methods and is found in many biofilm-producing environmental bacteria (17). The sorting signal retains the architectural description “signature motif, then TM helix, then arginine-rich cluster, always at the protein carboxyl terminus,” but the signature motif in PEP-CTERM proteins is Pro-Glu-Pro or PEP. The PEP-CTERM domain nearly always occurs multiple times per genome, if it occurs at all. As with LPXTG, all PEP-CTERM proteins have an N-terminal signal peptide to direct transit across the plasma membrane, and PEP-CTERM proteins in general lack discernible homology to each other between these regions. In contrast to LPXTG proteins, however, most PEP-CTERM proteins lack detectable homology to known enzymes or to other proteins with characterized globular domains. Instead, most PEP-CTERM proteins have extensive regions of low-complexity sequence, usually rich in residues that could suggest extensive glycosylation, either O linked (Thr, Ser) or N linked (Asn) (17).
All genomes encoding cohorts of PEP-CTERM proteins also encode a multiple-membrane-spanning protein related to EpsH from Methylobacillus sp. strain 12S (17). EpsH is encoded within an extended locus for producing methanolan, a species-specific extracellular polymeric substance (EPS) (38). EpsH, now called exosortase, lacks any detectable homology to the smaller soluble enzyme sortase, but its four nearly invariant residues are oriented toward the extracellular face, and three of these, Cys, Arg, and His, on three consecutive helices, match the highly conserved known catalytic triad of the sortase superfamily. The architectural similarity between the LPXTG target sequences that co-occur with sortases and PEP-CTERM sequences that co-occur with exosortases and their similar many-or-none distributions across species suggest analogous functions. PEP-CTERM, like LPXTG, may be the recognition sequence for protein-processing functions that could include protein modification, cleavage, sorting, and attachment. Unlike sortases, exosortases were found to occur mostly in metabolic contexts containing EPS biosynthesis enzymes and usually within the EPS biosynthesis gene neighborhood itself (17).
Since the last published study of PEP-CTERM and exosortase, many additional genomes have been sequenced, providing much broader coverage of bacterial and archaeal lineages. Several microbial genomes now available encode over 60 PEP-CTERM proteins; Opitutaceae bacterium TAV2, an extreme case, encodes 194 members, amounting to 4% of its 4,826 proteins. Exosortases, as defined by TIGRFAMs (30) hidden Markov model (HMM) TIGR02602, commonly occur once per genome. However, a significant number of bacterial genomes encode additional exosortase paralogs, and these may be divergent enough to score well below the trusted cutoff score provided with model TIGR02602. Distant homologs of exosortase occur in species that lack canonical PEP-CTERM sequences, that is, those recognized by TIGRFAMs HMM TIGR02595. The lack suggests that new subfamilies from the broader superfamily that includes exosortase may serve as processing enzymes that recognize different forms of target sequence. Such specialization and diversity would parallel the observation that sortase homologs evolutionarily distant from sortase A may recognize sorting signals that differ from the canonical LPXTG while performing tasks at least as variable as cell wall attachment and pilin cross-linking (8, 26).
The numbers of complete and high-quality draft reference genomes are now large enough to support a broad new investigation of the comparative genomics of exosortase-related proteins and their probable targets. The work that follows identifies several novel classes of putative protein-sorting enzymes, their corresponding proposed target sequences, and in several cases additional associated protein families. Several systems to which these molecular markers belong occur in the archaea, or in Gram-positive bacteria, with cell envelopes structured very differently from those of Gram-negative species with PEP-CTERM proteins. Multiple different systems can coexist in the same organism. Rather than being associated primarily with EPS biosynthesis, and by implication with biofilm formation, these novel systems seem to point to a variety of cellular processes that require protein export and processing. For one archaeal system, the list of targets identified includes a protein that has been partially characterized. The S-layer-forming major cell surface glycoprotein in various halophilic archaea is highly expressed, has multiple types of carbohydrates attached (23, 31), and has a large C-terminal lipid modification that could serve as a membrane anchor (21).
Host cell envelope structures, the types of proteins bearing C-terminal recognition sequences, and additional partner proteins when these are found differ greatly among the new systems we describe. The commonalities that remain, therefore, may shed light on the reason that such widely different lineages carry systems with such similar designs. The target proteins, in general, have recognizable N-terminal signal peptides and therefore transit the plasma membrane during their biogenesis. Many also either show homology to known prokaryotic glycoproteins or carry low-complexity sequence regions that suggest extensive glycosylation. Several systems that include exosortase homologs include identified carbohydrate attachment enzymes. The variety of specialized systems in which exosortase homologs apparently have coevolved with cognate C-terminal protein-sorting signals suggests that there is a common enzymatic processing step, conserved from bacteria to archaea, that helps orchestrate the maturation of exported and heavily posttranslationally modified proteins.
MATERIALS AND METHODS
Defining novel families of exosortase-related proteins.
Novel exosortase family proteins were detected by performing iterative PSI-BLAST searches at NCBI (2) using the option “composition-based statistics” (28) to prevent false-positive detection of apparent homology that instead reflects simply the compositional similarity of one multiple-membrane-spanning sequence to another. Inclusion thresholds were adjusted during PSI-BLAST iteration to favor conservation of the proposed active-site residues Cys, Arg, and His. This criterion resulted in the exclusion of all sequences of eukaryotic origin; there appear to be no eukaryotic members of the exosortase superfamily. Multiple trials with different starting sequences retrieved similar protein sets. From the resulting sequence collection, members of known exosortase subfamilies (30) were removed, the remaining sequences were aligned by CLUSTAL W (33) or MUSCLE (11), and trees constructed by both the neighbor-joining method and the unweighted-pair group method using average linkages were examined. Deeply branched subgroups with sufficient numbers of members were treated as candidate new, functionally distinct protein families. Members were realigned by MUSCLE and made nonredundant to less than 80% pairwise identity. HMMs to represent these candidate new protein families were constructed using HMMER3 (19) and deposited in the TIGRFAMs database (30). The HMMs discussed in this study are described in Table 1. Full descriptions and multiple-sequence alignments are available at http://www.jcvi.org/cgi-bin/tigrfams/Listing.cgi.
Table 1.
HMMs used in this study
| Protein family | Gene | System | HMM |
|---|---|---|---|
| Exosortase/archaeosortase domain | Exosortase family signature | TIGR04178 | |
| PGF-CTERM | PGF-CTERM/archaeosortase A | TIGR04126 | |
| Archaeosortase A | artA | PGF-CTERM/archaeosortase A | TIGR04125 |
| Oligosaccharyl transferase | PGF-CTERM/archaeosortase A | TIGR04154 | |
| VPXXXP-CTERM | VPXXXP-CTERM/archaeosortase B | TIGR04143 | |
| Archaeosortase B | artB | VPXXXP-CTERM/archaeosortase B | TIGR04144 |
| PEF-CTERM | PEF-CTERM/archaeosortase C | TIGR03024 | |
| Archaeosortase C | artC | PEF-CTERM/archaeosortase C | TIGR03762 |
| PIP-CTERM | PIP-CTERM/archaeosortase D | TIGR04173 | |
| Archaeosortase D | artD | PIP-CTERM/archaeosortase D | TIGR04175 |
| Archaeosortase family protein ArtE | artE | Unassigned | TIGR04124 |
| VPDSG-CTERM | VPDSG-CTERM/exosortase C | TIGR03778 | |
| Exosortase C | xrtC | VPDSG-CTERM/exosortase C | TIGR04151 |
| VPLPA-CTERM | VPLPA-CTERM/exosortase D | TIGR03370 | |
| Exosortase D | xrtD | VPLPA-CTERM/exosortase D | TIGR04152 |
| VPEID-CTERM | VPEID-CTERM/exosortase E | TIGR04161 | |
| Exosortase E/CAAX protease | xrtE | VPEID-CTERM/exosortase E | TIGR04162 |
| Exosortase family protein XrtF | xrtF | XrtF system (Flavobacteria) | TIGR04128 |
| Exosortase F-associated protein | XrtF system (Flavobacteria) | TIGR04127 | |
| Gpos-CTERM | XrtG system (Gram-positive bacteria) | TIGR04145 | |
| Exosortase family protein XrtG | xrtG | XrtG system (Gram-positive bacteria) | TIGR03110 |
| Putative glycosyltransferase, TIGR03111 family | XrtG system (Gram-positive bacteria) | TIGR03111 | |
| 6-Pyruvoyl tetrahydropterin synthase related | XrtG system (Gram-positive bacteria) | TIGR03112 | |
| Integral membrane protein | XrtG system (Gram-positive bacteria) | TIGR03766 | |
| IPTL-CTERM | IPTLxxWG-CTERM/exosortase H | TIGR04174 | |
| Exosortase H | xrtH | IPTLxxWG-CTERM/exosortase H | TIGR04177 |
| PEP-CTERM, cyanoexosortase subclass | Cyanoexosortase system | TIGR04155 | |
| Cyanoexosortase A | crtA | Cyanoexosortase system | TIGR03763 |
| Cyanoexosortase B | crtB | Cyanoexosortase system | TIGR04156 |
| Cyanoexosortase-associated protein | Cyanoexosortase system | TIGR04153 |
Counting C-terminal sorting signals encoded per genome with iterated models.
To count the full cohorts within specific lineages for proteins containing a sorting signal such as PEP-CTERM (TIGR02595), member sequences detected (that is, that scored above the provided trusted cutoff) by the original TIGRFAMs HMM were collected and a new HMM was built from the multiple-sequence alignment of their C-terminal regions. The species-specific model was then used to search the target genome again to recruit proteins with more divergent forms of the putative sorting signal until iteration of the process recruited no more new sequences. The iterative refinement of lineage-specific models and sorting signal region multiple-sequence alignments that resulted were manually reviewed at each step to confirm conservation through the signature motif, integrity of the TM helix, presence of the cluster of basic residues, and location at the protein C terminus.
Defining novel families of C-terminal protein-sorting signals. (i) Method 1.
Protein C-terminal regions with architectures similar to PEP-CTERM and LPXTG, that is, “signature motif, TM helix, basic residues,” may score weakly to the PEP-CTERM model TIGR02595 itself, well below the trusted cutoff. However, proteins with such weakly matched sequences may include several from a single genome with strong mutual sequence similarity in their C-terminal regions. Iterative refinement of prospective new C-terminal protein-sorting domain HMMs was performed as described above for lineage-specific counting of previously defined sorting signals.
(ii) Method 2.
Genes for targets of sortase-like enzymes often are clustered with genes for their cognate enzymes (26). In order to find new classes of sorting signals, member proteins of candidate novel families of exosortase-like enzymes define sets of gene neighborhoods to explore. For a given set, the collection of all neighboring proteins encoded up to three genes away was collected and aligned by CLUSTAL W to find proteins recurringly present in the neighborhoods of exosortase genes. The resulting multiple-sequence alignments were inspected to find cohorts of proteins sharing homologous C-terminal regions with PEP-CTERM-like architecture.
PPP.
Partial phylogenetic profiling (PPP) (17) was performed using ProPhylo on a release based on 1,466 complete and high-quality draft prokaryotic reference genomes (5). Profiles, that is, lists of genomes marked according to whether or not a particular marker is found in each one, were constructed according to results from a given HMM and used with PPP to find additional well-correlated protein families.
Protein family construction and availability.
HMM constructions for protein families or for candidate protein-sorting domains were performed with HMMER3. Seed alignments were generated by MUSCLE or by CLUSTAL W, inspected, culled of misaligned sequences, and realigned as necessary. HMM search results were inspected manually to find scoring thresholds that delineate complete sets of sequences derived from a single ancestral node in neighbor-joining molecular phylogenetic trees estimated from multiple-sequence alignments.
Typically, these boundaries mark sharp falloffs in scores from sequences within versus sequences outside the subfamily, by changes in the protein architecture and the genomic contexts of matching proteins, and by the greater taxonomic distances of the species of origin for the next most closely matching proteins. Where possible, two or more protein families from the same proposed system were constructed together and taxonomic co-occurrence was used as a further criterion for fine-tuning of protein family boundaries. Seed alignments, descriptions, and HMMs for newly defined protein families were deposited in the TIGRFAMs protein family database (30).
Proteomics.
Proteomic data for Methanospirillum hungatei, Methanosarcina barkeri, Halorhabdus utahensis, and Cyanothece sp. were generated at the Pacific Northwest National Laboratory, in the lab of Richard D. Smith, using standard protocols (9). Thermo RAW files were converted to mzXML and processed with the prokaryotic proteogenomics pipeline (27). Data were filtered using the MS-GF (http://proteomics.ucsd.edu/Software/MSGeneratingFunction.html) program's spectrum probability of 1e-10, resulting in a 0.1% false-discovery rate at the peptide level. Peptides presented in this work derive from a reanalysis of previous experiments and are limited to proteins that are targets of the putative sorting/modification systems discussed.
Signal peptide prediction.
Proteins with predicted C-terminal recognition sites for archaeosortase/exosortase proteases or transpeptidases were examined for the presence of signal peptides using signalP (6) and examining the first 70 amino acids only. All three taxonomy options (euk, Gram+, and Gram−) were tried. Proteins with a PGF-CTERM motif but no predicted signal peptide were aligned with PGF-CTERM proteins with a successfully predicted signal peptide in order to identify genes with apparently faulty translation start site predictions.
RESULTS
Homology extends through TM segments in PEP-CTERM-like regions of a particular class.
Alignment of multiple C-terminal putative protein-sorting signals from a single species often shows apparent sequence homology that runs through the TM domain and is not limited to the signature motif for which the domain is named. This particularly high sequence similarity running through hydrophobic TM segments is not typical for other sorting signals with TM segments such as type I signal peptide, lipoprotein signal peptide, or even for C-terminal LPXTG tail regions of proteins processed by the (soluble) enzyme sortase. For example, among the 26 PEP-CTERM proteins of the human gut commensal bacterium Akkermansia mucinophila (36), only three pairs of proteins can be found sharing over 40% sequence identity in their N-terminal signal peptide regions. In stark contrast, their PEP-CTERM regions show a high level of sequence identity, with 15 of 24 positions having a single amino acid conserved through at least 80% of the member sequences and with no gaps, evidence for homology. Strong conservation of residues located mid-span in TM regions is not visible in alignments of PEP-CTERM regions from widely divergent organisms but becomes apparent in collections limited to paralogs from a single organism or from related organisms with closely related exosortases. This observation appears to provide evidence that each lineage-specific cohort of targets has coevolved with its corresponding processing protein. One implication is that regions of protein-protein interaction with exosortases (proteins that span the membrane multiple times) include much of the hydrophobic TM helix of the sorting signal and not just its signature motif. Different forms therefore behave as homology domains throughout their lengths and may be discriminated readily by HMMs. We therefore looked for distinctive subfamilies of putative C-terminal protein-processing signals, for novel subfamilies of exosortase-like enzymes, and for conserved relationships between the two.
Identification of remote homologs of exosortase.
PSI-BLAST starting with the remote exosortase homolog AF2046 from the archaeon Archaeoglobus fulgidus and ending with convergence retrieved 427 proteins, all prokaryotic. PSI-BLAST starting from different initial sequences retrieved very similar sets of proteins. In the set retrieved, 181 were canonical bacterial exosortases (captured by TIGRFAMs model TIGR02602). The broader-hitting model PF09721, derived by Pfam from TIGR02602, identifies 413 of the 427. From the set of proteins collected by PSI-BLAST, a multiple alignment was constructed, culled, edited, trimmed to a core homology region of about 97 amino acids, and used to construct a signature HMM for the broader family (TIGR04178) of all exosortase homologs. Searches with this new model confirmed the apparent absence of any eukaryotic member of the exosortase/archaeosortase family. In a collection of 1,466 prokaryotic reference genomes, this model finds 326 proteins in 240 genomes, an average count of 1.36 per genome in the 16.4% of the genomes that have a member. The significant rate at which paralogs occur suggests that some lineages encode related yet functionally distinct and nonoverlapping systems that operate in parallel.
The archaeosortase A/PGF-CTERM system.
A distinctive subfamily of exosortase-like proteins occurs in 29 out of 57 reference genomes from the phylum Euryarchaeota and nowhere else. Because of its restriction to the archaea and its remote homology to exosortase, the family is designated archaeosortase A, with the gene designation artA, and is modeled by TIGRFAMs HMM TIGR04125 (Table 1 lists all of the new HMMs described in this work). The HMM always identifies exactly one archaeosortase A gene per genome, and it shows a very sharp separation between the scores of members (scores of >180) and nonmembers (scores of <40). Genomes containing this protein form a monophyletic group descended from a single node according to a phylogenetic tree for the phylum Euryarchaeota inferred from a fusion analysis of 53 ribosomal proteins (3); members occur in the order Archaeoglobales, the order Halobacteriales, and the class II methanogens, which include the orders Methanomicrobiales and Methanosarcinales. However, the family is absent from the order Thermoplasmatales, the family Thermococcaceae, and the class I methanogens. The protein family, however, would not be monophyletic according to a more recently proposed archaeal phylogeny based on gene order (24), one that suggests a revised definition for the class II methanogens.
Searching proteins encoded in the immediate vicinity of artA (method 2; see Materials and Methods) for C-terminal regions with architecture similar to that of PEP-CTERM (but with a different signature motif) found six species among the members of the class Halobacteria in which a protein with such a region was encoded by the adjacent gene (four species) or separated by no more than two intervening genes (two additional species). Iterative alignment, HMM construction, and searching for additional proteins with similar C-terminal sequence regions led to the identification of a sequence homology domain whose tripartite architecture echoes that of PEP-CTERM. Its signature motif, however, is Pro-Gly-Phe (PGF), and the region described by the resulting model (TIGR04126) therefore is named PGF-CTERM. Panel A in Fig. 1 shows the sequence logo for this putative protein-sorting/processing signal. The PGF-CTERM model finds its best matches (including all matches that exceed the trusted cutoff of the HMM) in exactly the same 29 genomes as the archaeosortase A model. This perfect taxonomic codistribution, the genomic colocalization in the class Halobacteria, and paired homology relationships (exosortase/archaeosortase and PEP-CTERM/PGF-CTERM) strongly suggest that the pair represents an ancient conserved protein-processing system in the domain Archaea.
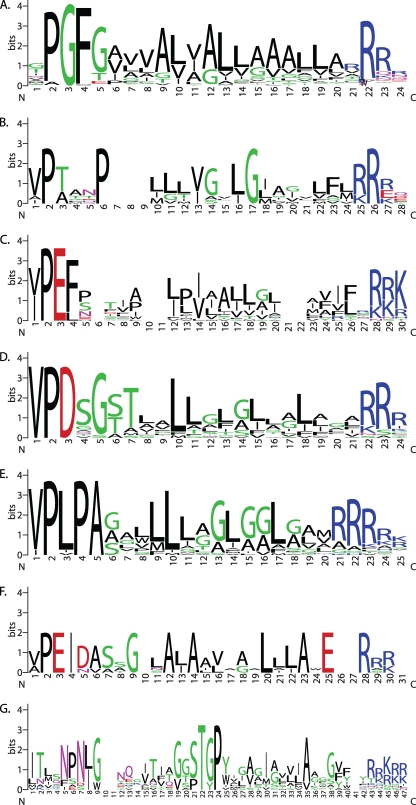
Fig 1.
Sequence logos for C-terminal protein-sorting domains associated with exosortase/archaeosortase family proteins. The putative sorting signals shown are as follows: panel A, PGF-CTERM (TIGR04126), cognate sequence for archaeosortase A; panel B, VPXXXP-CTERM (TIGR04143), cognate sequence for archaeosortase B; panel C, PEF-CTERM (TIGR03024), cognate sequence for archaeosortase C; panel D, VPDSG-CTERM (TIGR04151), cognate sequence for exosortase C; panel E, VPLPA-CTERM (TIGR04152), cognate sequence for exosortase D; panel F, VPEID-CTERM (TIGR04161), cognate sequence for exosortase E/CAAX prenyl protease; panel G, Firmcu-CTERM domain (TIGR04145), candidate cognate sequence for exosortase G.
Because the PGF-CTERM model describes a region of only 28 amino acids, with few positions that are particularly strongly conserved across broad taxonomic ranges, any cutoff set for the model to avoid false positives necessarily will miss some true examples. However, panels of customized models can be developed by iterative refinement to more precisely represent lineage-specific forms of the PGF-CTERM sorting signal. Table 2 shows species with archaeosortase A, the number of PGF-CTERM proteins found by iterated models derived from the PGF-CTERM model TIGR04126, and any additional exosortase homologs that may be found. As shown in Table 2, the number exceeds 50 putatively archaeosortase-processed proteins in certain genomes. The full list of identified PGF-CTERM proteins, with current RefSeq annotations, is presented in Table S1 in the supplemental material.
Table 2.
Archaeosortase A-encoding species and PGF-CTERM domain counts
| Strain | No. of ArtA targets | Additional archeaosortase (no. of targets) |
|---|---|---|
| Archaeoglobus fulgidus DSM 4304 | 11 | |
| Archaeoglobus profundus DSM 5631 | 8 | |
| Ferroglobus placidus DSM 10642 | 11 | ArtC (3) |
| Halalkalicoccus jeotgali B3 | 3 | |
| Haloarcula marismortui ATCC 43049 | 13 | |
| Halobacterium salinarum R1 | 6 | |
| Haloferax volcanii DS2 | 8 | |
| Halogeometricum borinquense DSM 11551 | 20 | |
| Halomicrobium mukohataei DSM 12286 | 10 | |
| Haloquadratum walsbyi DSM 16790 | 3 | |
| Halorhabdus utahensis DSM 12940 | 10 | |
| Halorubrum lacusprofundi ATCC 49239 | 10 | |
| Haloterrigena turkmenica DSM 5511 | 23 | |
| Methanocella paludicola SANAE | 12 | |
| Methanococcoides burtonii DSM 6242 | 25 | ArtC (10) |
| Methanocorpusculum labreanum Z | 3 | |
| Methanoculleus marisnigri JR1 | 11 | |
| Methanohalobium evestigatum Z-7303 | 14 | ArtB (9) |
| Methanohalophilus mahii DSM 5219 | 17 | ArtB (5) |
| “Candidatus Methanoregula boonei” 6A8 | 8 | |
| Methanosaeta thermophila PT | 3 | |
| Methanosarcina acetivorans C2A | 52 | |
| Methanosarcina barkeri Fusaro | 52 | |
| Methanosarcina mazei Go1 | 33 | ArtC (5) |
| “Candidatus Methanosphaerula palustris” E1-9c | 2 | |
| Methanospirillum hungatei JF-1 | 2 | |
| Natrialba magadii ATCC 43099 | 17 | |
| Natronomonas pharaonis DSM 2160 | 9 | |
| Uncultured methanogenic archaeon RC-I | 22 |
The set of PGF-CTERM proteins was examined for probable signal peptide regions at the N terminus. Among these 418 proteins, 26 appeared truncated in their predicted gene models based on pairwise alignments to their closest sequence matches in the cohort, and another 14 appeared improperly extended past a start site, similarly supported by pairwise homology, that would lead to signal peptide prediction. Of the remainder, only 25 had no signal peptide prediction by signalP. These “signal orphans,” with obvious C-terminal signal sequences but no clear N-terminal signal peptides, may represent a combination of faulty gene models for PGF-CTERM proteins lacking full-length homologs, false negatives in predictions made when signalP is applied to archaeal sequences, nonfunctional genes, or unrecognized alternative methods of protein translocation.
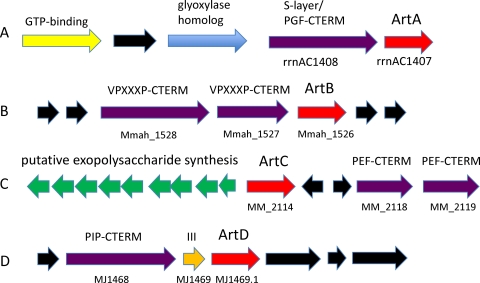
Proteins with the PGF-CTERM domain include ones whose annotations suggest enzymatic function, existence as a glycoprotein, or a structural role in S-layer formation. The list includes a cell surface glycoprotein from several species of Halobacterium. This abundant protein forms an S-layer. It is known to be heavily glycosylated near its C terminus, with both complex N-linked carbohydrates on Asn residues and O-linked disaccharides on Thr residues (23), and to be modified somewhere in the C-terminal region by diphytanylglyceryl phosphate (21); charge-to-mass ratios for several characteristic fragments of a diphytanylglycerol-derived prenyl group were identified by mass spectrometry. The actual site and nature of the attachment are unknown, but the extreme C-terminal region covered by our PGF-CTERM model was recognized as a potentially important feature (21). The diphytanylglyceryl-type modification has been described only in halophilic archaea. A gene neighborhood from Haloarcula marismortui ATCC 43049, shown in Fig. 2A, shows the major cell surface glycoprotein gene next to artA and to other genes conserved as neighbors of artA in the class Halobacteria.
Fig 2.
Gene neighborhoods of archaeosortases. Genes encoding members of the archaeosortase/exosortase family are shown in red. Genes for their putative substrates with the corresponding C-terminal TM domains are shown in purple. (A) Tandem organization of the major cell surface glycoprotein of H. marismortui with its putative processing enzyme, archaeosortase A. A similar gene neighborhood, with a GTP-binding protein (yellow), a conserved hypothetical gene (black), a glyoxylase homolog, and artA, occurs in both Halomicrobium mukohataei DSM 12286 (with the S-layer protein gene adjacent to artA) and Haloquadratum walsbyi DSM 16790. (B) Three tandem genes from M. mahii DSM 5219 code for two VPXXP-CTERM proteins and their putative cognate sorting enzyme, archaeosortase B. An artB gene and a VPXXP-CTERM gene are also in tandem in M. evestigatum Z-7303. (C) The artC gene of M. mazei Go1 sits between a nine-gene predicted exopolysaccharide locus and a pair of PEF-CTERM-containing putative targets. (D) The invariant gene neighborhood of PIP-CTERM/archaeosortase D systems consists of the lone putative target, a very small protein with an archaeal type III signal peptide, and artD. Several surrounding uncharacterized proteins appear also to be part of the conserved gene neighborhood.
Two additional proteins, the orthologous pair MA0829 from Methanosarcina acetivorans C2A and MM1976 from Methanosarcina mazei Go1, were shown to be surface-exposed S-layer proteins (15). Although unrelated to the halobacterial glycoproteins, except in the PGF-CTERM region, these proteins likewise exist as glycoproteins, with confirmed N-linked modifications and with anomalous migrations as if 30 to 60 kDa larger than predicted from the polypeptide sequence. In these species and several other methanogens, artA is not next to any PGF-CTERM gene. Instead, intriguingly, it is next to the gene inferred to encode CDP-2,3-di-O-geranylgeranyl-sn-glycerol:l-serine O-archaetidyltransferase (archaetidylserine synthase) (25), a membrane lipid biosynthesis enzyme that transfers a large prenyl group from its CDP carrier onto a serine. In M. hungatei JF-1, another archaeal species with an S-layer (13), two proteins contain the PGF-CTERM motif. The upper portion of Fig. 3 shows peptide coverage results for YP_503687 from a global proteomic assay. No peptides are found overlapping the extreme N-terminal region, an expected finding since the predicted signal peptide would be cleaved off in the process of transiting the cytoplasmic membrane. However, despite proteomic evidence so extensive that most residues are covered by 15 or more different peptides, the final 70 residues have no coverage at all. This region consists of a Thr-rich stretch (a probable O-linked glycosylation region), followed immediately by the putative PGF-CTERM sorting/processing signal region. This gap in the proteomic coverage of a highly expressed protein strongly supports the hypothesis that the C-terminal region either is cleaved and absent from the mature protein or is modified in such a way that no part of the region is detectable in the mass spectrometry results. We observe similar results for protein YP_305280 from M. barkeri strain Fusaro, as shown in the lower portion of Fig. 3; there is extensive proteomic coverage yet none in the Thr-rich or PGF-CTERM segment at the C terminus. All additional PGF-CTERM proteins from M. hungatei (1 additional protein), Halogeometricum borinquense (8 proteins with up to 97 unique peptides), and Halorhabdus utahensis (4 proteins with up to 53 unique peptides) that have a least some proteomic coverage likewise have no peptides overlapping their PGF-CTERM regions.
Fig 3.
Peptides observed by mass spectrometry analysis. For M. hungatei protein YP_503687 (top) and M. barkeri protein YP_305280 (bottom), peptides assigned by mass spectrometry are shown as mapped to the respective full-length precursor sequences. For YP_503687, the 809 observed peptides cover nearly every residue between the N-terminal signal peptide and the final 70 residues. M. barkeri protein YP_305280, shown in the lower picture, had 104 observed peptides, although none overlapping the final 89 residues. An alignment of the respective C-terminal regions lacking peptide coverage is shown between the two proteomic coverage graphics. The threonine-rich regions are shown in olive, with Thr (T) residues in black boldface. The PGF-CTERM regions are in red.
In M. barkeri, 52 proteins have PGF-CTERM signals, a minority with the variant form PSF or PAF replacing the more common PGF motif. Of these 52, 17 have proteomic evidence and 16 of these lack any peptide that overlaps any part of the PGF-CTERM region. YP_303678, in which PSF replaces PGF, is the lone exception. This 1,165-amino-acid-long protein has 78 possible N-linked glycosylation motifs, N-X-S/T (where X is not proline), although just 1 in the 8 regions (a total of 122 residues) spanned by proteomic evidence. However, proteomic analysis finds consecutive peptides near the C terminus, separated by a tryptic cleavage site, SFIFEDVKPYIQANSLAEVLR, followed by NPPKLPSFLL. The last five residues of the latter peptide, shown in bold, represent the start of a “PSF” motif variant of a putative PGF-CTERM domain. This peptide ends not at a tryptic site but where the expected continuation, GFAVTLLIGFAVLRKKK-COOH, runs through the rest of the predicted TM segment to the end of the protein. Such a location suggests intramembrane cleavage, analogous to cleavage by a rhomboid family serine protease (35), and therefore suggests that the cleavage site may be offset by several residues from the signature motif. Other possible explanations, however, include defective processing for this deviation from the canonical PGF motif, detection of an incompletely processed form, and false identification of a peptide (false discovery is benchmarked at <0.1% of the peptides reported) by the proteomic analysis pipeline.
For all 418 proteins identified as PGF-CTERM proteins in archaeal reference genomes, we examined the last 50 residues leading up to, but not including, the PGF-CTERM region. Sixty percent of these 50-residue regions contained low-complexity sequence, as identified by seg (37), where 15 to 20% is typical for the last 50 residues in archaea. Analysis of the amino acid composition in these regions showed Thr to be the most abundant at 14.0%, followed by Glu at 12.4% and Ser at 10.5%; Thr and Ser side chains are targets for O-linked glycosylation. The abundance of Thr is dramatically lower both in archaeal proteins overall and also in the subset that have a predicted N-terminal signal peptide. In this same 50-residue region, a remarkable 40.8% of the Asn residues occurred in the motif N-X-S or N-X-T (where X is any residue but Pro). This overrepresentation of putative N-linked glycosylation sites (1) suggests that many of these sites are functional and undergo N-linked glycosylation on the asparagine side chain. For 80% of the PGF-CTERM proteins, signalP identified a probable signal peptide within the first 70 amino acids. The remaining 20% may represent a combination of genes with start sites assigned in error too far upstream, or too far downstream, sequences truncated because of sequencing errors, nonfunctional genes, and proteins with mechanisms other than a signal peptide for insertion into the membrane.
We identified an additional protein subfamily whose members span exactly the same 29 genomes as archaeosortase A and PGF-CTERM. This is a subfamily of AglB (archaeal glycosylation B) (18), a homolog of the oligosaccharyltransferase complex STT3 subunit from the endoplasmic reticulum of eukaryotes, and is defined by model TIGR04154. Because of the apparent monophyly of the PGF-CTERM-encoding species, BLAST cutoffs can be chosen to select proteins from exactly that species list, for an average of 30 (but as few as 6) proteins per genome. Many from the remainder of the list are obvious housekeeping proteins such as ribosomal proteins and translation factors, DNA and RNA polymerase subunits, tRNA ligases, ATP synthase subunits, signal peptidases, and proteins of unknown function. This AglB subfamily, however, is interesting among these because it is the only family to have undergone a significant expansion, with 46 members occurring among the set of 29 genomes with archaeosortase A as measured by PPP (17) search results. Its HMM produces sharply lower scores for even the top-scoring homolog outside this set of genomes. Many proteins with PGF-CTERM regions, such as S-layer glycoproteins, are known or thought to be targets of N-glycosylation involving AglB (18).
The archaeosortase B/VPXXXP-CTERM system.
Within the archaea, we detected additional sets of exosortase remote homologs that are mutually closely related but distinct from archaeosortase A (Table 1).
For several of these subfamilies, the neighborhoods were examined for member proteins with conserved C-terminal sequences architecturally similar to LPXTG and PEP-CTERM (method 2). Methanohalophilus mahii DSM 5219 and Methanohalobium evestigatum Z-7303 each have a member of the exosortase-related subfamily we now refer to as archaeosortase B (ArtB, TIGR0414). In the genomes of both of these species, archaeosortase B is one of two members of the larger exosortase/archaeosortase domain family (TIGR04178), since the more common archaeosortase A/PGF-CTERM system is also present. In each genome, a protein encoded next to artB shows a C-terminal sequence region structurally similar to PEP-CTERM and PGF-CTERM, that is, ending with a TM region and a cluster of basic residues, although not similar to known sorting signals in the signature motif region. A search for proteins with similar C-terminal sequence regions shows five C-terminally homologous proteins in M. mahii and nine in M. evestigatum with the signature motif VPXXXP. The homology domain was defined as VPXXXP-CTERM (TIGR04143). No additional examples of VPXXXP-CTERM putative protein-sorting domains can be found in any other complete microbial genome. A sequence logo representation based on the seed alignment of model TIGR04143 is shown in Fig. 1B. The genomic region from M. mahii is shown in Fig. 2B.
The archaeosortase C/PEF-CTERM system.
A search for proteins with C-terminal regions weakly similar to PEP-CTERM (method 1, see Materials and Methods) identified a new variant putative protein-sorting signal. The domain occurs in 5 proteins in M. mazei Go1, 10 in Methanococcoides burtonii DSM 6242, 12 in Aciduliprofundum boonei T469, and 3 in Ferroglobus placidus DSM 10642. Its signature motif is either Val or Ile, followed by Pro, Glu, and Phe (in two cases Leu), so the domain is designated PEF-CTERM. It is recognized by model TIGR03024. An exosortase-related subfamily, archaeosortase C (ArtC), co-occurs within all genomes with PEF-CTERM and is described by model TIGR03762. It finds the only exosortase/archaeosortase family member in A. boonei and the only member other than archaeosortase A in M. mazei, M. burtonii, and F. placidus. In F. placidus, the putative sorting enzyme and one of its apparent targets are encoded just seven genes apart. The sequence logo for the PEF-CTERM system is shown in Fig. 1C. An illustrative genomic region from M. mazei is shown in Fig. 2C, where artC is situated between an EPS biosynthesis region and two PEF-CTERM genes.
The archaeosortase D/PIP-CTERM system.
An exosortase/archaeosortase domain subfamily restricted to five species of methanococci was identified and is designated archaeosortase D (ArtD), modeled by TIGRFAMs entry TIGR04175. Its apparent corresponding recognition domain, PIP-CTERM (TIGR04173), occurs in a single protein per genome, encoded just one gene away as part of a conserved cassette, as is typical for dedicated systems (one enzyme, one target). The gene separating the two encodes a small protein with an archaeal class III signal peptide that suggests it is the target of a prepilin peptidase (32). Figure 2D shows the typical genomic region, including flanking conserved hypothetical proteins that are not analyzed further here.
Exosortase/PEP-CTERM system subtypes: XrtA and XrtB.
In our previous work reporting the discovery of the PEP-CTERM domain and exosortase enzyme, we noted that exosortase enzymes frequently occur in EPS (exopolysaccharide or extracellular polymeric substance) biosynthesis regions (17). Subsequent to publication, we noted that various glycosyltransferases, polysaccharide deacetylases, polysaccharide chain length determinants, ATPases, and proteins with various other functions not only coclustered with exosortase genes but belonged to protein subfamilies whose species distributions closely matched those of particular exosortase subfamilies (J. D. Selengut, poster, 9th Annu. Conf. Comput. Genomics, 28 to 31 October 2006, Baltimore, MD). Exosortase A (previously designated exosortase 1 in the TIGRFAMs library) is modeled by TIGR03109. This form regularly co-occurs with histidine kinase PrsK (TIGR02916), the DNA-binding response regulator PrsR (TIGR02915), a chain length determinant family (TIGR03007), a polysaccharide deacetylase family (TIGR03006), and a glycosyltransferase family (TIGR03013), among others. Exosortase B (XrtB), previously designated exosortase 2, is described by model TIGR03113. XrtB has several different correlated protein families, including EpsL (TIGR03014) and a different chain length determinant family, TIGR03017. Species with exosortase A or exosortase B all have cohorts of proteins with canonical PEP-CTERM domains recognized by model TIGR02595, although these may represent different subgroups within the canonical PEP-CTERM domain family. Five species (Nitrosospira multiformis, Thiobacillus denitrificans, Hahella chejuensis, Accumulibacter phosphatis, and Herbaspirillum seropedicae) have both XrtA and XrtB.
The cyanoexosortase system.
In nearly every non-Prochlorococcus cyanobacterial reference genome, a PEP-CTERM system is present with either of two exosortase subfamilies present, designated cyanoexosortase A (CrtA) and cyanoexosortase B (CrtB). Pairwise sequence identities are high within each family but barely 20% between members of the two different families. An uncharacterized protein usually encoded next to a cyanoexosortase gene and occasionally fused to it has been observed and is modeled by TIGR04153. A variant form of the PEP-CTERM domain in which the extreme C terminus is extended, lysine containing, and less purely basic is observed in these organisms and is modeled by TIGR04155. Several residues mid-span in the TM region are strongly conserved not only within paralogous domain families from individual cyanobacteria but between species as well. This observation further supports the notion that central portions of the TM region participate in specific protein-protein interaction.
There is considerable diversity among the remaining exosortases that recognize canonical PEP-CTERM sequences (those that can be identified by model TIGR02595) but do not belong to the XrtA, XrtB, CrtA, or CrtB subfamily. Additional cataloguing and protein family definition could be performed for recognizable subclasses of canonical PEP-CTERM domains and their corresponding exosortases, but that work is outside the scope of this paper. However, various exosortase/archaeosortase family proteins in bacteria recognize sequences that differ from the canonical PEP-CTERM sequence.
The exosortase C/VPDSG-CTERM system.
In Verrucomicrobiae bacterium DG1235, only one exosortase homolog occurs and no canonical PEP-CTERM targets are observed. The same form of exosortase occurs also in Verrucomicrobiae bacterium Ellin514. Members of this subfamily are defined as exosortase C or XrtC (model TIGR04151). In Verrucomicrobiae bacterium DG1235, a set of 20 genes share a PEP-CTERM-like C-terminal region in which the consensus for the signature motif is VPDSG; its sequence logo is shown in Fig. 1D. A VPDSG-CTERM gene (model TIGR03778) occurs just one gene away from its cognate exosortase gene in bacterium DG1235 and three genes away in bacterium Ellin514. The latter bacterium has a second exosortase and has nine PEP-CTERM proteins in addition to its three VPDSG-CTERM proteins.
The exosortase D/VPLPA-CTERM system.
A search for variant PEP-CTERM-like motifs (method 1) identified a subfamily with the consensus sequence VPLPA (model TIGR03370). This form occurs in a number of alpha- and deltaproteobacteria, including Roseobacter (three species), Geobacter metallireducens, Oceanospirillum sp. strain MED92, Desulfovibrio salexigens DSM 2638, etc. We identified and defined an exosortase variant family, exosortase D (XrtD), modeled by TIGR04152, that always occurs in these same genomes, often as one of two or three paralogous exosortases. The logo is shown in Fig. 1E.
The exosortase E/VPEID-CTERM system.
An exosortase variant subfamily, exosortase E (XrtE), that appears so far only in some alphaproteobacteria, is described by model TIGR04162. It is unusual in being fused to a C-terminal region homologous to eukaryotic CAAX box prenyl proteases. By method 2, we identified its presumed corresponding target region, the VPEID-CTERM domain, described by model TIGR04161 (Fig. 1F). XrtE/VPEID-CTERM appears to be a dedicated system, processing only one target protein per genome. The target protein usually is encoded next to the xrtE gene (e.g., Phaeobacter gallaeciensis BS107 and Ruegeria sp. strain R11). Members are among the smallest of any proposed exosortase or archeaosortase targets, averaging fewer than 50 residues from the end of the signal peptide to the start of the VPEID motif. Most are rich in Gly-Asn dipeptide repeats, rather than Thr and Ser residues. Under the hypothesis that PEP-CTERM domains signal C-terminal protein processing (cleavage and/or lipid attachment) in order to accommodate glycosylation, this asparagine richness suggests N-linked glycosylation. Additional proteins of a conserved gene neighborhood show homology to RND family efflux transporter subunits.
The exosortase G system.
The Firmicutes, like the Archaea, lack an outer membrane and lack members of the bacterial exosortase family TIGR02602. However, a distinct clade of remote homologs of exosortase, restricted to the Firmicutes, is designated XrtG, the exosortase family protein of Gram-positive bacteria. It is described by model TIGR03110. The 14 reference genomes with this system include members of the genera Lactobacillus (4), Clostridium (3), Oenococcus (2), Coprococcus, Eubacterium, Leuconostoc, Pediococcus, and Syntrophomonas. The corresponding phylogenetic profile is somewhat information rich because the distribution of the marker, found in about 1% of the reference genomes, is sufficiently sporadic within the Firmicutes. PPP finds a group of four regularly co-occurring protein families, usually encoded as a cassette. One member is XrtG. Another is a putative glycosyltransferase (TIGR03111), further demonstrating correlations between the exosortase/archaeosortase family and posttranslational modifications such as glycosylation. The third is a putative 6-pyruvoyl-tetrahydropterin synthase (EC 4.2.3.12) by homology (TIGR03112). This third family represents a possible protein function not normally expected to be associated with protein modification. The fourth and final family occurs in 10 of the 14 genomes that contain the other three markers, although in one case, the gene is not reported because of an internal stop codon that may represent either a nonsense mutation or a sequencing error. These proteins show full-length homology but various sequence lengths because some members have long, low-complexity inserts rich in either Ser or Thr, residues associated with O-linked carbohydrate attachment. The proteins all end, however, with a TM segment and a cluster of basic residues, just like PEP-CTERM and LPXTG domain proteins. Preceding the TM region is a motif with consensus sequence GSTGPY. This C-terminal region may represent a target for protein sorting and posttranslational modification, although it is quite different from any other proposed exosortase/archaeosortase recognition sequence. Family TIGR04145 models an extended C-terminal region, including the GSTGPY motif region, for this Ser/Thr-rich protein. The corresponding sequence logo is shown in Fig. 1G for completeness. In the Firmicutes, where LPXTG/sortase-like systems are expected to dominate protein-sorting and attachment systems, the unusual finding of an exosortase family protein in a cassette with a glycosyltransferase, an unexpected synthase, and a putative surface glycoprotein suggests a great plasticity of biological roles in the exosortase/archaeosortase superfamily.
The exosortase F system in the class Flavobacteria.
TIGRFAMs model TIGR04128 describes XrtF, an exosortase family protein found in 15 species of the class Flavobacteria: Gramella forsetii KT0803, Flavobacterium psychrophilum JIP02/86, Dokdonia donghaensis MED134, etc. Members of this family mark a gene neighborhood with at least two other regularly occurring protein families, including a highly hydrophobic protein family TIGR04127. However, no members of these neighborhoods have C-terminal sequences with any PEP-CTERM-like architecture. PPP suggests a relationship between this system and several different families of large, repetitive flavobacterial proteins. Several of these potentially linked families share a C-terminal region even if other regions are not detectably similar by BLAST. This region may represent the recognition sequence for XrtF, although it shows little resemblance to the PEP-CTERM/LPXTG architecture, and its correspondence to the XrtF family is not exact. Alternatively, XrtF may have another target that is neither similar enough to PEP-CTERM to detect by method 1 nor coclustered with the genes of the XrtF system to allow detection by method 2.
PEP-CTERM proteins from Cyanothece are not detected by proteomics.
Proteomic data for four Cyanothece strains (PCC 7822, PCC 8801, PCC 8802, and ATCC 51142) with 2 or 3 exosortases and 35 to 87 PEP-CTERM proteins each failed to identify any peptides for either set of proteins. PEP-CTERM proteins are likely to be glycosylated at multiple sites, which might mask their presence in standard proteomic analyses, but the lack of data for the exosortases suggests very low abundance or absence from the cell under these growth conditions. This situation contrasts with the PGF-CTERM-bearing surface glycoproteins in the archaea, where dense proteomic coverage over much of the protein length suggests that these are among the most abundant proteins in archaeal cells.
Archaeosortase/exosortase-independent putative sorting and/or anchoring signals.
A carboxyl-terminal domain structured like PEP-CTERM is observed occasionally in species that lack any protein with detectable homology to either sortases or exosortases. Several members of the order Myxococcales have no sortase or exosortase homolog yet share a C-terminal homology region whose distinctive motif is a GC pair, often in a longer GCxC motif (model TIGR03901). The motif is separated by a short spacer region from the TM helix, an arrangement more similar to the LPXTG domain structure than to the PEP-CTERM domain. From 2 (Anaeromyxobacter dehalogenans 2CP-C) to over 60 (Plesiocystis pacifica SIR-1) member proteins are found. This region occurs in all eight members of the order Myxococcales included in our set of 1,466 reference genomes but in no other genome. The combination of rarity (only eight species) and monophyletic distribution (there is a last common ancestor of these eight species and no others) is unfavorable for comparative genomics. Therefore, we have not been able to identify a sorting enzyme unequivocally through comparative genomic methods such as PPP. Many protein families, or clades within larger protein families, will have exactly that distribution. We therefore do not make any prediction as to what cellular machinery may process these proteins.
In Myxococcus xanthus, 34 proteins have this TIGR03901 Cys-containing C-terminal domain. A recent proteomic study targeting M. xanthus (20) identified peptides from <3% of the proteins in vesicles prepared from outer membrane, but this set included 7 (∼20%) of the 34 proteins with TIGR03901-matching tail regions. From 2 to 25 peptides were identified per protein. None of the peptides found by proteomics, however, overlap tail region segments matched by TIGR03901. The absence of proven peptides from the tail region could represent chance, absence of those residues from the mature protein, or posttranslational modifications that interfere with the proteomic work, either chemically or computationally.
In several archaea, no member of the sortase or exosortase/archaeosortase family is found, yet several proteins share C-terminal regions with TM regions followed by clusters of basic residues. Examples include the proven surface glycoprotein SSO1273 of Sulfolobus solfataricus (16) and its bidirectional best-hit homolog in Aeropyrum pernix. These and additional proteins from these two species with sequence similarity in the C-terminal TM region tend to have regions extremely rich in threonine adjacent to the TM region, signifying a probable region of O-linked glycosylation. In these features, they resemble a number of PGF-CTERM proteins. However, we cannot identify any obvious motif in the manner of the LPXTG or PEP motif of sortase and exosortase targets or any sortase or exosortase homolog in the respective genomes. This joint lack of any putative recognition motif and any archaeosortase/exosortase homolog suggests different processing from what occurs in species with archaeosortase A, perhaps without cleavage of the TM region and without a C-terminal lipid modification.
DISCUSSION
Broad taxonomic distribution of archaeosortase/exosortase-like systems.
Sequence homologies among paralogous domains architecturally similar to PEP-CTERM yet different in the key signature motifs make it possible to find new examples of such protein-sorting signals, build HMMs to define them, discriminate among them, and use comparative genomic techniques to find or verify corresponding exosortase processing enzymes. In this paper, we have found a number of distinctive C-terminal putative protein-processing domains and identified archaeosortase/exosortase family multi-TM proteins likely to partner with them. The systems described share remote homologies but occur in widely varied genomics contexts, with variable sets of additional partner proteins. Two or more such systems, likely performing specific, nonredundant functions, can be encoded within the same genome. More than 16% of prokaryotic genomes have at least one such system.
A common link of exosortases and archaeosortases to glycosylation of membrane-transiting proteins.
In our earlier paper reporting the discovery of exosortase (Xrt) and its presumed target sequence, PEP-CTERM, we noted the strong association between exosortases and loci involved in EPS production, as well as a phylogenetic correlation with biofilm formation (17). Xrt always occurred in species with an outer membrane; eventual transit across the outer membrane seemed a likely processing step for PEP-CTERM proteins. The Ser/Thr-rich composition of most, but not all, PEP-CTERM proteins strongly suggested glycosylation, as did the glycosyltransferases frequently neighboring the exosortase gene. The idea of multiple protein-carbohydrate linkages supported the notion that poorly conserved, nonglobular PEP-CTERM proteins played a structural role in the formation of biofilm matrix. Regulatory motifs suggesting tight regulation preceded many bacterial PEP-CTERM genes, implying their expression might be toggled on and off to produce different consistencies of biofilm matrix. In the present paper, we describe homologous though only very distantly related archaeal systems whose targets include highly expressed glycoproteins involved in S-layer formation. The common thread that links the targets of exosortases and archaeosortases, therefore, is that prokaryotic glycoproteins seem to be the common targets.
C-terminal cleavage hypothesis for exosortase supported by C-terminal lipid modification of PGF-CTERM proteins.
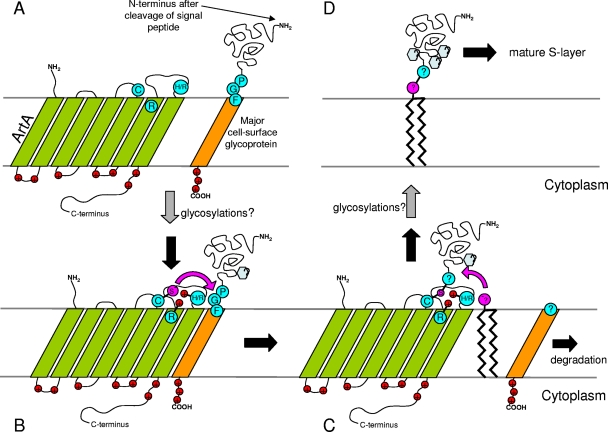
There has been no reported experimental characterization of PEP-CTERM proteins in bacteria, perhaps because the system is still unknown in any animal pathogen. This work, however, strongly implicates the PGF-CTERM domain as the recognition sequence for archaeosortase A, which is important because some study of posttranslational modification of a PGF-CTERM protein had already been achieved. The surprising earlier finding was a large prenyl group attached in the C-terminal region. It was not clear why a protein with a C-terminal TM helix anchor would need a C-terminal lipid anchor. Our study, however, pairs PGF-CTERM proteins with a multi-TM putative protease and therefore suggests that the TM helix may be removed prior to (or in concert with) prenylation. We previously argued in two ways that exosortase (and by extension archaeosortase) is a protease or transpeptidase. First was the clear parallel with the LPXTG/sortase system, in which proteins are cleaved in order to be attached covalently to the cell wall. Second was the intriguing fact that the invariant residues Cys, Arg, and His shared by nearly all exosortase homologs match the known catalytic triad of sortases. We have now extended this observation further, identifying additional exosortase/archaeosortase family members out to the limits of homology detection. The Cys, Arg, and His residues remain invariant in the new families described in this paper, although some additional homologs are seen that may represent variant or inactivated forms. For protein-processing systems involving proteins of the archaeosortase/exosortase superfamily, attachment to a large lipid group could prove to be a common feature that accompanies the removal of PGF-CTERM-like C-terminal TM domains (Fig. 4).
Fig 4.
Model of archaeosortase A processing of the halobacterial major cell surface glycoprotein. Panel A shows the archaeosortase (green) as a multiple-membrane-spanning protein with the presumed active-site Cys oriented toward the extracellular face. The putative target protein is shown with its signal peptide already removed and with the helix of the C-terminal PGF-CTERM domain also spanning the membrane (orange). Panel B shows physical association of the PGF-CTERM helix with the bundle of archaeosortase A TM helices. The active-site residues are positioned to cleave the glycoprotein. At this point, several posttranslational modifications, such as glycosylations, may already have happened. The exact target site is unknown. Panel C shows the target protein, after cleavage, transiently attached to the conserved Cys residue of the archaeosortase/exosortase family, following the mechanism of sortase and the model of exosortase. Release of the glycoprotein is shown by transfer onto diphytanylglyceryl phosphate (transpeptidation), although another possibility is transfer to water (hydrolysis), with the lipid moiety being attached elsewhere. Meanwhile, the removed C-terminal peptide is headed for degradation. Panel D shows the glycoprotein after its interaction with archaeosortase, able to undergo (further) glycosylation and entry into the S-layer in mature form.
Cleavage as an ordered step in a maturation sequence for surface glycoproteins.
The conceptual model, to this point, suggests that many archaeosortase/exosortase targets are heavily glycosylated, at least in some cases with multiple types of carbohydrate, that the C-terminal TM helix anchor is removed, and that in some systems a lipid anchor may be added. This represents a fairly complex sequence of events. Pulse-chase studies of Haloferax suggest that glycosylation and lipid modification both follow translocation across the plasma membrane (22). In several well-investigated archaeal systems, protein glycosylation is understood to occur on the extracytoplasmic face, requiring that both enzymes and donor groups become positioned appropriately (12). Removal of the C-terminal helix and attachment instead to a lipid anchor would likely change the relative positioning of the target relative to the carbohydrate attachment enzymes and other posttranslational modification enzymes. The change might enable glycosylation, permit a shift in processing from one type of glycosylation to another, or release the nascent glycoprotein from its interaction with glycosylation machinery, marking the end of a defined stage in posttranslational processing. Further processing steps may occur after cleavage, such as transit across the outer membrane in Gram-negative bacteria.
An embarrassment of exosortases and archaeosortases.
One of the puzzles in earlier work on the PEP-CTERM system was the occurrence of several exosortase paralogs in some species. At the time, there were not sufficient numbers of prokaryotic reference genomes to allow clear recognition of new classes of sorting signals and to associate these with particular subtypes and variants of exosortase. The new findings reported here mirror the “embarrassment of sortases” and corresponding diversity of targets seen in bacteria (26). They may point to multiple protein modification chemistries that can coexist in a single cell. Archaeal attachment to mevalonate-derived lipid ethers (22) may be only the first of many types of modification catalyzed. Further characterizations of exosortases, archaeosortases, and related systems will afford a better understanding of surface structures and biofilm formation in prokaryotes and perhaps offer opportunities for rational reengineering of microbial cell surfaces to serve a variety of industrial applications.
Implications of positive (archaeal) and negative (bacterial) proteomic results.
A number of factors could contribute to the difficulties in seeing bacterial PEP-CTERM proteins in proteomic analyses. There may be no appreciable expression of PEP-CTERM proteins under the growth conditions used for the mass spectrometry analysis. The combination of N-terminal and C-terminal TM domain region removal with extensive glycosylation may prevent sufficiently long unmodified peptides from being available to the analysis. The EPS that forms the matrix of a biofilm often contains proteinaceous material (14); PEP-CTERM proteins in some species may become such material, unavailable for analysis because of its recalcitrance to protein purification. Several archaeal PGF-CTERM proteins, in contrast, show extensive sets of proven peptides in proteomic studies. The lessons learned from them may bear on bacterial systems. The proteomic results clearly suggest that the PGF-CTERM TM anchor is removed, fitting our previous speculation that exosortase may be a transpeptidase, together with the published characterization of a PGF-CTERM protein that describes a novel C-terminal acylation. In bacteria as well, processing by exosortase/archaeosortase family proteins may feature cleavage of the recognition sequence and perhaps also the attachment of a large acyl group. PEP-CTERM proteins seem likely to transit the outer membrane, as well as the plasma membrane, so replacement of a C-terminal TM helix with a lipid anchor would have strong implications for the mechanisms of further processing during export.
The possibility of additional nonhomologous C-terminal domain processing enzymes.
Sortases are soluble enzymes in their catalytic regions, while archaeo- and exosortases are deeply embedded integral membrane proteins. It is therefore reasonable to believe, since they share no detectable similarity at the sequence level, that their similar functions have arisen through convergent evolution. We have observed, in genomes lacking detectable sortase or archaeo- and exosortase homologs, C-terminal domains having the tripartite structure described in this work and furthermore that these domains are found in multiple proteins in those genomes. This seems to suggest that additional nonhomologous processing enzymes remain to be discovered.
While this work was under review, Craig et al. (10) reported that a previously uncharacterized family of acyltransferases regularly found in bacteria in gene neighborhoods shared with exosortases attach long-chain acyl groups to free amino acids. Because acyltransferase activity was characterized in heterologous systems possessing neither exosortase nor its putative PEP-CTERM targets, this work leaves open the possibility that these acyltransferases are involved primarily in PEP-CTERM protein modification, rather than in the production of a bioactive small molecule, as the article suggests, or in the modification of extracellular polysaccharides.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 HGO0488. S.H.P. was funded by an NSF grant (EF-0949047). Data from the Pacific Northwest National Laboratory were obtained in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy/Biological and Environmental Research national scientific user facility. The Pacific Northwest National Laboratory is operated for the DOE by Battelle under contract DE-AC05-76RLO 1830.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print 28 October 2011
REFERENCES
- 1. Abu-Qarn M, Eichler J. 2007. An analysis of amino acid sequences surrounding archaeal glycoprotein sequons. Archaea 2:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bapteste E, Brochier C, Boucher Y. 2005. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea 1:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnett TC, Patel AR, Scott JR. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 186:5865–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basu MK, Selengut JD, Haft DH. 2011. ProPhylo: partial phylogenetic profiling to guide protein family construction and assignment of biological process. BMC Bioinformatics 12:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 7. Bierne H, et al. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Budzik JM, Oh SY, Schneewind O. 2009. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J. Biol. Chem. 284:12989–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Callister SJ, et al. 2008. Comparative bacterial proteomics: analysis of the core genome concept. PLoS One 3:e1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Craig JW, Cherry MA, Brady SF. 2011. Long-chain N-acyl amino acid synthases are linked to the putative PEP-CTERM/exosortase protein-sorting system in gram-negative bacteria. J. Bacteriol. 193:5707–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eichler J, Adams MW. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69:393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Firtel M, Southam G, Harauz G, Beveridge TJ. 1993. Characterization of the cell wall of the sheathed methanogen Methanospirillum hungatei GP1 as an S layer. J. Bacteriol. 175:7550–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 15. Francoleon DR, et al. 2009. S-layer, surface-accessible, and concanavalin A binding proteins of Methanosarcina acetivorans and Methanosarcina mazei. J. Proteome Res. 8:1972–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gogliettino M, et al. 2010. A highly selective oligopeptide binding protein from the archaeon Sulfolobus solfataricus. J. Bacteriol. 192:3123–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haft DH, Paulsen IT, Ward N, Selengut JD. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jarrell KF, Jones GM, Nair DB. 2010. Biosynthesis and role of N-linked glycosylation in cell surface structures of archaea with a focus on flagella and S layers. Int. J. Microbiol. 2010:470138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kahnt J, et al. 2010. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J. Proteome Res. 9:5197–5208 [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi A, Sagami H, Ogura K. 1999. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J. Biol. Chem. 274:18011–18016 [DOI] [PubMed] [Google Scholar]
- 22. Konrad Z, Eichler J. 2002. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochem. J. 366:959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lechner J, Sumper M. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262:9724–9729 [PubMed] [Google Scholar]
- 24. Luo H, et al. 2009. Gene order phylogeny and the evolution of methanogens. PLoS One 4:e6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morii H, Koga Y. 2003. CDP-2,3-di-O-geranylgeranyl-sn-glycerol:l-serine O-archaetidyltransferase (archaetidylserine synthase) in the methanogenic archaeon Methanothermobacter thermautotrophicus. J. Bacteriol. 185:1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pallen MJ, Lam AC, Antonio M, Dunbar K. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97–102 [DOI] [PubMed] [Google Scholar]
- 27. Payne SH, Huang ST, Pieper R. 2010. A proteogenomic update to Yersinia: enhancing genome annotation. BMC Genomics 11:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schäffer AA, et al. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 12:4803–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Selengut JD, et al. 2007. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35:D260–D264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sumper M, Berg E, Mengele R, Strobel I. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szabó Z, et al. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. U. S. A. 96:12424–12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urban S. 2006. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 20:3054–3068 [DOI] [PubMed] [Google Scholar]
- 36. van Passel MW, et al. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wootton JC, Federhen S. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266:554–571 [DOI] [PubMed] [Google Scholar]
- 38. Yoshida T, et al. 2003. Genes involved in the synthesis of the exopolysaccharide methanolan by the obligate methylotroph Methylobacillus sp strain 12S. Microbiology 149:431–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.