Abstract
Mycoplasma mobile glides on solid surfaces by the repeated binding of leg structures to sialylated oligosaccharide fixed on a solid surface. To obtain information about the propulsion caused by the leg, we made elongated and stiff cells using a detergent. Within 30 min after the cells were treated with 0.1% Tween 60, the cells were elongated from 0.8 μm to 2.2 μm in length while maintaining their gliding activity. Fluorescence and electron microscopy showed that a part of the cytoskeletal structure was elongated, while the localization of proteins involved in the gliding was not modified significantly. The elongated cells glided with repeated pivoting around the cellular position of gliding machinery by 10 degrees of amplitude at a frequency of 2 to 3 times per second, suggesting that the propulsion in a line perpendicular to the cell axis can occur with different timings. The pivoting speed decreased as the cell length increased, probably from the load generated by the friction. The torque required to achieve the actual pivoting increased with the cell length without saturation, reaching 54.7 pN nm at 4.3 μm in cell length.
INTRODUCTION
Mycoplasmas are commensal and occasionally parasitic bacteria that lack peptidoglycan layers and have small genomes (29). Mycoplasma mobile, a fish pathogen, forms a membrane protrusion at one pole and exhibits gliding motility in the direction of the protrusion (16–18). The average speed is 2.0 to 4.5 μm/s, or three to seven times the length of the cell per second, exerting a force of up to 27 pN (7, 20, 23). This motility, combined with the ability to adhere to the host cell surface, likely plays a role in pathogenesis, as has been suggested for another species, Mycoplasma pneumoniae (11, 12, 17, 18). This motility is shown not to be related to other known mechanisms of bacterial movement, nor does it involve motor proteins known to be involved in eukaryotic cell motility (9).
Thus far, we have identified huge proteins, Gli123, Gli349, and Gli521, with masses of 123, 349, and 521 kDa, respectively, involved in this gliding mechanism that are localized at the so-called cell neck, the base of the membrane protrusion (13, 28, 32, 34, 36); we have also visualized by microscopy the putative machinery and the component proteins (1, 15, 19, 27) and identified both the direct energy source used and the direct binding target (8, 24, 35). The force generated by the gliding machinery may be supported from inside the cell by a cytoskeletal “jellyfish” structure (25). On the basis of these results, we proposed a working model, called the centipede or power-stroke model, where cells are propelled by “legs” composed of Gli349 that through repeated cycles catch and release sialic acids fixed on the glass surface (6, 17, 18, 37). However, the actual movements of the legs have not been detected.
Previously, we showed that the gliding force is generated at the head-like structure by analyzing the behaviors of cells elongated by prolonged cultivation (22). This may allow us to obtain additional information about force generation if we analyze the behavior of cells that have another morphology. In studies of liposomes, detergents have been shown to change the liposome morphology by the insertion of lipids and proteins into the lipid bilayer (3, 30). Such a methodology may be applicable to the induction of changes in the cell morphology of mycoplasmas whose membranes are not supported by a peptidoglycan layer. In the present study, we used Tween 60 detergent to make elongated and stiff M. mobile cells, analyzed the pivoting of the elongated cells, and discussed the leg movements involved in gliding.
MATERIALS AND METHODS
Strains and culture conditions.
The wild type and mutant M. mobile strain 163K (ATCC 43663) were grown in Aluotto medium at 25°C (2, 37). Nonbinding and nongliding mutants m12, m13, and m9, which were missing by a nonsense mutation intact proteins Gli123, Gli349, and Gli521, respectively, were reported previously (23, 32, 34, 36). Mutants with the mutations gli521 (P476R), gli521 (S859R), and gli349 (S1362W) modified for gliding were reported previously (37).
Optical microscopy and movement analyses.
Mycoplasma cells were collected by centrifugation at 12,000 × g for 4 min at room temperature (RT) and suspended in Aluotto medium. The suspension was poured into a tunnel assembled by taping a coverslip cleaned with saturated ethanolic KOH onto a glass slide (26, 37). After incubation at RT for 10 min, the floating cells were removed by replacing the medium. Cell elongation was induced by adding detergents in phosphate-buffered saline with glucose (PBS/G) solution consisting of 75 mM sodium phosphate (pH 7.3), 68 mM NaCl, and 10 mM glucose. To stop cell gliding in control experiments, 0.1% Nile red in PBS/G was added to gliding cells in the tunnel slide. After incubation for 1 min at RT, the cells were irradiated for 1 s through a CY3-4040C filter (Semrock, Rochester, NY) using a USH-102D mercury lamp (USHIO, Tokyo, Japan) set in a lamp house (Olympus, Tokyo, Japan). Cell behavior was observed under a BX50 phase-contrast microscope (Olympus, Tokyo, Japan) and recorded as previously described (26). All video data were analyzed by Image J (version 1.41o) software (http://rsb.info.nih.gov/ij/) as previously described (26). To trace the detailed movements of individual cells, cell axes were detected by the fitting of ellipses. Immunofluorescence microscopy was performed as previously described (25, 34).
EM.
The cells bound to grids were stained by 2% (wt/vol) ammonium molybdate and observed by transmission electron microscopy (EM; H-7000; Hitachi, Tokyo, Japan) as previously described (25, 34). Cell elongation was induced with the treatment of 0.1% Tween 60 in PBS/G on the grid. Mycoplasma cells were chemically fixed by 3% paraformaldehyde and 0.1% glutaraldehyde in PBS for 10 min at RT and were washed three times by PBS. The EM images were captured by a FastScan-F214 (T) charge-coupled-device (CCD) camera (TVIPS, Gauting, Germany). For immunogold EM, the cells bound to EM grids were chemically fixed and labeled with 5-nm gold particles using the previously isolated antibodies and then stained with 2% ammonium molybdate as previously described (13, 25, 32, 34).
Calculation of torque produced by cell pivoting.
The torque (T) for cell pivoting was estimated by multiplying its rotational frictional drag coefficient (f) by the angular velocity (ω), as shown in the following equation and as reported previously (4, 5):
where L is the distance from the gliding machinery to the tail end measured by phase-contrast microscopy, S is the cell width, and η is the viscosity of the buffer, 8.9 × 10−4 Ns/m2 (Pa) (14). ω was estimated as the standard deviation in the distribution of pivoting angle speed (radian/s).
RESULTS
Morphological changes induced by detergents.
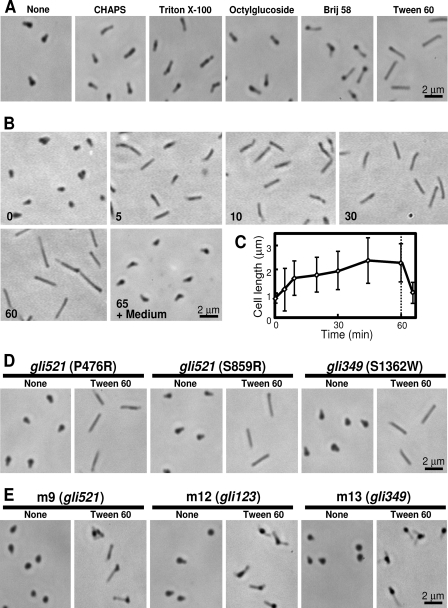
Previously, we used 0.01% Triton X-100 to damage the M. mobile cell membrane (25, 35). In the present study, we treated M. mobile cells with 0.003% Triton X-100 to change the cell shape (Fig. 1A). The cells were elongated 1.4-fold along the axis through the Triton X-100 treatment for 30 min. To determine the most effective detergent, we tried five other detergents, including 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), Tween 80, octylglucoside, Brij 58, and Tween 60. We decided the critical concentration to lyse the cells for each detergent and examined the effects of the detergents on cells at concentrations three times lower than the critical ones. The cells were obviously not damaged under those concentrations, and all detergents elongated the cells after incubation for 30 min. As Tween 60 was the most effective (Fig. 1A), it was used in the subsequent experiments. The morphological change caused by the addition of 0.1% Tween 60 was traced by phase-contrast microscopy (Fig. 1B and C). The cells became elongated with incubation time and reached 2.2 μm, 2.5 times longer than their original length (0.8 μm), at 45 min, with gliding; meanwhile, the cell width changed from 0.45 μm to 0.25 μm. Cell size (both length and width) and gliding speed returned to normal within 5 min after the detergent-containing buffer was replaced with fresh growth medium. To examine whether or not gliding motility is required for cell elongation, the same treatments were applied to mutants with various gliding properties (23, 37). Elongation occurred in mutants of modified gliding and nongliding, nonbinding cells (Fig. 1D and E), suggesting that gliding motility is not essential for cell elongation.
Fig 1.
Changes in cell morphology induced by detergents. (A) Wild-type cell images after treatment with various detergents for 30 min. CHAPS, Triton X-100, octylglucoside, Brij 58, and Tween 60 were applied at concentrations of 0.1%, 0.003%, 0.1%, 0.001%, and 0.1%, respectively. (B) Phase-contrast images of M. mobile cells after the addition of 0.1% Tween 60. The time (in minutes) after the addition of Tween 60 is indicated in the lower left of each panel. At 60 min, the buffer containing Tween 60 was replaced with fresh medium. (C) Change in cell length after addition of 0.1% Tween 60. Long axes of 50 cells at each time point were averaged. The buffer containing Tween 60 was replaced with fresh medium at the time marked by a dashed line. (D) Cell images of mutants with characteristic gliding after treatment with 0.1% Tween 60 for 30 min. (E) Cell images of nonbinding, nongliding mutants m9 (lacking gli521), m12 (lacking gli123), and m13 (lacking gli349) after treatment with 0.1% Tween 60 for 30 min.
Protein localization after morphological changes.
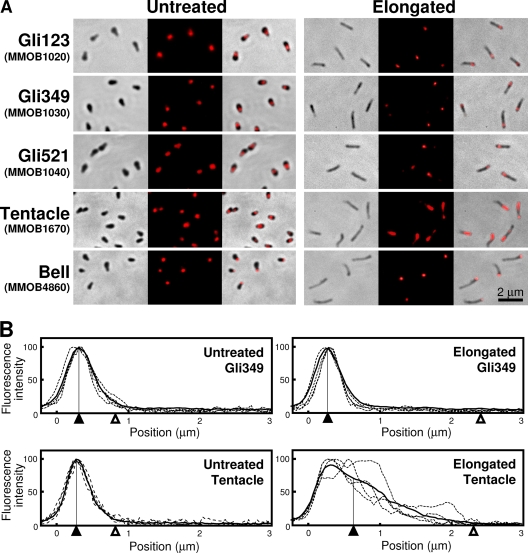
To elucidate the structural changes that occur with cell elongation, the localization of protein components was examined by immunofluorescence microscopy (Fig. 2A). The cells were examined for three component proteins of the gliding machinery—Gli123, Gli349, and Gli521—as well as the jellyfish tentacle protein coded MMOB1670 and the jellyfish bell protein coded MMOB4860. All of these proteins were localized in the untreated cells, as reported previously (Fig. 2A) (25, 36). In all elongated cells found in any field, the component proteins of the gliding machinery, Gli123, Gli349, and Gli521, were localized at similar distances from the front end of the cell. The Gli349 fluorescence intensity profile showed that this protein is localized about 0.33 μm from the front in the untreated cell and 0.30 μm from the front in the cells treated for 20 min with Tween 60 (Fig. 2B). Cell elongation did not significantly change the subcellular localization of the bell proteins relative to the cell front. However, elongation did significantly change the localization of the tentacle protein, whose fluorescence intensity profile showed that it is localized at an average distance of 0.31 μm from the front in the untreated cells and is distributed more diffusely, with an average position of 0.54 μm from the front in the elongated cells.
Fig 2.
Subcellular localization of proteins involved in gliding machinery and cytoskeletal jellyfish structure examined in elongated cells. (A) Immunofluorescence microscopy of untreated (left) and elongated (right) cells. Phase-contrast, fluorescence, and merged images are presented in the left, center, and right columns, respectively, for each cell type. Cell elongation was induced by 0.1% Tween 60 for 30 min. Gli123, Gli349, and Gli521, which are involved in the gliding machinery, were labeled after the cells were chemically fixed. The component proteins of jellyfish structure coded MMOB1670 and MMOB4860 were labeled after the cells were chemically fixed and permeabilized by 1% Triton X-100 (25). The labeled proteins are indicated at the left with their open reading frame identifiers in parentheses. (B) Sectional profiles of fluorescence intensities for Gli349 and the tentacle protein coded MMOB1670. The profiles of four independent cells were traced by broken lines, and 10 traces were averaged into a thick line with the peak position marked by a filled triangle. The maxima of the average trace and background levels were assigned 100% and 0%, respectively. The distance from the front end of the cell is shown as the x axis, and the average position of the tail end of the cell is indicated by an open triangle.
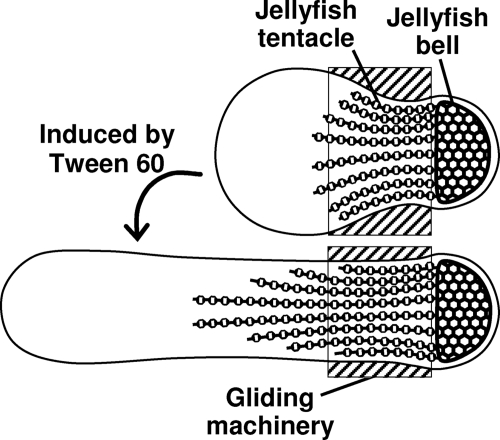
Next, we observed the elongated cells by negative-staining EM (Fig. 3). The widths of fully elongated cells ranged from 150 to 250 nm, slightly shorter than those observed in phase-contrast microscopy. The feature of a bulgy and angular outline at the tapered end was observed in an untreated cell, as reported previously (22, 23, 25, 33), and was also found at a pole of an elongated cell (Fig. 3A). The component proteins of the gliding machinery were detected just behind the bulgy and angular part under immunogold EM (Fig. 3B), as was the case with the untreated cells (32, 34); this is consistent with the results of immunofluorescence microscopy (Fig. 2). The jellyfish tentacle has been reported to be recognized sometimes on the basis of its filamentous patterns along the cell axis under EM (25). Line patterns were found more clearly on elongated cells, in the area corresponding to that labeled for the tentacle protein in immunofluorescence microscopy (Fig. 3C). These observations show that the tentacle of the jellyfish structure was elongated by Tween 60 treatment, whereas the jellyfish bell and the gliding machinery were not modified (Fig. 4). Next, we observed the surface structures under negative-staining EM. On the surface of elongated cells, filamentous structures can be seen more clearly than on that of untreated cells (Fig. 3A) (25).
Fig 3.
Cell structures examined by EM. (A) Structural changes from untreated to elongated cells. The left and right blocks of images present untreated and elongated cells, respectively. Cell shapes, a magnified cell image, and surface structures are presented in the upper left, lower, and upper right panels, respectively, in each block. Bulgy and angular outlines at a pole are marked by a white triangle in the lower panels. The regions in the lower panels identified by dashed boxes are magnified in the upper right panels. Surface filamentous structures are marked by white arrows. (B) Immunogold EM in cells elongated by 0.1% Tween 60. The cells were chemically fixed and labeled by the antibodies against the proteins indicated at the upper left. The dashed box region of the image for Gli123 is magnified in the inset and overlaid by artificial dots at the positions of immunogold particles. (C) The linear patterns corresponding to the jellyfish tentacles observed in an elongated cell. The density profiles of five positions are shown in the right panels. The peaks showing the linear patterns are marked by asterisks.
Fig 4.
Illustration of cell elongation induced by 0.1% Tween 60. Cells are shown with the cytoskeletal jellyfish structure. Localization of component proteins of the gliding machinery is presented by a hatched box. In the elongated cells, the localization of gliding machinery is not modified, although the jellyfish tentacles are expanded.
Gliding motility after morphological changes.
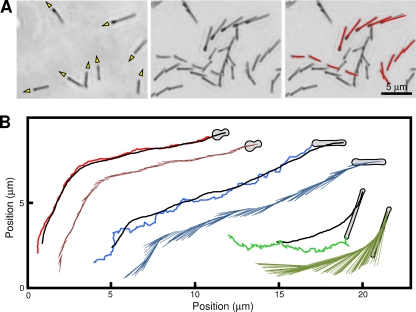
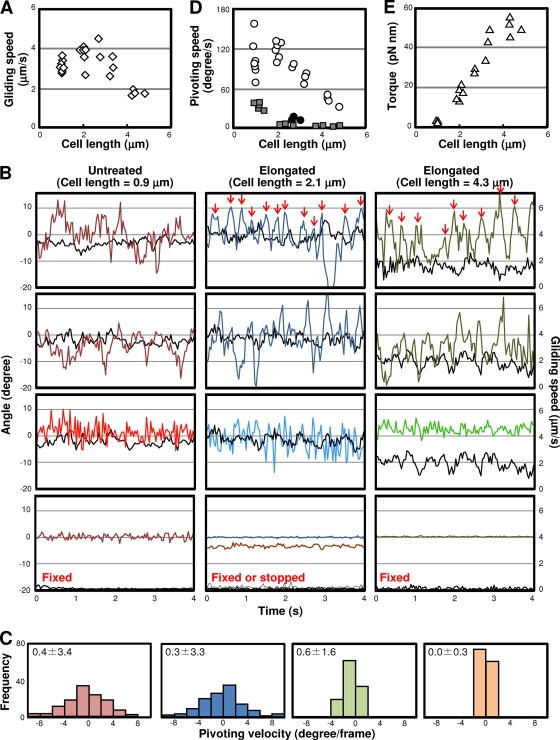
Although the morphology was changed through the elongation, the cells continued to glide, and we analyzed their motility in detail (Fig. 5A; see Videos S1 and S2 in the supplemental material). The continuous observation of cells during the morphological change showed that the cells were always gliding in the direction of the tapered end of untreated cells and the corresponding direction of elongated cells. As the cells had some stiffness, their images and behaviors could be portrayed by lines connecting the gliding machinery and the tail end (Fig. 5). On the basis of the results of fluorescence microscopy, the gliding machinery was uniformly assumed to be positioned at 300 nm from the front end of the cell (Fig. 2B). The displacements of gliding machinery and the angle of the cell axis relative to the glass surface were analyzed for cells of various lengths treated by Tween 60 for various times. The average gliding speed, measured by tracing the gliding machinery for 4 s, was 3.5 μm/s for the untreated cells, and the speed was reduced in elongated cells, i.e., 1.8 μm/s at a 4.5-μm cell length (Fig. 6A). The integrated video image showed that the tail end fluctuated around the gliding machinery, that is, the pivoting of the cell (Fig. 5B). To analyze the pivoting movements, we plotted the direction of the cell axis for every 0.033 s. As the direction of the cell axis changed continuously with gliding, as shown in Fig. 5, we normalized the angle of the cell axis by subtracting the direction of the preceding movement, to set the neutral angle of cell pivoting near 0 degree. We tried various durations to decide the preceding moving direction of gliding machinery and found that the neutral direction of the cell axis departed from 0 degree with the duration. Then, we adopted 0.167 s, the longest duration where the neutral angle of cell pivoting did not significantly depart from 0 degree (Fig. 6B). The cells were pivoting by 10 degrees of amplitude at a frequency of 2 to 3 times per s; this movement was clearer in the elongated cells than in the untreated ones (see Videos S1 and S2 in the supplemental material).
Fig 5.
(A) Tracing gliding motility of elongated cells. The cell movements were observed by phase-contrast microcopy. (Left) Single frame of video image. The directions of movement are shown with yellow triangles. (Center) Integrated video image. Movements for 4 s are presented by five integrated video frames. (Right) Cell images in each video frame were fitted with red lines and integrated into a single image. (B) Typical traces of cells having various lengths. The cells 0.9, 2.1, and 4.3 μm in length are presented by red, blue, and green traces, respectively. The positions of the gliding machinery and the tail end of the cell for 4 s at 0.033-s intervals are plotted and traced. The position of the gliding machinery and the tail end at each time point are connected by a line and integrated as shown on the lower side of each trace. The cell images of the end frame are presented schematically.
Fig 6.
Detailed analyses of gliding motility of elongated cells. (A) Relationship between gliding speed and cell length. The gliding speeds over 0.167 s were calculated every 0.033 s and averaged for 300-s traces of 15 cells. (B) Direction of cell axis (colored) and gliding speed (black). (Top two rows) The angle of the cell axis relative to the direction of movement in the preceding 0.167 s is plotted for every 0.033 s. The gliding speed over 0.167 s is plotted for every 0.033 s. Two independent cells are presented for each cell length. The pivoting points are marked by red arrows for traces of elongated cells. (Row third from the top) The change in cell axis direction is shown every 0.033 s for the same traces as in the neighboring upper graphs with their gliding speeds. (Bottom row) To check the accuracy of measurements, cells of various lengths were fixed chemically onto glass and analyzed for the direction of cell axis and moving speed. The gliding speeds are traced as black lines. As a control for nongliding cells, the axes and speeds of cells stopped by irradiation were traced as shown by the blue and gray lines, respectively, in the center graph. (C) Distributions of angles pivoting in a video frame every 0.033 s for 4 s. The standard deviations are shown in the upper left of each graph. (D) Relationships between mean pivoting speed and cell length. The mean pivoting speed is defined as the standard deviation in the distribution of pivoting velocity and plotted as an open circle. The fake pivoting speeds caused by noise detected on cells fixed to a glass surface are plotted as gray squares. The pivoting speeds of nongliding cells stopped by irradiation are plotted as filled circles. (E) Relationship between the torque needed for actual pivoting and the cell length.
To evaluate the accuracy of measurements, we traced the axes of cells chemically fixed onto the glass surface. As the cells were fixed on the solid surface, the fluctuation in the trace should be the noise from the system. Such noise was much reduced in the elongated cells, probably because of the advantages in cell-axis determination (Fig. 6B and D). To determine whether the pivoting was actually caused by the active movements in gliding machinery and not by Brownian motion, the changes in cell axes were also analyzed for nongliding cells. We stopped the gliding motility by applying the method of strong irradiation on a fluorescent dye. The radicals produced by irradiation of a dye inserted into the cell membrane are known to damage the protein or membrane and, finally, to cause cell death (10). In the present study, a dye, 0.1% Nile red, was added to gliding mycoplasmas. After 1 min, we irradiated the cells by a strong light at wavelengths ranging from 510 to 550 nm for 1 s. The irradiation immediately stopped cells from gliding; the cells pivoted slightly by 0.5 to 1 degree (Fig. 6B, bottom center). This range was much smaller than that observed in the gliding cells, showing that the pivoting of gliding cells was generated by the active movements of the machinery.
Next, we analyzed the angle speed of pivoting for cells of various lengths. The change in the direction of the cell axis was calculated for every 0.033 s without normalization, as presented in the third row of graphs from the top of Fig. 6B; their distributions are summarized in Fig. 6C. The distributions did not show obvious asymmetry. The mean pivoting speeds, estimated to be the standard deviations of the distributions, were 120 degrees/s at a 2.0-μm cell length, decreased as cell length increased, and reached 30 degrees/s at a 4.3-μm cell length (Fig. 6D). The mean pivoting speeds of cells shorter than 2.0 μm were more distributed than those of longer cells, probably because the cell axis was determined less precisely for shorter cells, as shown by the bottom row of graphs of Fig. 6B. As a rod-shaped cell pivoted around the gliding machinery, the torque can be estimated as it is for the torque generated around the flagellum motor in swimming bacteria (4, 5). In the present study, we estimated the torque required for the pivoting observed by applying the calculation method used for bacterial flagellum motors, with slight modifications. The calculated torque increased with the cell length, reached 54.7 pN nm at a cell length of 4.3 μm, and did not show saturation under the present conditions (Fig. 6E).
DISCUSSION
Mechanism of cell elongation.
In the present study, M. mobile cells were elongated by treatment with detergents (Fig. 1). The lipid bilayer of liposomes is known to be extended by the insertion of detergents (3, 30). The elongation of M. mobile is probably caused by a similar mechanism since there is no peptidoglycan layer, and morphological changes in mycoplasma cells can easily be caused by modifying the membrane area. The volumes of elongated cell were estimated to be about 0.4 μm3 as a cylinder. This number is similar to that for untreated cells of about 0.4 μm3 estimated as two spheres combined. The treatment by detergents probably caused the membrane expansion with retention of the cell volume, resulting in cell elongation. The axis of cell elongation agreed with the original one, which is guided by the protein meshwork of the gliding machinery and the cytoskeletal jellyfish structure (18, 25). This mechanism can explain the quick recovery after replacement of Tween 60 with fresh growth medium (Fig. 1B and C), because the albumin included in the growth medium is known to adsorb lipids (31). Albumin should remove the detergents in the medium. Actually, the cell morphology of Mycoplasma capricolum is known to change from rod shaped to spherical by the depletion of lipid in 2 h (21, 31). It was consistent with the above-described assumption that Tween 60 treatment did not significantly change the protein profile of M. mobile cells in SDS-PAGE (data not shown). However, the mechanism responsible for the elongation of the tentacle part of the jellyfish structure is unclear (Fig. 2 and 3). The membrane expansion by the detergent might have significant effects on the arrangements of component proteins, because some jellyfish-tentacle proteins have transmembrane segments in their sequences (25). Previously, elongated M. mobile cells were also induced by prolonged cultivation (22). However, the elongated cells in that study appeared to be much more flexible than those observed in the present study, suggesting that the cells were elongated through different mechanisms.
Surface structures of elongated cells.
We observed the filamentous structures on the surface more frequently and clearly in the elongated cells (Fig. 3A) than in the untreated cells. We attribute this to the difference in thickness, because fewer gliding units are laid in the surfaces of elongated and thinner cells. The integrated gliding machinery has not been well visualized, although it has been partially visualized in rapid freeze and fracture EM (19). Complete imaging of the gliding machinery may be easier with elongated cells with thinner necks.
Gliding mechanism suggested by pivoting.
In the unique mechanism of M. mobile gliding, legs sticking out from the gliding machinery propel the cell with repeated binding to sialyllactose fixed on a solid surface (6, 17, 18, 37). Previously, M. mobile was estimated to have 450 units of gliding machinery localizing around the cell neck, of which 100 units may participate in propulsion, assuming that a quarter of the units are aligned facing the solid surface (18, 36). Then, the information about the timing of propulsion by individual units is critical to elucidate this gliding mechanism. However, no experimental data have been available. The pivoting movements observed here should reflect the active propulsion of 100 units of the gliding machinery, because the stopped cells did not pivot (Fig. 6). The pivoting also suggests that the units of gliding machinery from a line perpendicular to the cell axis do not always propel the cell simultaneously (Fig. 7).
Fig 7.
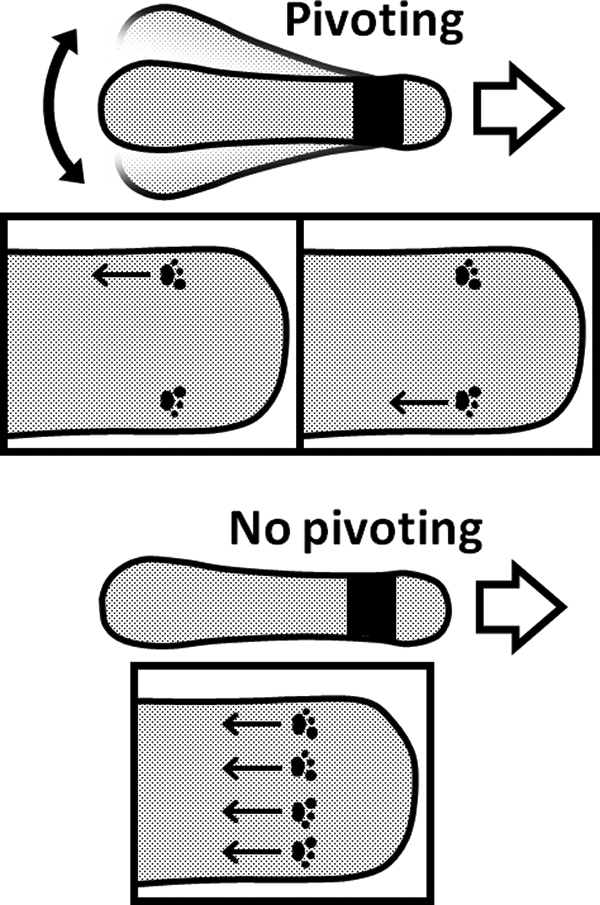
Proposed mechanism underlying cell pivoting. The black area shows the gliding machinery from which 450 legs are sticking out. The moving direction of the cell is indicated by open arrows. The footmarks present the foot at the tip of a leg bound to the target on the solid surface. The movements of the foot relative to the cell axis generate the propulsive force, as indicated by arrows. The cell pivoting suggests that feet in a line do not propel the cell with the same timing (upper). If the feet in a line always propel the cell with the same timing, the cell cannot pivot (lower).
The gliding speed in the elongated cells was reduced by as much as 50% (Fig. 6A). This decrease may suggest damage to individual gliding units or to disorder in leg movements. However, pivoting movements likely occur normally in untreated cells, because the traces of untreated cells also suggested such movements, although the pivoting was traced less precisely than the case of elongated cells (Fig. 5B and 6B).
The torque generated around the gliding machinery increased with cell length, reaching 54.7 pN nm at 4.3 μm, but did not show saturation (Fig. 6E). This observation suggests that the pivoting is not at the equilibrium between the torque and the friction; in other words, the gliding machinery can generate more torque. The number 54.7 pN nm is about 5% of the torque reported for the bacterial flagellum motor (4, 5). Considering the width of the whole gliding machinery, 150 nm, the average distance between two legs may be 75 nm. Then, the average of the minimal force needed for pivoting can be calculated to be 0.73 pN. This number is 37-fold smaller than the maximal stall force, 27 pN, suggesting that the stall force is generated by less than 37 units for usual gliding (20). In these estimations, we did not assume other loads, including friction on glass from other legs and other cell surface structures. Therefore, the possibility remains that the torque generated by the machinery may be larger than the value estimated here.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research on the priority areas system cell engineering by multiscale manipulation and innovative nanoscience of supermolecular motor proteins working in biomembrane (to M.M.); by a Grant-in-Aid for Scientific Research (A) (to M.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; by a grant from the Institution for Fermentation Osaka (to M.M.); and by a grant from JSPS Research Fellowships for Young Scientists (to D.N.).
Footnotes
Published ahead of print 14 October 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adan-Kubo J, Uenoyama A, Arata T, Miyata M. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding revealed by rotary-shadowing electron microscopy. J. Bacteriol. 188:2821–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aluotto BB, Wittler RG, Williams CO, Faber JE. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35–58 [Google Scholar]
- 3. Baumgart T, Hess ST, Webb WW. 2003. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425:821–824 [DOI] [PubMed] [Google Scholar]
- 4. Berg HC. 1993. Random walks in biology. Princeton University Press, Princeton, NJ [Google Scholar]
- 5. Che YS, Nakamura S, Kojima S, Kami-ike N, Namba K, Minamino T. 2008. Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 190:6660–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J, Neu J, Miyata M, Oster G. 2009. Motor-substrate interactions in mycoplasma motility explains non-Arrhenius temperature dependence. Biophys. J. 97:2930–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hiratsuka Y, Miyata M, Tada T, Uyeda TQP. 2006. A microrotary motor powered by bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaffe JD, Miyata M, Berg HC. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaffe JD, et al. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jori G, et al. 2006. Photodynamic therapy in the treatment of microbial infections: basic principles and perspective applications. Lasers Surg. Med. 38:468–481 [DOI] [PubMed] [Google Scholar]
- 11. Kirchhoff H. 1992. Motility, p 289–306 In Maniloff J, McElhaney RN, Finch LR, Baseman JB. (ed), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC [Google Scholar]
- 12. Krunkosky TM, Jordan JL, Chambers E, Krause DC. 2007. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 42:98–103 [DOI] [PubMed] [Google Scholar]
- 13. Kusumoto A, Seto S, Jaffe JD, Miyata M. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001–4008 [DOI] [PubMed] [Google Scholar]
- 14. Magariyama Y, Kudo S. 2002. A mathematical explanation of an increase in bacterial swimming speed with viscosity in linear-polymer solutions. Biophys. J. 83:733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Metsugi S, et al. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 1:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyata M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6–12 [DOI] [PubMed] [Google Scholar]
- 17. Miyata M. 2007. Molecular mechanism of mycoplasma gliding-a novel cell motility system, p 137–175 In Lenz P. (ed), Cell motility. Springer, New York, NY [Google Scholar]
- 18. Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64:519–537 [DOI] [PubMed] [Google Scholar]
- 19. Miyata M, Petersen J. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyata M, Ryu WS, Berg HC. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyata M, Seto S. 1999. Cell reproduction cycle of mycoplasma. Biochimie 81:873–878 [DOI] [PubMed] [Google Scholar]
- 22. Miyata M, Uenoyama A. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285–289 [DOI] [PubMed] [Google Scholar]
- 23. Miyata M, et al. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311–1320 [DOI] [PubMed] [Google Scholar]
- 24. Nagai R, Miyata M. 2006. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J. Bacteriol. 188:6469–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakane D, Miyata M. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 104:19518–19523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakane D, Miyata M. 2009. Cytoskeletal asymmetrical-dumbbell structure of a gliding mycoplasma, Mycoplasma gallisepticum, revealed by negative-staining electron microscopy. J. Bacteriol. 191:3256–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nonaka T, Adan-Kubo J, Miyata M. 2010. Triskelion structure of the Gli521 protein, involved in the gliding mechanism of Mycoplasma mobile. J. Bacteriol. 192:636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohtani N, Miyata M. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 403:71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seddon AM, Curnow P, Booth PJ. 2004. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 1666:105–117 [DOI] [PubMed] [Google Scholar]
- 31. Seto S, Miyata M. 1999. Partitioning, movement, and positioning of nucleoids in Mycoplasma capricolum. J. Bacteriol. 181:6073–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seto S, Uenoyama A, Miyata M. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimizu T, Miyata M. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431–434 [DOI] [PubMed] [Google Scholar]
- 34. Uenoyama A, Kusumoto A, Miyata M. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uenoyama A, Miyata M. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 102:12754–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uenoyama A, Miyata M. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uenoyama A, Seto S, Nakane D, Miyata M. 2009. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery suggested by inhibitory antibodies and mutants. J. Bacteriol. 191:1982–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.