Abstract
Sortases catalyze the covalent anchoring of proteins to the cell surface on Gram-positive bacteria. Bioinformatic analysis suggests the presence of structural genes encoding sortases and their substrates in the Bacillus subtilis genome. In this study, a β-lactamase reporter was fused to the cell wall anchoring domain from a putative sortase substrate, YhcR. Covalent anchoring of this fusion protein to the cell wall was confirmed by using the eight-protease-deficient B. subtilis strain WB800 as the host. Inactivation of yhcS abolished the cell wall anchoring reaction. The amounts of fusion protein anchored to the cell wall were proportional to the levels of YhcS. These data demonstrate that YhcS and YhcR are the sortase and sortase substrate, respectively, in B. subtilis. Furthermore, yhcS is not essential for the survival of B. subtilis under the cultivation condition tested. YhcR fusions were distributed helically in the lateral cell wall. Interestingly, when viewed with an epifluorescence microscope, YhcS also appeared to form short helical arcs. This is the first report to illustrate such distribution of sortases in a rod-shaped bacterium. Models for the spatial distribution of both the sortase and its substrate are discussed. The amount of the reporters displayed on the surface was unambiguously quantified via a unique strategy. Under optimal conditions with the overproduction of YhcS, 47,300 YhcR fusions could be displayed per cell. Displayed reporters were biologically functional and surface accessible. Characterization of the sortase-substrate system allowed the successful development of a YhcR-based covalent surface display system. This system may have various biotechnological applications.
INTRODUCTION
Bacteria possess a wide range of proteins displayed on the cell surface that interact with molecules and cells in the environment. To display proteins on the cell surface, several mechanisms are used in Gram-positive bacteria (17). Surface proteins are either covalently or noncovalently attached to peptidoglycan. Some even bind noncovalently to secondary cell wall polymers (42).
Sortase is a membrane-bound enzyme that is responsible for the covalent attachment of proteins to the peptidoglycan of Gram-positive bacteria (39). These wall-anchored surface proteins contain two essential elements. First, an N-terminal signal peptide is required to direct these proteins to the secretory pathway. Second, a C-terminal cell wall anchoring domain (CWAD) is required for cell wall anchoring. CWADs have three crucial features, which are an LPXTG motif (where X can be any amino acid), a hydrophobic transmembrane domain, and a tail with positively charged amino acid residues (38, 39).
In Bacillus subtilis, two putative sortases (YhcS and YwpE) and putative substrates (YhcR and YfkN) have been identified by bioinformatic analyses (10, 20, 46). Although YhcR and YfkN have been identified as an endonuclease (45) and a trifunctional nucleotidase (8), respectively, conclusive demonstration that these proteins are covalently anchored to the cell wall has proven to be difficult. This is primarily due to the presence of high levels of extracellular proteases produced by B. subtilis that degrade these wall-bound proteins. With the first discovery of the structural gene encoding sortase in Staphylococcus aureus more than a decade ago (40) and the subsequent discovery of many sortases and their substrates in many Gram-positive bacteria (46), it is of interest to determine whether any sortase and sortase substrate exist in B. subtilis, one of the best characterized Gram-positive bacteria.
In this study, we conclusively demonstrate that yhcS and yhcR encode a sortase and a sortase substrate, respectively. YhcS was found to be responsible for anchoring YhcR to the cell wall in a covalent manner. To overcome the significant proteolytic degradation of surface proteins, the large nuclease domain (1,085 residues, 118.5 kDa) of YhcR was replaced with a small reporter (β-lactamase) and the fusion protein was produced in an eight-protease-deficient B. subtilis strain (WB800). The amounts of the fusion protein anchored to the cell wall were proportional to the levels of sortase present in the cell. Interestingly, visualization using an epifluorescence microscope showed that wall-bound reporters were distributed in a helical fashion, while green fluorescent protein (GFP)-sortase fusions seem to localize in helical arcs or tracks. To our knowledge, this is the first time that the distribution of a sortase in rod-shaped bacteria has been elucidated. Notably, a strategy to quantify the number of wall-bound reporters in an accurate manner was also developed. This covalent surface display system can display not only an enzyme (e.g., β-lactamase) but also has the potential to display giant enzyme complexes (e.g., cellulosomes) for biofuel conversion.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacillus subtilis strain WB800 (62) was used as the expression host for cloning and protein production unless stated otherwise. In this strain, a total of seven extracellular protease genes and a wall-bound protease gene (wprA) have been deleted. Inactivation of wprA is essential for the successful display of wall-bound proteins. All B. subtilis cells were propagated at 30°C on tryptose blood agar base (TBAB) plates, and Escherichia coli cells were propagated at 37°C on LB agar plates. For protein production, cells were cultured in superrich medium (SRM) (Bacto tryptose, 25 g/liter; yeast extract, 20 g/liter; dipotassium phosphate, 3 g/liter; glucose, 4.5 g/liter; pH 7.5) at 30°C for 14 h and harvested for analysis. The following antibiotics were used for selection: kanamycin (10 μg/ml), spectinomycin (250 μg/ml), ampicillin (75 μg/ml), erythromycin (5 μg/ml), and lincomycin (5 μg/ml). B. subtilis transformation was performed by the Spizizen competent cell method (55). Table S1 in the supplemental material lists the bacterial strains used in this study.
Construction of expression vectors and strains.
To allow covalent anchoring of the recombinant B. subtilis sortase substrates to the cell wall, an expression vector, pWB980-BLA-CWADYhcR, was constructed (see Fig. S1 in the supplemental material). This vector contains the coding region for the YhcR cell wall anchoring domain (CWAD) inserted in the expression vector pWB980-BLA-L56-LytE (9) which produces a β-lactamase (BLA) fusion protein. At the C-terminal end of BLA-L56-LytE is a cell wall binding module derived from LytE, a B. subtilis cell wall hydrolase, for binding to the cell wall in a noncovalent manner. There is a 56-amino-acid linker (L56) located between BLA and the LytE cell wall binding domain. The cell wall binding module of LytE was replaced by the CWAD derived from YhcR (CWADYhcR) by the following process. A 438-bp DNA fragment encoding CWADYhcR was amplified by PCR using B. subtilis WB800 chromosomal DNA as the template and two primers, BSCYHCRF and BSCYHCRB (see Table S2 in the supplemental material). The PCR product was digested with BglII and NheI and inserted into the BclI/NheI-digested pWB980-BLA-L56-LytE vector to generate pWB980-BLA-CWADYhcR. To produce another fusion protein, BLA-CWADYhcRMO2 (Fig. S1), the expression vector pWB980-BLA-CWADYhcRMO2 was constructed in a two-step process. First, sequence encoding CWADYhcR was modified to eliminate the termination codon and to introduce a NotI site at the 3′ end. This modified sequence was generated by PCR with pWB980-BLA-CWADYhcR as the template and BSSacBF/YHCRCTMB as the primer pair (Table S2). The 1,439-bp PCR product was then digested with BstEII and NheI and ligated to the BstEII/NheI-digested pWB980-BLA-CWADYhcR to generate pWB980-BLA-CWADYhcRCTM, which contained a modified 3′ end at the coding sequence of CWADYhcR for the insertion of the monomeric orange fluorescent protein 2 (MO2) gene. In the second stage of construction, the plasmid pBSK-KCMO2ST3 (Table S1), an E. coli Bluescript plasmid carrying a synthetic gene encoding MO2, was used. This synthetic gene was ordered from Epoch Biolabs (Missouri City, TX). The mo2 nucleotide sequence was redesigned for optimal expression in B. subtilis based on the previously reported protein sequence for MO2 (51). pBSK-KCMO2ST3 was double digested with NotI and NheI to release the mo2 insert, which was then inserted into the NotI/NheI double digested pWB980-BLA-CWADYhcRCTM to generate pWB980-BLA-CWADYhcRMO2. This vector allows the production of a fusion protein with β-lactamase at the N-terminal region, the CWADYhcR domain in the middle, and MO2 at the C-terminal end.
Plasmid pWB980-YhcS is an expression vector for the production of the wild-type full-length sortase, YhcS. The 716-bp full-length yhcS was generated by PCR amplification of the B. subtilis WB800 chromosomal DNA with primers BSYHCSF and BSYHCSB (see Table S2 in the supplemental material). The PCR product was digested with KpnI and NheI and inserted into KpnI/NheI-digested pWB980 (61) to generate pWB980-YhcS. For the simultaneous production of both YhcS and BLA-CWADYhcRMO2 in the same expression host, a pWB980-pE18 binary vector system was used (61). One plasmid (pWB980-BLA-CWADYhcRMO2, kanamycin resistant) overproduces BLA-CWADYhcRMO2. The other plasmid (pE18-YhcS, erythromycin resistant) overproduces YhcS. These two plasmids can coexist in the same B. subtilis host. To construct pE18-YhcS, yhcS was subcloned into pE18-P43 (61) by first cutting pWB980-YhcS with NheI, blunt ended using T4 DNA polymerase I and subsequently cut with BlpI to release the insert carrying yhcS. This fragment was inserted into SmaI/BlpI-digested pE18-P43 to generate pE18-YhcS.
YhcS has an N-terminal transmembrane segment followed by a sortase catalytic domain. To purify the non-membrane-bound form of sortase for injection into mice to obtain antisortase antibodies, a glutathione S-transferase (GST) sortase fusion construct (GST-YhcSNM) was made. YhcSNM is a truncated version of YhcS with its N-terminal transmembrane segment deleted (NM designates the non-membrane-bound version). GST-YhcSNM is soluble and can be affinity purified. Plasmid pGEX2T-YhcSNM was made in a two-step process. The first step is to generate pWB980-YhcSNM. A PCR product encoding YhcS was generated by using B. subtilis WB800 genomic DNA as the template and BSYHCSF and BSYHCSB (see Table S2 in the supplemental material) as the primers. This fragment was digested with both ClaI and NheI and inserted into the ClaI/NheI-digested pWB980-XylA. This generates pWB980-YhcSNM. In the second step, the DNA fragment encoding YhcSNM was amplified by PCR using pWB980-YhcSNM as the template and yhcSNMfrGSTF and yhcSNMfrGSTR as primers (Table S2). The pGEX2T plasmid and the PCR product (647 bp) were digested with BamHI and NheI. The PCR product was then inserted into pGEX2T to generate pGEX2T-YhcSNM. pGEX2T-YhcSNM was transformed into E. coli DH5α for protein production.
Generation of a sortase knockout mutant (WB800Srt−) using the Cre/lox- and PCR-based method.
B. subtilis WB800Srt− was constructed by the replacement of yhcS in strain WB800 with an antibiotic cassette using a genome engineering method developed by Yan et al. (63). Briefly, the spectinomycin resistance (Spcr) cassette (1,178 bp) was amplified from vector p7S6 (Bacillus Genetic Stock Center) (Table S1) with primers Lox71F and Lox66R (see Table S2 in the supplemental material). Two primer pairs, 1kbyhcRF/1kbyhcRR (front flanking region) and 1kbyhcTF/1kbyhcTR (back flanking region), were used to amplify two 1-kb DNA fragments flanking the upstream and downstream yhcS sequence, respectively. Extensions of 21 nucleotides that were complementary to the 5′ and 3′ ends of the amplified Spcr cassette were added to the 5′ ends of both the reverse and forward primers of the upstream and downstream flanking regions. All three fragments were amplified using Vent polymerase, and the products were used for a two-step PCR fusion amplification. Step A (10 cycles) included 73 μl water, 10 μl ThermoPol buffer (10×), 10 μl deoxynucleoside triphosphate (dNTP) mix (2.5 mM each), 2 μl of MgSO4 (100×), 1 μl (100 ng) of the upstream flanking fragment, 2 μl (200 ng) of marker cassette fragment, 1 μl (100 ng) of the downstream flanking fragment, and 1 μl of Vent polymerase. Step B (30 cycles) included 73 μl water, 10 μl ThermoPol buffer (10×), 10 μl dNTP mix, 1 μl MgSO4 (100×), 2 μl (10 mM) forward primer of the upstream flanking fragment, 1 μl (10 mM) reverse primer of the downstream flanking fragment, 1 μl of the unpurified PCR product from step A, and 1 μl of Vent polymerase. The resulting PCR product was gel purified and transformed into B. subtilis WB800. The transformants were selected on TBAB plates containing 250 μg/ml spectinomycin. The successful generation of the yhcS knockout strains was confirmed by PCR screening of the spectinomycin-resistant transformants. One knockout strain (WB800Srt−) was kept for further studies. To remove the antibiotic Spcr marker, pTSC (Bacillus Genetic Stock Center) (see Table S1 in the supplemental material) carrying a constitutively expressed Cre recombinase gene was introduced into the yhcS knockout strain via transformation. Erythromycin-resistant transformants were selected. These transformants were then grown on antibiotic-free LB liquid medium for 16 h and plated on LB plates. The removal of the Spcr marker was confirmed by the inability of these cells to grow on TBAB plates containing 250 μg/ml spectinomycin. To remove the pTSC plasmid, the cells were grown overnight in LB without erythromycin at 51°C for 16 h, as pTSC has a temperature-sensitive replicon and is unable to replicate at 51°C. The resulting erythromycin-sensitive WB800Srt− strain was used for the construction of WB800Srt− amyE::pxyl-gfp-yhcS.
Construction of WB800Srt− amyE::pxyl-gfp-yhcS.
To construct a green fluorescent protein-sortase (GFP-YhcS) fusion strain, pSG1729 (5) was used. This plasmid was obtained from the Bacillus Genetic Stock Center. The full-length sortase gene yhcS was PCR amplified with the primer pairs GFPYHCSF and GFPYHCSB (see Table S2 in the supplemental material) using pWB980-YhcS as the template to generate a 695-bp PCR product which was subsequently digested by both BamHI and NheI and subcloned into the BamHI- and NheI-digested pSG1729. The resulting clone contains yhcS inserted in the positive orientation downstream of the green fluorescent protein (GFP) sequence and was designated pSG1729-GFP-YhcS. The mut1 variant (64) of the green fluorescent protein gene was fused to the N-terminal end of YhcS at the gene level, and the resulting gfp-yhcS gene was placed under the control of a xylose-inducible promoter (pxyl) of the pSG1729 plasmid that replicates only in E. coli. GFP was fused to the N terminus of YhcS to preserve the proper topology and avoid the secretion of GFP. GFP is not normally functional when translocated to the extracellular side of the membrane in bacteria (23, 41).
The pSG1729 plasmid contains the 5′ and 3′ ends of the B. subtilis amyE sequence for integration into the B. subtilis amyE locus via a double-crossover event and a spectinomycin antibiotic marker for selection. A short flexible linker (14 amino acids) between GFP and YhcS was included with the intention to separate the two domains and to allow both domains to fold independently. Plasmid pSG1729-GFP-YhcS was integrated into the genome of the spectinomycin-sensitive WB800Srt− strain to generate WB800Srt− amyE::pxyl-gfp-yhcS containing a single copy of the inducible GFP-sortase fusion gene. The integration of pxyl-gfp-yhcS into the amyE locus was confirmed by PCR with primers diGFPyhcSMR1 and diGFPyhcSMR2 (see Table S2 in the supplemental material). The sortase knockout strain WB800Srt− was chosen to ensure that the localization signal of GFP-YhcS was not displaced by the presence of wild-type YhcS.
Release of wall-bound forms of BLA-CWADYhcR and BLA-CWADYhcRMO2 by lysozyme.
Profiling of sortase substrates was examined by subjecting B. subtilis WB800(pWB980-BLA-CWADYhcR) (for BLA-CWADYhcR) and WB800(pWB980-BLA-CWADYhcRMO2) (for BLA-CWADYhrRMO2) to lysozyme digestion. Washed cells were resuspended in 150 μl SET buffer (20% sucrose, 50 mM Tris, 50 mM EDTA [pH 7.6]). Lysozyme and phenylmethylsulfonyl fluoride (PMSF) were added to the cell suspensions to a final concentration of 8 mg/ml and 1 mM, respectively. Samples were incubated at 37°C for 30 min. Protoplast formation was confirmed by using a phase-contrast microscope. Protoplasts were separated from the digested cell wall by centrifugation using a microcentrifuge at 5,400 × g for 7 min. The resulting protoplast pellet was resuspended with 150 μl of SET buffer. Both the protoplast and cell wall fractions were used for Western blotting.
Immunofluorescence microscopy.
Localization of wall-bound BLA-CWADYhcRMO2 on the bacterial surface was examined by using an epifluorescence microscope as previously described (9) with the following changes. Briefly, cells cultured overnight in SRM were harvested and fixed with 4% paraformaldehyde for 30 min at 4°C after they were washed with phosphate-buffered saline (PBS). Cell samples were then blocked with 1% (vol/vol) bovine serum albumin (BSA) for 1 h before they were incubated with rabbit primary antibodies against β-lactamase in PBS in a microcentrifuge tube with gentle agitation for 2 h. The cells were washed three times with PBS containing 0.1% Tween 20 (vol/vol) (5 min per wash) before binding with Alexa Fluor 488-conjugated goat anti-rabbit antibodies and Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) for another 2 h in microcentrifuge tubes with gentle agitation. WGA binds N-acetylglucosamine in the peptidoglycan. The cells were washed with PBS containing 0.1% Tween 20 again before 4′,6′-diamidino-2-phenylindole (DAPI), a fluorescent dye for DNA, was added. The cells were then spotted onto glass microscope slides, and SlowFade antifade reagent was subsequently added to prevent photobleaching. The stained cells were visualized.
As the excitation and emission wavelengths of Alexa Fluor 488 coincide with those of GFP, it would not be possible to use the same secondary antibodies for the colocalization studies of BLA-CWADYhcRMO2 with GFP-YhcS. B. subtilis WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) was probed with Alexa Fluor 568-conjugated rabbit anti-FLA antibodies after the binding of the primary antibodies. The cells were washed again and spotted onto glass microscope slides for visualization using the fluorescence microscope. Optical z-stacks were taken. Images were saved as ZVI files for deconvolution software processing. Deconvolution was applied to images using Huygen's deconvolution software (Scientific Volume Imaging, Hilversum, Netherlands). The Carl Zeiss Axioimager Z1 fluorescence compound microscope was used for imaging. The Zeiss 100× objective was used for image capture.
Other methods.
Cell wall from a late-log-phase B. subtilis culture was isolated as described previously (9). Purified GST-YhcSNM was used as the antigen to generate polyclonal antibodies in mice according to the previously published procedures (58). GST-YhcSNM was purified using glutathione-Sepharose columns as recommended by the manufacturer. Rabbit antibodies against β-lactamase generated previously (59) were used in this study. The numbers of wall-bound BLA-CWADYhcRMO2 in B. subtilis WB800(pWB980-BLA-CWADYhcRMO2) and WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) were estimated by Western blotting using anti-BLA antibodies as previously described (9) with the following changes. Known numbers of bacterial cells were treated with lysozyme before separating the digested cell wall from protoplasts by centrifugation. The soluble supernatant containing the digested cell wall was separated by SDS-PAGE for Western blotting. The activity of BLA was assayed as previously described (9) with the following modifications. The assay used 7-(thienyl-2-acetamido)-3-[2-(4-N,N-dimethyl-amino-phenylazo) pyridinium-methyl]-3-cephem-4-carboxylic acid (PADAC) as the substrate and was carried out using 96-well microtiter plates. The total volume of each assay mix (consisting of PADAC and PBS) per well was 100 µl. The amount of PBS containing PADAC (PADAC-PBS) was adjusted such that the initial optical density at 595 nm (OD595) observed was 1.00 (±0.05) before the start of the assay. Washed cells that were resuspended in PBS were pipetted to the assay mixture to begin the assay, and readings at OD595 were recorded at 1-min intervals for a total of 10 min. The total volume of the cell samples added to PADAC-PBS for the assay was adjusted such that the rate of product formation was linear. Enzymatic activities were calculated using the molar extinction coefficient of 52,700 M−1 cm−1.
RESULTS
Design, production, and characterization of BLA-CWADYhcR and BLA-CWADYhcRMO2.
To demonstrate the anchoring of the putative sortase substrate YhcR to the cell wall using the B. subtilis endogenous sortase, a β-lactamase fusion (BLA-CWADYhcR) was constructed. This fusion can be divided into three parts. The N-terminal portion is the β-lactamase reporter. Its C-terminal end consists of the cell wall anchoring domain derived from YhcR (CWADYhcR). This domain comprises the LPDTS sorting motif, a transmembrane segment, and a positively charged tail (Fig. 1A). Between these two domains is a 152-amino-acid linker region. The first 50 amino acids form a designer linker which is composed of mainly glycine, proline, serine, and threonine residues. It is then followed by 102 amino acids that are naturally present upstream of the LPDTS sorting motif of CWADYhcR. Since a linker of ∼123 amino acids is sufficient to penetrate through the thick B. subtilis cell wall and project the reporter to the cell surface (44), this 152-amino-acid sequence should be long enough for its function as a linker.
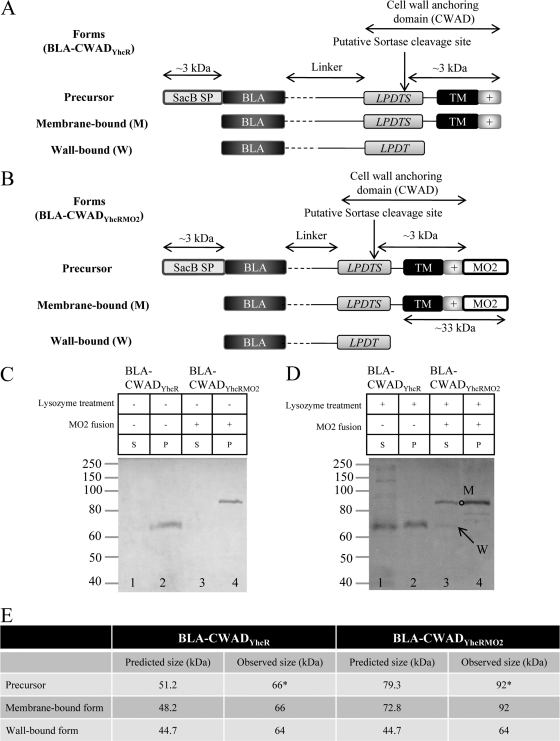
Fig 1.
Rational design of the reporter fusions, BLA-CWADYhcR and BLA-CWADYhcRMO2, and their production. (A) Illustration depicting the construction and properties of the BLA-CWADYhcR reporter fusion. The reporter β-lactamase (BLA) was fused to the cell wall anchoring domain of YhcR at the gene level. The precursor protein is directed to the Sec-based secretory pathway by the B. subtilis SacB signal peptide (3 kDa). The SacB signal peptide (SP) is removed during/after translocation and the membrane-bound form (M) of the fusion is recognized by sortase. Sortase (YhcS) is likely to cleave between the threonine and serine residues of the LPDTS motif and allows anchoring of BLA-CWADYhcR to peptidoglycan to generate the wall-bound form (W) of the reporter. The transmembrane segment (TM) and the positively charged C-terminal tail in the cell wall anchoring domain (+) are indicated. In the linker region, the 50-amino-acid designer linker (broken line) and the 102-amino-acid linker naturally present in YhcR (solid black line) are indicated. (B) Illustration depicting the addition of the monomeric orange fluorescent protein 2 (MO2) to generate the BLA-CWADYhcRMO2 reporter fusion. MO2 was introduced into the C-terminal end of BLA-CWADYhcR. With this addition, the transmembrane segment and the positively charged tail of CWADYhcR would acquire a 28.1-kDa MO2 which would shift the membrane-bound form (M) of BLA-CWADYhcRMO2 to a higher-molecular-mass position in SDS-polyacrylamide gels. In contrast, the wall-bound form (W) of BLA-CWADYhcRMO2 would have the same molecular mass as the initial BLA-CWADYhcR construct. (C) Western blot analysis of the cellular distribution of BLA fusions from B. subtilis without lysozyme treatment (−). B. subtilis strains WB800(pWB980-BLA-CWADYhcR) (lanes 1 and 2) and WB800(pWB980-BLA-CWADYhcRMO2) (lanes 3 and 4) were analyzed. A centrifugation step was performed to separate the soluble and cell fractions before electrophoresis and immunoblotting. The soluble supernatant fraction (S) and the whole-cell pellet fraction (P) are indicated above the gel. The amounts of samples loaded were normalized to cell density. Rabbit anti-BLA was used to probe for the reporter fusion with a 1:2,000 dilution. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (D) Western blot analysis of the cellular distribution of BLA fusions from B. subtilis with the lysozyme treatment. Strains WB800(pWB980-BLA-CWADYhcR) (lanes 1 and 2) and WB800(pWB980-BLA-CWADYhcRMO2) (lanes 3 and 4) were treated with lysozyme (+) to release the wall-bound reporters. A centrifugation step was performed to separate wall-bound and membrane-bound forms before electrophoresis and immunoblotting. The soluble supernatant fraction (S) and the protoplast fraction (P) are indicated above the gel. The membrane-bound form (M) (open circle) and the wall-bound form (W) (arrow) of BLA-CWADYhcRMO2 are shown. The amounts of samples loaded were normalized to cell density. Rabbit anti-BLA was used to probe for the reporter fusion with a 1:2,000 dilution. (E) Predicted and observed molecular masses of BLA-CWADYhcR and BLA-CWADYhcRMO2. The observed molecular masses of the fusion proteins on SDS-polyacrylamide gels are constantly larger than the predicted values because of the presence of the unstructured designer linker region. A detailed explanation is provided in reference 9. The precursor and membrane-bound forms of the fusion proteins have similar molecular masses. The precursors may comigrate with the membrane-bound form of the fusions on SDS-polyacrylamide gels. Asterisks indicate the observed molecular masses of the precursors if they exist.
Cell wall sorting of BLA-CWADYhcR was examined by two approaches. The first approach was Western blot analysis (with anti-BLA antibodies) of the lysozyme-treated B. subtilis WB800(pWB980-BLA-CWADYhcR) (Fig. 1D) with the nontreated WB800(pWB980-BLA-CWADYhcR) (Fig. 1C) as the negative control. A total of three forms (the full-length precursor, the membrane-bound intermediate, and the wall-bound form) of BLA-CWADYhcR (Fig. 1A) would be generated during the sorting process. The wall-bound form of BLA-CWADYhcR was present in the supernatant fraction of the lysozyme-treated sample (Fig. 1D, lane 1) and was absent in the untreated control (Fig. 1C, lane 1). The membrane-bound and precursor forms of BLA-CWADYhcR should be present in the protoplast fraction (Fig. 1C and D). Since the three forms of protein had relatively similar molecular masses (Fig. 1A), it was not a surprise to observe that the mobility of the wall-bound form of BLA-CWADYhcR was similar to that of the membrane-bound form and/or the precursor form of BLA-CWADYhcR (Fig. 1D, lanes 1 and 2). This property can impose a problem in the quantification of the wall-bound form of BLA-CWADYhcR. Any lysis of the protoplasts during the lysozyme treatment would release the membrane/precursor forms of BLA-CWADYhcR to the supernatant fraction. Consequently, the band intensity observed in the supernatant fraction might not accurately reflect the quantity of the wall-bound form of BLA-CWADYhcR released from the lysozyme treatment. To address this concern, a 28-kDa monomeric orange fluorescent protein 2 (MO2) was added to the C-terminal end of CWADYhcR to generate BLA-CWADYhcRMO2 (Fig. 1B). After the sortase-mediated processing, the wall-bound form of BLA-CWADYhcRMO2 would have a much smaller molecular mass with reference to both the membrane and precursor forms.
Analysis of the lysozyme-treated samples of B. subtilis WB800(pWB980-BLA-CWADYhcRMO2) indicated that the wall-bound form of BLA-CWADYhcRMO2 was present in the supernatant fraction (Fig. 1D, lane 3, arrow) in small quantities. A form of BLA-CWADYhcRMO2 with higher molecular weight (Fig. 1D, lane 3, open circle) was also detected in the supernatant fraction. Observation of this higher-molecular-weight form in the supernatant was likely due to low levels of cell lysis occurring during lysozyme treatment. Without the ability to separate the membrane-bound form from the wall-bound form, the amount of the wall-bound form may easily be overestimated by five times or more. Together, these results suggest that BLA-CWADYhcRMO2 was covalently anchored to the B. subtilis cell wall. A discrepancy between the expected and observed molecular weights of BLA-CWADYhcR and BLA-CWADYhcRMO2 was observed. This was due to the presence of the unstructured designer linker, which results in a slower migration of proteins in an electrophoretic run (9).
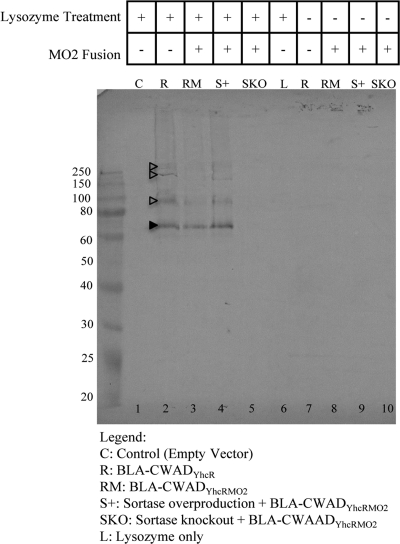
To unambiguously confirm the covalent anchoring of BLA-CWADYhcR and BLA-CWADYhcRMO2 to the peptidoglycan, a second approach was taken. The cell wall fractions of B. subtilis WB800(pWB980-BLA-CWADYhcR) and WB800(pWB980-BLA-CWADYhcRMO2) were prepared and boiled in the presence of SDS to remove any noncovalent wall-bound proteins. Western blot analysis reveal the presence of the 64-kDa reporter fusion in the cell wall fractions prepared from WB800(pWB980-BLA-CWADYhcR) and WB800(pWB980-BLA-CWADYhcRMO2) (Fig. 2, lanes 2 and 3). The protein bands with molecular masses greater than 64 kDa were likely the wall-bound proteins linked to the cell wall fragments which were not completely digested by lysozyme. The presence of deacetylated glucosamine (2) and attachment of teichoic acid and covalently linked BLA-CWADYhcR/BLA-CWADYhcRMO2 in B. subtilis peptidoglycan can potentially prevent the complete digestion of the cell wall by lysozyme. The cell wall from the negative-control strain WB800(pWB980) showed no detectable β-lactamase fusion bands (Fig. 2, lane 1). Cell wall samples that were not treated with lysozyme also did not show any β-lactamase bands (Fig. 2, lanes 7 to 9).
Fig 2.
Western blot analysis of the covalently linked wall-bound β-lactamase from the purified B. subtilis cell wall preparations. Purified cell wall samples from B. subtilis strains WB800(pWB980) (control [C]), WB800(pWB980-BLA-CWADYhcR) (R), WB800(pWB980-BLA-CWADYhcRMO2)(RM), WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) (sortase overproduction [S+]), and WB800Srt−(pWB980-BLA-CWADYhcRMO2) (sortase knockout [SKO]) were probed with anti-BLA antibodies in the Western blot analysis. Samples in lanes 1 to 5 were treated with lysozyme (+) to release BLA-CWADYhcR, and samples in lanes 7 to 10 were not treated with lysozyme (−). Lane 1, cell wall from B. subtilis WB800(pWB980) which has the expression vector without any inserts; lane 6, lysozyme in buffer (L). The samples in both lanes 1 and 6 are negative controls. The filled arrowhead marks the expected wall-bound form of BLA. The open arrowheads mark the wall-bound BLA anchored to the incompletely digested cell wall. The amounts of cell wall loaded were normalized against the cell density. The only exception is the sample from strain WB800Srt−(pWB980-BLA-CWADYhcRMO2). The amount of this sample (SKO) loaded is 10 times more than the normalized amount of cell wall from other samples.
Cell wall anchoring of BLA-CWADYhcRMO2 is mediated by the YhcS sortase.
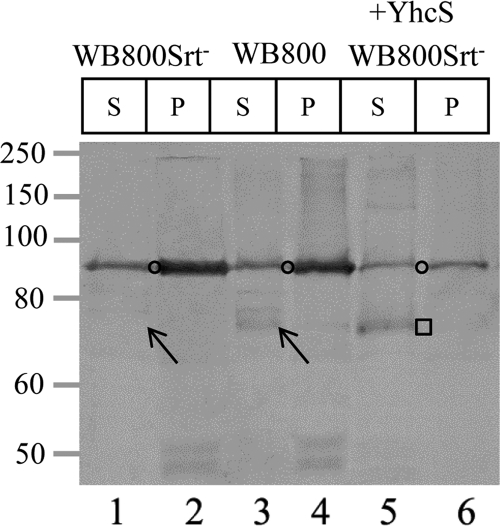
Since yhcS (10, 20, 46) encodes a putative sortase and is located next to yhcR in the B. subtilis genome, it is of interest to determine whether YhcS is responsible for anchoring BLA-CWADYhcRMO2 to the cell wall. A yhcS knockout strain (WB800Srt−) was constructed (see Fig. S2 in the supplemental material) and confirmed by genomic PCR analysis (data not shown). The effect of knocking out the yhcS gene in the WB800Srt− strain on cell wall protein anchoring was examined in comparison to the positive-control strain by Western blotting. A wall-bound form of BLA-CWADYhcRMO2 was observed in B. subtilis WB800(pWB980-BLA-CWADYhcRMO2) (Fig. 3, lane 3, arrow). This band was not detected in B. subtilis WB800Srt−(pWB980-BLA-CWADYhcRMO2) (Fig. 3, lane 1, arrow). Similar findings were observed using purified cell wall fractions from these strains (Fig. 2, lane 3 versus lane 5). Loading 10 times more purified cell wall prepared from WB800Srt−(pWB980-BLA-CWADYhcRMO2) still did not allow the detection of the wall-bound BLA-CWADYhcRMO2 (Fig. 2, lane 5). In contrast, the ability to anchor BLA-CWADYhcRMO2 covalently to the cell wall (Fig. 2, lane 4; Fig. 3, square) could be restored by introducing yhcS back into the Srt− strain [i.e., WB800Srt−(pE18-YhcS, pWB980-BLA-CWADYhcRMO2)]. No wall-bound BLA-CWADYhcRMO2 could be detected from WB800Srt−(pE18, pWB980-BLA-CWADYhcRMO2) (data not shown). These data indicate that knocking out yhcS abrogates the sorting of BLA-CWADYhcRMO2 to the cell wall and that YhcS is the sortase responsible for anchoring BLA-CWADYhcRMO2 to the peptidoglycan. Furthermore, yhcS is not an essential gene required for the survival of B. subtilis when cells are cultivated in the superrich medium.
Fig 3.
Knockout of yhcS abrogates cell wall anchoring of BLA-CWADYhcRMO2. Western blot analysis of the wall-bound BLA-CWADYhcRMO2 was performed. B. subtilis strains WB800Srt−(pWB980-BLA-CWADYhcRMO2), WB800(pWB980-BLA-CWADYhcRMO2), and WB800Srt−(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) were treated with lysozyme. BLA-CWADYhcRMO2 molecules produced from strain WB800(pWB980-BLA-CWADYhcRMO2) were used as the positive control. S, the supernatant fraction; P, the protoplast pellet. The membrane-bound form of BLA-CWADYhcRMO2 is marked by the open circle. The arrow in lane 1 highlights the loss of the wall-bound form of BLA-CWADYhcRMO2 in strain WB800Srt−(pWB980-BLA-CWADYhcRMO2). The square depicts the presence of wall-bound BLA-CWADYhcRMO2 with the rescue of sortase activity. The amounts of samples loaded were normalized to cell density. Rabbit anti-BLA was used to probe for the reporter fusion with a 1:2,000 dilution.
Sortase levels determine the amounts of BLA-CWADYhcRMO2 anchored to the cell wall.
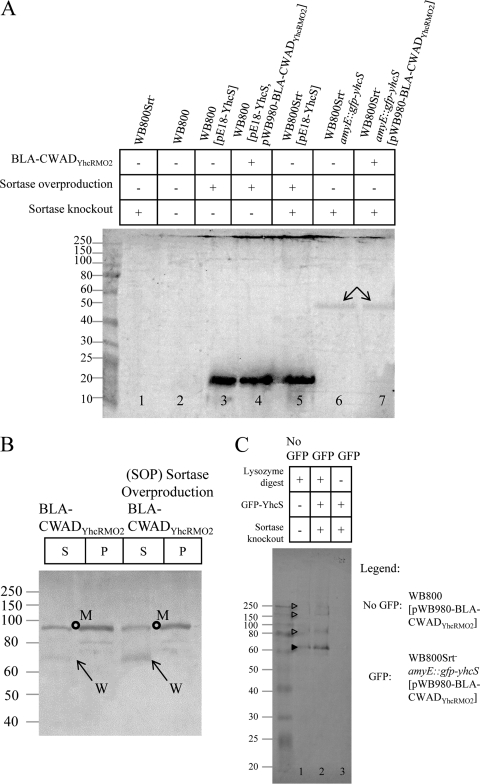
The physiological level of YhcS in B. subtilis WB800 was very low (Fig. 4A, lane 2). Even though the sample was concentrated 6 times, no YhcS could be detected by Western blotting. Introducing the pE18-YhcS plasmid into B. subtilis strains WB800, WB800(pWB980-BLA-CWADYhcRMO2), and WB800Srt− resulted in overproduction of YhcS in these strains. Higher levels of cellular YhcS led to increases in the amounts of the wall-bound BLA-CWADYhcRMO2 (Fig. 4B, arrows; see Fig. S4C in the supplemental material) in WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2). These results show that the sortase level acts as a “bottleneck” step which limits the number of BLA-CWADYhcRMO2 that can be anchored to peptidoglycan.
Fig 4.
Wall-bound BLA-CWADYhcRMO2 levels in relation to the cellular levels of YhcS and YhcS-GFP. (A) Comparison of sortase levels in different B. subtilis strains by using Western blots probed with anti-YhcS antibodies. Cells of B. subtilis strains WB800, WB800Srt−, WB800(pE18-YhcS), WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2), WB800Srt−(pE18-YhcS), WB800Srt− amyE::pxyl-gfp-yhcS, and WB800Srt− amyE:: pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) were treated with lysozyme and sonicated for immunoblotting. Samples from WB800Srt− (lane 1) and WB800 (lane 2) were concentrated six times for this assay. As there were no discernible differences between 0.5 and 2.0% xylose induction, 1% xylose was used to induce GFP-YhcS throughout this experiment. The arrows indicate the GFP-YhcS produced by xylose induction. YhcS (21 kDa) is observed in lanes 3 to 5. The amounts of samples loaded were normalized to cell density. Mouse anti-YhcS was used to probe for the reporter fusion with a 1:750 dilution. (B) Sortase overproduction increases the wall-bound BLA-CWADYhcRMO2 level. Strains WB800(pWB980-BLA-CWADYhcRMO2) (control) and WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) (sortase overproduction strain) were treated with lysozyme. The soluble supernatant fraction (S) of the lysozyme-treated samples and the protoplast fraction (P) are indicated above the gel. BLA-CWADYhcRMO2 levels in these fractions were analyzed by Western blotting. The membrane-bound form (M) (open circle) and the wall-bound form (W) (arrow) of BLA-CWADYhcRMO2 are indicated. The amounts of samples loaded were normalized to cell density. Rabbit anti-BLA was used to probe for the reporter fusion with a 1:2,000 dilution. (C) Detection of BLA-CWADYhcRMO2 from the purified cell wall of B. subtilis WB800Srt− amyE::pxyl-gfp-yhcS. Strain WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) was induced with 1% xylose. Purified cell wall samples from strains WB800(pWB980-BLA-CWADYhcRMO2) and WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) were used for Western blotting. The cell wall samples in lanes 1 and 2 were treated with lysozyme to release BLA-CWADYhcRMO2, and the cell wall sample in lane 3 was not treated with lysozyme. The expected wall-bound form of BLA (filled arrowhead) and the wall-bound BLA anchored to the incompletely digested cell wall (open arrowheads) are indicated.
Immunofluorescence visualization of BLA-CWADYhcRMO2.
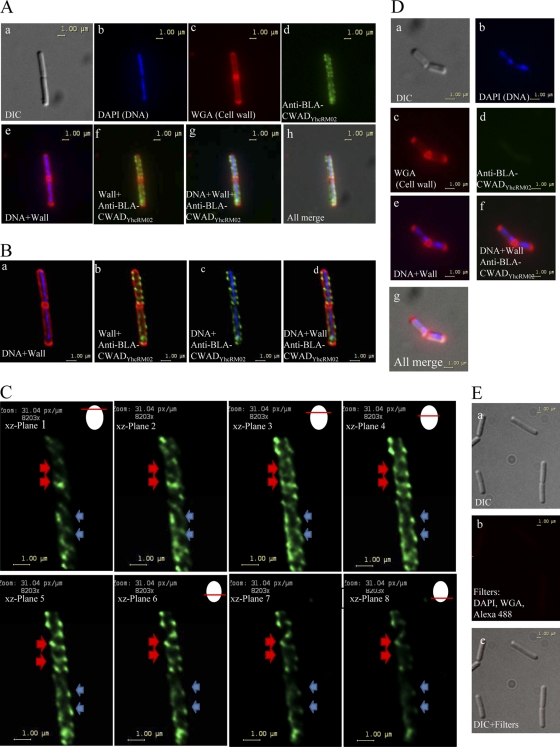
The surface accessibility and distribution profile of wall-bound BLA-CWADYhcRMO2 were examined by using immunofluorescence microscopy. B. subtilis WB800(pWB980-BLA-CWADYhcRMO2) was stained with a DNA-specific fluorescent dye (DAPI [blue]), a peptidoglycan binding agent, Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) (red) and rabbit antibodies against BLA (probed with Alexa Fluor 488-conjugated secondary antibodies [green]) (Fig. 5A, panels a to d). Superimposing the three different colored images suggests that BLA-CWADYhcRMO2 was displayed on the cell surface (Fig. 5A, panels e to h).
Fig 5.
Visualization of BLA-CWADYhcRMO2 on Bacillus subtilis WB800(pWB980-BLA-CWADYhcRMO2) cell surface by immunofluorescence microscopy. (A) Localization of BLA-CWADYhcRMO2 using an epifluorescence microscope. The cells were probed with rabbit anti-BLA antibodies (followed by Alexa Fluor 488-conjugated anti-rabbit antibodies as the secondary antibody [green]), Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) (red), and DAPI (blue). (a) A differential interference contrast (DIC) filter was applied to enhance the contrast of Bacillus subtilis cells for easier visualization. (b to d) Cells stained with DAPI, WGA, or anti-BLA antibodies, respectively. (e to h) Composite images generated by merging images obtained from panels a to d. (B) Application of the deconvolution software to reduce the optical distortion and noise obtained in the epifluorescence microscope image. The image used for this application was the same cell as shown in Fig. 4A. (a to d) Composite images showing the localization of BLA-CWADYhcRMO2 with respect to DNA and cell wall (WGA). (C) Z-stack images of BLA-CWADYhcRMO2 through the same cell in panel A. Eight planes of focus are shown. The red arrows indicate helical filaments observable through slices 3 to 8 (from gradually visible in slices 1 to 3 to optimally visible in slices 4 to 6 to gradually invisible in slices 7 and 8). The blue arrows indicate helical filaments observable through slices 1 to 6. Illustrations showing the current optical focus of the microscope used to observe the different planes of the cell is shown on the upper right corner. (D) WB800(pWB980) cells stained with DAPI for DNA (blue in panel b), Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) (red in panel c), and rabbit anti-BLA antibodies (followed by Alexa Fluor 488-conjugated goat anti-rabbit antibodies, green in panel d). (e to g) Composite images generated by merging images obtained from panels a to d. (E) WB800(pWB980-BLA-CWADYhcRMO2) cells probed with Alexa Fluor 488-conjugated antibodies against rabbit anti-BLA antibodies (green) only without preprobing with the anti-BLA antibodies. (a) DIC image; (b) cells observed with blue, green, and red filters; (c) composite image generated by merging images obtained by using DAPI, WGA, and Alexa Fluor 488.
Interestingly, the images reveal that BLA molecules were distributed nonuniformly in a helical manner along the lateral wall. To acquire clear fluorescence signals and sharper images, z-stacks that were transverse to the short axis of the cell were obtained, and a deconvolution algorithm was applied (Fig. 5B). Examining individual image sections obtained under different z-planes for the green channel (Fig. 5C) reveals a helical pattern of localization of BLA-CWADYhcRMO2 along the cell wall (5, 14, 49). These helices tracked along the entire cell length and were tilted in comparison to the longitudinal axis of the cell (Fig. 5C). The nonuniform distribution of BLA-CWADYhcRMO2 was also observed in B. subtilis WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) (data not shown).
In the control strain, WB800(pWB980), which was stained similarly, no binding of the anti-BLA antibodies was observed (Fig. 5D). Furthermore, WB800(pWB980-BLA-CWADYhcRMO2) cells probed only with Alexa Fluor 488-conjugated rabbit anti-BLA antibodies also did not show any fluorescence under the green, red, or blue channels used (Fig. 5E). All these data demonstrate that (i) BLA-CWADYhcRMO2 molecules were surface accessible and (ii) BLA-CWADYhcRMO2 molecules distribute in a helical manner.
Production of GFP-YhcS and its biological activity.
Since BLA-CWADYhcRMO2 distributed helically on the cell wall, it would be of great interest to determine the distribution of YhcS in B. subtilis. A GFP-YhcS strain (WB800Srt− amyE::pxyl-gfp-yhcS) was constructed. Production of GFP-YhcS in this strain could be observed (see Fig. S3 in the supplemental material) only under induction conditions. In both strains (WB800Srt− amyE::pxyl-gfp-yhcS with or without pWB980-BLA-CWADYhcRMO2), induction with xylose did not reveal any differences in the GFP-YhcS level when the xylose level was varied from 0.5% to 2.0% (Fig. S3B and S3C, lanes 2 to 5 and 7 to 10). However, the amount of GFP-YhcS seemed to be slightly lower in strain WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) than in strain WB800Srt− amyE::pxyl-gfp-yhcS. This is not unexpected, since a portion of the cellular energy is consumed for the production of the BLA fusion in WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2).
To confirm that YhcS is functional in a GFP fusion format, sorting of BLA-CWADYhcRMO2 to the cell wall in B. subtilis WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) was examined. The amount of wall-bound BLA-CWADYhcRMO2 produced from WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) was even more than that produced from WB800(pWB980-BLA-CWADYhcRMO2) (Fig. 4C, lane 2 versus lane 1). This demonstrates that some, if not all, of the GFP-YhcS fusion proteins are active and able to anchor BLA-CWADYhcRMO2 to the peptidoglycan. Higher levels of wall-bound BLA-CWADYhcRMO2 produced from WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) suggests that the xylose pro-moter under induction conditions is stronger than the natural yhcS promoter.
Distribution of the GFP-YhcS sortase in B. subtilis.
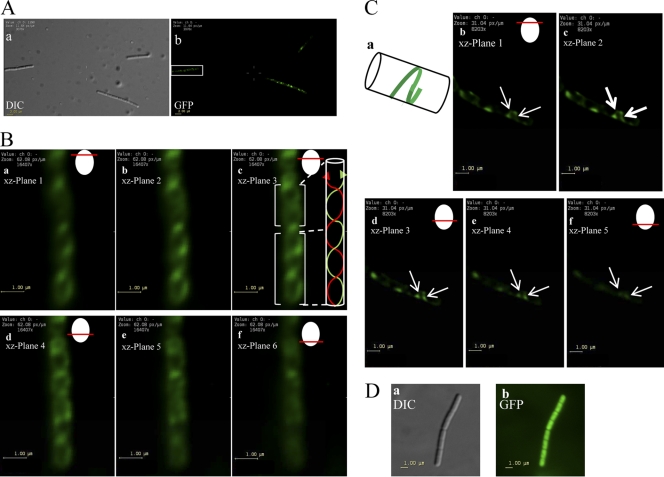
To investigate the spatial distribution of GFP-YhcS, xylose-induced WB800Srt− amyE::pxyl-gfp-yhcS cells were examined by using epifluorescence microscopy. The fluorescent foci of GFP-YhcS were observed to be nonuniformly distributed around the cell periphery in the form of patches or short arcs (Fig. 6). The fluorescence signals in these cells were much more diffuse than those observed for the wall-bound BLA-CWADYhcRMO2.
Fig 6.
GFP-YhcS distribution as observed by fluorescence microscopy. (A) Localization of GFP-YhcS in B. subtilis WB800Srt− amyE::pxyl-gfp-yhcS cultivated in SRM supplemented with 1% xylose. (a) Differential interference contrast (DIC) filter to enhance the contrast in Bacillus subtilis cells for easier visualization. (b) Observation of the same cells viewed with a GFP filter with deconvolution. As z-stacks were obtained for deconvolution, only one optical plane of focus is presented here in panel b. The cell highlighted in the white box was used to show the intertwining ribbons observed under different optical sections (in panel B). (B) Intertwining ribbons of GFP-YhcS discernible by assembling a series of z-stack images (a to f) taken in successive planes following by sharpening using deconvolution. Optical sections of the cell from panel A (white box) was used for deconvolution. Six xz planes were used to show the development of the intertwining ribbons as the image is moved downward in the z plane. An illustration showing the current optical plane of focus used to observe GFP-YhcS in the cell is shown in the top right corner. (Inset in panel c) Illustration showing the direction of ribbons observed in panel c. (C) Helices observed in strain WB800Srt− amyE::pxyl-gfp-yhcS through z-stack images. The cell observed here is a different cell from the one shown in panels A and B. The cells were grown overnight in SRM supplemented with 1% xylose. An illustration showing the current optical plane of focus used to observe GFP-YhcS in the cell is shown in the top right corner. (a) Illustration showing the half-helix observed in the images in panels b to f. (b to f) Z-stack images demonstrating the development of a half-helix as the optical section is moved downwards in the xz plane. The white arrows indicate the development of half-helix through slices 1 to 5. (D) Distribution of GFP in strain WB800Srt− amyE::pxyl-gfp. WB800Srt− amyE::pxyl-gfp cells were grown overnight in SRM supplemented with 1% xylose. The cells were washed twice with PBS before they were spotted onto a microscope slide and observed using the DIC filter and the GFP filter.
To further investigate whether the arcs would track laterally along the membrane, the fluorescent pattern of GFP-YhcS was observed in stacks of optical sections. Two different cells are presented here to illustrate the different helices observed in B. subtilis WB800Srt− amyE::pxyl-gfp-yhcS (Fig. 6B and C). In some cells, some of these structures were observed to consist of tracks that resemble intertwining ribbons (Fig. 6B, panel c) while in other cells, discrete arcs were seen to localize to the cell periphery in different planes (Fig. 6C, panels c to e). The optical sections through successive planes reveal that these helical tracks traverse about half of a helical turn (Fig. 6C, white arrows). These helices were observed to be tilted relative to the long axis of the cell. Although Fig. 6B reveals an intersecting helix, complete helices were rarely detected in most cells. In distinct contrast, the control producing only GFP (WB800Srt− amyE::pxyl-gfp) gives rise to a uniform green fluorescence located throughout the cell's cytoplasm (Fig. 6D). This indicates that GFP by itself does not localize to the cell periphery.
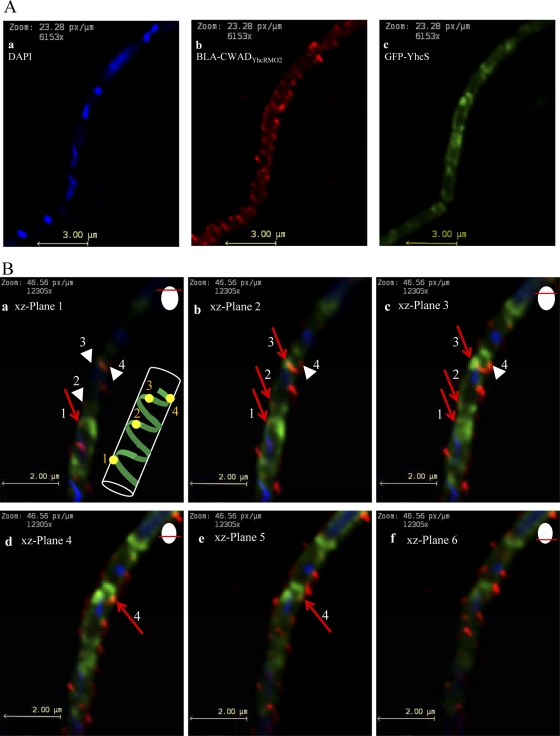
Interaction between BLA-CWADYhcRMO2 and GFP-YhcS is necessary for sorting BLA-CWADYhcRMO2 to the cell surface. Therefore, it is logical to predict that there should be some degree of transient colocalization of BLA-CWADYhcRMO2 with GFP-YhcS. Xylose-induced WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) cells were probed with anti-BLA antibodies for fluorescence visualization studies (Fig. 7A). Some of the BLA-CWADYhcRMO2 signals overlapped with a short segment of a GFP-YhcS arc (Fig. 7B, red arrows). The observed colocalized fluorescent signals from both proteins suggest that some GFP-YhcS and BLA-CWADYhcRMO2 are in close proximity to one another. When viewing progressively along the z-stack plane (i.e., downwards through the cell) (Fig. 7B, panels a to f), colocalization of the two proteins appears to occur progressively (red arrow 1 in Fig. 7B in panel a is followed by red arrows 2 and 3 in panels b and c before the appearance of red arrow 4 in panels d and e). The appearance of these colocalization spots is consistent with the idea that BLA-CWADYhcRMO2 molecules are inserted into the cell wall following the GFP-YhcS path at a given moment.
Fig 7.
Colocalization of BLA-CWADYhcRMO2 with GFP-YhcS observed by fluorescence microscopy. (A) Localization of DNA, BLA-CWADYhcRMO2, and GFP-YhcS using an epifluorescence microscope. WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) cells were probed with rabbit anti-BLA antibodies (followed by Alexa Fluor 568-conjugated goat anti-rabbit antibodies as the secondary antibody) and DAPI. (a to c) Observation of strain WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) with different filters. DNA was observed under the DAPI (blue) filter, BLA-CWADYhcRMO2 was observed under the Alexa Fluor 568 (red) filter, and GFP-YhcS was observed with the GFP filter (green). (B) Z-stacks of a composite image generated by merging individual images obtained from panel A under DAPI (blue), GFP (green), and Alexa Fluor 568 (red) filters. Each composite image represents one optical section of the xz plane as the plane is shifted downwards (panels a to f). Red arrows indicate spots of colocalization (yellow) of BLA-CWADYhcRMO2 and GFP-YhcS seen by overlapping of red- and green-colored regions. White arrowheads indicate the regions where colocalization will develop when the z plane of view is moved downwards through the cell. (Inset panel a) Cartoon illustrating the observed colocalization spots (yellow) which appear progressively in z planes a to f. Yellow spots 1 to 4 in the cartoon correspond to arrows 1 to 4 in the panel.
Comparison of sortase production levels in wild-type and recombinant B. subtilis strains.
To systematically compare the sortase levels of these strains, whole cells from B. subtilis WB800, WB800Srt−, WB800(pE18-YhcS), WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2), WB800Srt−(pE18-YhcS), WB800Srt− amyE::pxyl-gfp-yhcS, and WB800Srt− amyE::pxyl-gfp-yhcS(pWB980-BLA-CWADYhcRMO2) were used in this study. Strains WB800Srt− and WB800 were concentrated six times based on cell density measurement for this study.
No sortase was detected in B. subtilis WB800Srt− (Fig. 4A, lane 1). Unfortunately, no sortase could be detected from strain WB800 either (Fig. 4A, lane 2). Despite repeated efforts to use concentrated cell samples (six times more concentrated) (for WB800Srt−and WB800) and more anti-YhcS antibodies, immunoblotting did not give any detectable signal for these samples. This is likely attributed to a low expression level of yhcS in strain WB800. In comparison, significantly larger amounts of sortase were found in all three strains which had the pE18-YhcS plasmid [i.e., WB800(pE18-YhcS), WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2), and WB800Srt−(pE18-YhcS)]. The levels of sortase detected from these three strains were comparable (Fig. 4A, lanes 3 to 5, a 21-kDa protein band which matches the expected size of YhcS). Interestingly, induction of the single copy of gfp-yhcS under the control of the pxyl promoter produced a larger amount of sortase than was produced from the wild-type sortase gene under the control of its own natural promoter in strain WB800 (Fig. 4A, lanes 6 and 7). Presumably, pxyl is a stronger promoter than the endogenous sortase promoter. The production level of GFP-YhcS (Fig. 4A, lanes 6 and 7) is much lower than the YhcS level from the strains carrying pE18-YhcS (Fig. 4A, lanes 3 to 5). This is not unexpected, since the pE18 vector used in this study has a cop6 mutation that increases the copy number of this plasmid in the host cell (61).
Quantification of the number of the wall-bound BLA-CWADYhcRMO2 in B. subtilis strains.
To quantify the number of BLA-CWADYhcRMO2 molecules that can be displayed on the cell surface, the amounts of BLA molecules anchored to the cell wall in B. subtilis WB800(pWB980-BLA-CWADYhcRMO2) and WB800(pE18-YhcS, pWB980-BLA-CWADYhcRMO2) were estimated by densitometric analysis of immunoblots (see Fig. S4A and S4B in the supplemental material). The numbers of BLA-CWADYhcRMO2 molecules were determined to be 3,770 and 47,300 molecules in strain WB800 and the sortase overproduction strain, respectively (Fig. S4C). This observation indicates that higher levels of BLA-CWADYhcRMO2 can be displayed by increasing the levels of sortase.
Functional activity of BLA-CWADYhcRMO2.
To examine whether the displayed BLA molecules are biologically active, the enzymatic activity of β-lactamase was measured by using whole cells from the following three strains: WB800(pWB980-BLA-CWADYhcRMO2), WB800(pWB980) (negative control), and WB800(pWB980-BLA), a strain that secretes BLA into the growth medium. The background level in the absence of any β-lactamase was also measured by measuring buffer without any cells added. Although extremely low activity (<5 units) was detected in either the buffer or the washed cell fraction of the negative control (WB800 carrying the empty vector), enzymatic activity (∼18 units) from the washed cells of WB800(pWB980-BLA) was significantly higher. This activity could likely be attributed to BLA molecules trapped nonspecifically in the cell wall, as the β-lactamase substrate (PADAC) is small enough to diffuse into the peptidoglycan. In contrast, the enzymatic activity detected from strain WB800(pWB980-BLA-CWADYhcRMO2) was significantly higher with 35 units. Both the wall-bound and membrane-bound forms of BLA-CWADYhcRMO2 can contribute to this observed activity. These data indicate that the β-lactamase reporter fused to CWADYhcRMO2 was biologically active.
DISCUSSION
Characterization of YhcR by Oussenko and coworkers (45) suggests that YhcR is a nuclease that may anchor to the cell wall covalently. In this study, B. subtilis yhcS and the sequence encoding the cell wall anchoring domain of YhcR were characterized. This allows us to present conclusive evidence for the presence of a biologically functional sortase system in B. subtilis with YhcS as the sortase and YhcR as the sortase substrate.
Spiral-like distribution of the sortase substrate YhcR.
Immunofluorescence visualization of BLA-CWADYhcRMO2 on the surface of B. subtilis revealed that the sortase substrate was distributed in helices around the surface of the cell (Fig. 5). A similar observation was also made by Nguyen and Schumann with their engineered B. subtilis surface display system (44). Why do BLA-CWADYhcRMO2 molecules distribute themselves in a helical manner? Previous studies of B. subtilis have suggested that nascent glycan strands are inserted helically (11, 28) into the lateral cell wall at several dispersed sites (1, 29, 54). During cell elongation, these maturing helical glycan cables will develop a rotational torque, allowing it to sweep across the cell surface (6, 7). Since BLA-CWADYhcRMO2 molecules are covalently anchored to the peptidoglycan strands, it is not too unexpected to see the spiral-like distribution of BLA-CWADYhcRMO2.
Short helical arcs of YhcS and implications for cell wall biosynthesis.
As demonstrated in this study, the sorting of BLA-CWADYhcRMO2 to the cell wall is dependent on the catalytic activity of YhcS. It would be of interest to examine the distribution of YhcS for two reasons. First, sortases in cocci (Staphylococcus aureus and Streptococcus pyogenes) are found to localize in specific membrane foci (12, 47). However, the distribution of sortase(s) in rod-shaped bacteria has not been reported. Do sortases distribute randomly or in a highly organized manner? Second, are there any correlations between the distributions of the cell wall-anchored sortase substrates and the sortase? Our results show that GFP-YhcS localizes in certain spatially restricted zones near the lateral cell wall (Fig. 6). Some formed short arcs, helices, or patches. Although the levels of GFP-YhcS detected were higher than the physiological levels of YhcS, GFP-YhcS was not highly overproduced (Fig. 4A, lane 2 versus lanes 6 and 7). Therefore, the observed localization patterns were likely to represent the genuine physiological distribution of sortase in B. subtilis. Some spots in a short segment of the GFP-YhcS showed a certain degree of colocalization with BLA-CWADYhcRMO2 (Fig. 7). These colocalized sortases may be in the process of anchoring BLA-CWADYhcRMO2 to the peptidoglycan strand. We did not observe a long stretch of overlap between the BLA-CWADYhcRMO2 cables and the GFP-YhcS arcs. This may reflect the segregation of the BLA-CWADYhcRMO2 anchored peptidoglycan strands from the sortases. Any initial colocalization would be lost after the transient interactions between the maturing peptidoglycan strands and the sortase arcs during cell wall maturation and cell elongation processes.
The next notable question is how sortase, a membrane protein, would be localized to certain sites on the membrane without lateral diffusion. There are at least three possible models. For the first model, the localization pattern of GFP-YhcS resembles the helical patterns previously observed for a number of proteins such as the cytoplasmic shape-determining cytoskeletal proteins MreB/Mbl/MreBH (13, 24, 27, 31), enzymes (e.g., penicillin binding proteins [PBPs] and teichoic acid biosynthetic enzymes) involved in cell wall synthesis (16, 24, 25, 49, 50) during cell elongation, and the scaffold membrane proteins, MreC and MreD (18, 21, 24, 34, 57), that act as a bridge between the cytoskeletal proteins and the cell wall synthesizing proteins. These proteins assemble to form multienzyme complexes (known as holoenzymes) to mediate cell wall biosynthesis. MreB, MreBH, and Mbl have been suggested to form continuous helical cytoskeletal cables located underneath the cell membrane to guide the movement of the sidewall elongation machineries (including MreC, MreD, RodA, RodZ, and cell wall biosynthetic enzymes such as PBPs) along the sidewall. As sortase is responsible for covalently anchoring cell wall proteins, it is tempting to speculate that YhcS is associated with the cell wall biosynthetic machinery. This can account for the observed distribution of YhcS. Interestingly, new techniques, including the total internal reflection fluorescence microscopy and cryoelectron tomography (19, 26, 56) have recently been applied to study the dynamic distribution of B. subtilis MreB and other components in the sidewall elongation machineries during cell elongation. These findings suggest that MreB, MreBH, and Mbl form short segments or patches which exert a circumferential motion along the cell periphery. The sidewall biosynthetic enzyme complexes also have a similar dynamic motion. Because of the progressive motion of these MreB patches along the cell periphery and the increased depth of field in epifluorescence microscopy, MreB and components in the sidewall biosynthetic machinery could easily be considered to form continuous helical cables in previous studies (6, 24, 27, 31). In our current study, GFP-sortases were found to form short helical arcs in most cases. If YhcS is part of the sidewall biosynthetic enzyme complex, this observation can be explained well by the dynamic circumferential motion model.
For the second model, structures other than the cell wall biosynthetic complexes could also influence the helical distribution of YhcS. Recently, Barák and colleagues (3) have demonstrated that lipid spirals extend along the axis of B. subtilis and that the cell division protein MinD, which is attached to the lipid spirals, also forms spiral structures. Similarly, if YhcS attaches to these spirally distributed lipids, YhcS may distribute helically. Last, the Sec-dependent secretion apparatus in both B. subtilis and E. coli was shown to be distributed helically (5, 52). Evidence for the colocalization of sortase and the Sec apparatus in Streptococcus mutans has also been presented (30). It is possible that YhcS molecules released from the helically distributed Sec apparatus in B. subtilis distribute themselves in a helical manner. In this case, it is unclear how these short helical arcs of YhcS can maintain their distribution in the membrane without lateral diffusion. A combination of these models may also be possible.
Quantification of the wall-bound BLA-CWADYhcR.
Due to the small molecular mass difference, a clean separation of the wall-bound form of BLA-CWADYhcR from the membrane-bound/precursor forms of the reporter fusion by SDS-PAGE would be difficult. As a consequence, it was impossible to conclude that a yhcS knockout strain has been successfully constructed (compare Fig. 1D, lanes 1 and 2, to Fig. 3, lanes 1 and 3) and that YhcS is responsible for anchoring YhcR to the cell wall. Furthermore, the amount of BLA-CWADYhcR covalently linked to the cell wall would be overestimated by at least four- to fivefold (Fig. 1D, lanes 1 and 3). Our approach to overcome these problems by fusing a monomeric protein to the C-terminal tail of BLA-CWADYhcR can be universally applied to other sortase systems if serious quantification of the amounts of covalently wall-bound proteins is needed. The number of wall-bound BLA-CWADYhcRMO2 in this system is approximately 5 times lower than that of the display system reported by Nguyen and Schumann (44). This difference can be attributed to different factors. The system developed by Nguyen and Schumann is based on the sortase from Listeria monocytogenes, the cell wall anchoring domain from the S. aureus fibronectin binding protein B, and the α-amylase reporter from Bacillus amyloliquefaciens. The ability of the wall-bound fusion protein to be resistant to the protease activity from B. subtilis is an important factor in determining the number of molecules that can be successfully displayed. Protease resistance depends on the nature of the domains used in the fusion construction and the length of the linker. Linkers that are too short (94 amino acids) or too long (162 or more amino acids) can reduce the number of molecules that are displayed on the cell surface (44). It is possible that different sortase-based display systems can have a difference in the number of molecules that can be displayed on the surface. The other possible reason may be due to the use of the MO2 fusion approach in this study.
Development of a robust cell surface display system.
We showed that our reporter fusions are functional and surface accessible. The effective pore radius of the B. subtilis peptidoglycan is estimated to be ∼2.1 nm (15). Under these conditions, only globular proteins with molecular masses less than 50 kDa are predicted to be able to penetrate through the peptidoglycan layers. The immunoglobulin G molecules used in this study for the probing of the surface-displayed β-lactamase have molecular masses in the range of 150 kDa. Consequently, they should not be able to penetrate through the peptidoglycan layers. This assumption, in fact, has been demonstrated to be the case in Streptococcus pyogenes (47). Therefore, successful probing of β-lactamase by IgG reflects the surface accessibility of the wall-bound proteins. Successful display of functional enzymes to the microbial cell surface shows increasing promise in developing whole-cell biocatalysts, as our society progresses toward an industrialized and technologically dependent future (60). However, the major challenge of a microbial surface display at the moment is the development of a system that is sufficiently robust for industrial applications (4, 32, 37, 48). Fragility of the outer membrane of Gram-negative bacteria (e.g., E. coli) can be a major concern for surface display. This problem can potentially be circumvented in Gram-positive bacteria with the use of the cell wall as the display platform. There are at least two advantages of the YhcS-YhcR-based surface display system. First, in contrast to the noncovalent surface display systems using the autolysin-based cell wall binding domains for surface display, the CWADYhcR fusions are covalently anchored to the cell surface and will not be easily detached from the cell surface even in an environment with a high external shearing force. Second, the displayed molecules are distributed along the lateral cell wall and not concentrated at the septa. This is a distinct advantage compared to other display systems that are localized at the septa (43). Display of proteins along the lateral cell wall should allow optimal exposure of the displayed enzymes toward the substrate. Furthermore, this system which displays moderate levels of molecules (104 molecules per cell) on the cell surface can potentially be more appropriate for displaying giant enzyme complexes such as cellulosomes. The molecular masses of cellulosomes can be ∼2 × 106 Da or greater (53). Each cellulosome is composed of tens of subunits. Relative to other high-density surface display systems that display 106 to 107 molecules per cell (9, 33), this system may have a better chance to allow the displayed complexes to have sufficient room to interact with their binding targets without imposing steric hindrance between complexes.
Other putative B. subtilis sortase substrates and sortases.
YfkN, a 2′,3′ cyclic nucleotide phosphodiesterase (8), is the other putative sortase substrate with an LPDTA motif, a transmembrane segment, and a positively charged C-terminal tail (8). Several BLA-CWADYfkN fusions were constructed. However, the CWADYfkN domain seemed to be very susceptible to the residual proteases in B. subtilis WB800. Convincing evidence for these BLA-CWADYfkN fusions as the covalently linked wall-bound proteins could not be obtained. Recently, an enzyme known as LPXTGase which competes with sortase to cleave sortase substrates in S. pyogenes and S. aureus has been identified (35, 36). Since the sequence of this enzyme is unknown, it is not clear whether there is a B. subtilis LPXTGase-like enzyme that has a preference to cleave YfkN.
YwpE is the second putative sortase in B. subtilis. The open reading frame for ywpE, in fact, encodes a truncated sortase-like protein. The AUG codon annotated in the SubtiList web server as the initiation codon for this gene is incorrect for two reasons. First, there is no ribosome binding site located upstream of this codon. Even though the mRNA for this gene can be translated, the translated product will not have an N-terminal transmembrane segment to anchor this protein to the membrane as a functional sortase. Second, the suggested N-terminal methionine residue for YwpE is, in fact, a conserved internal methionine in the sortase family. If the coding sequence is extended upstream of this methionine codon, the 28 amino acid residues located immediately upstream of this so-called N-terminal methionine residue can align very well with the conserved sortase sequences. These findings suggest that ywpE is an incomplete gene. B. subtilis WB800 is a derivative of B. subtilis 168. Alignment of B. subtilis 168 YwpE with the translated gene products from the genomes of B. subtilis subsp. spizizenii strain W23, B. subtilis ATCC 6633, Bacillus licheniformis, and B. amyloliquefaciens indicates that there is a YwpE homolog in each of these strains and species. In reference to these YwpE homologs, the B. subtilis 168 version of YwpE misses the first 81- or 82-amino-acid sequence which includes the N-terminal transmembrane segment. Interestingly, the gene next to ywpE in B. subtilis subsp. spizizenii strain W23, B. subtilis ATCC 6633, B. licheniformis, and B. amyloliquefaciens encodes a protein with similarity to collagen binding proteins. This protein has a putative sortase-mediated cell wall anchoring motif. YwpE may be responsible for anchoring this putative collagen binding protein to the cell surface. In B. subtilis 168, this collagen binding protein gene and the coding sequence for the beginning of YwpE are absent. It is possible that this gene segment was deleted during evolution, and the ywpE gene in B. subtilis 168 is likely to be defective.
What is the rationale of immobilizing YhcR and YfkN to the cell surface if these proteins are released into the culture supernatant by proteases from B. subtilis?
Although YfkN cannot be shown to be a covalently linked wall-bound protein in this study, a recent paper (22) provides evidence that YfkN is a covalently linked wall-bound protein. In the aforementioned study, the amount of YfkN in the culture supernatant produced from a sortase mutant cultivated under phosphate starvation condition is two times higher than that produced from the wild-type strain. It is hypothesized that anchoring YfkN to the cell wall minimizes the proteolytic cleavage of the membrane-bound form of YfkN by B. subtilis proteases. Since both YhcR and YfkN can be detected in the culture medium (8, 22, 45), this observation poses an interesting question concerning the purpose of immobilizing these enzymes on the cell surface. Although there is no definite answer to this question, the presence of YhcR and YfkN in both the wall-bound forms and the proteolytic cleaved forms in the culture supernatant can possibly be considered a very elegant strategy employed by B. subtilis. Under phosphate starvation conditions, cells would like to optimize the local phosphate uptake from the immediate surrounding environment as much as possible. B. subtilis might be able to use its cell wall as a scaffold or platform to concentrate and localize YhcR and YfkN. Wall-bound YhcR initiates the cleavage of any available RNA from the local environment into nucleotides. YfkN nucleotidases which are in close proximity can subsequently capture these locally released 2′, 3′, or 5′ nucleotides and hydrolyze them to releases phosphates which can now be easily imported into the cells. Since colocalization of these enzymes on the cell surface can potentially allow RNA to be hydrolyzed to phosphates efficiently for subsequent uptake, it would be important to have these newly synthesized nucleases and nucleotidases immobilized to the cell surface at least transiently. In addition, the combined actions of both YhcR and YfkN in the culture medium allow the release of phosphates from RNA present in the environment. B. subtilis would be able to salvage phosphates from RNA whether these RNA molecules are far away from the cells or close to the cell surface. With the physiological functions of YhcR and YfkN in mind, the low YhcS activity and the inability to detect YhcS by Western blotting when cells were cultivated in a phosphate-rich superrich medium can potentially be explained. The YhcS level may be higher when cells are phosphate starved (22).
Supplementary Material
ACKNOWLEDGMENTS
We thank Lorie Kwang for the construction of pWB980-BLA-CWADYhcR, Chyi-Liang Chen for the purified β-lactamase, Dave Hansen for the use of the Carl Zeiss Axioimager Z1 fluorescence compound microscope, Xiao-Zhou Zhang from Virginia Polytechnic Institute and State University for the recommendation of the marker-free gene knockout system used in this study, Peter J. Lewis from the University of Newcastle for the helpful discussions of some technical aspects of using pSG1729, and the Bacillus Genetic Stock Center for pSG1729, p7S6, and pTSC.
The research work is supported by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Published ahead of print 21 October 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Archibald AR, Hancock IC, Harwood CR. 1993. Cell wall structure, synthesis and turnover, p 381–410 In Sonenshein A, Hoch J, Losick R. (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC [Google Scholar]
- 2. Atrih A, Bacher G, Allmaier G, Williamson MP, Foster SJ. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barák I, Muchová K, Wilkinson AJ, O'Toole PJ, Pavlendová N. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 68:1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benhar I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1–33 [DOI] [PubMed] [Google Scholar]
- 5. Campo N, et al. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583–1599 [DOI] [PubMed] [Google Scholar]
- 6. Carballido-López R, Errington J. 2003. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell 4:19–28 [DOI] [PubMed] [Google Scholar]
- 7. Carballido-López R, Errington J. 2003. A dynamic bacterial cytoskeleton. Trends Cell Biol. 13:577–583 [DOI] [PubMed] [Google Scholar]
- 8. Chambert R, Pereira Y, Petit-Glatron MF. 2003. Purification and characterization of YfkN, a trifunctional nucleotide phosphoesterase secreted by Bacillus subtilis. J. Biochem. 134:655–660 [DOI] [PubMed] [Google Scholar]
- 9. Chen CL, Wu SC, Tjia WM, Wang CLC, Wong SL. 2008. Development of a LytE-based high-density surface display system in Bacillus subtilis. Microb. Biotechnol. 1:177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Comfort D, Clubb RT. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 72:2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 12. DeDent AC, McAdow M, Schneewind O. 2007. Distribution of protein A on the surface of Staphylococcus aureus. J. Bacteriol. 189:4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Defeu Soufo HJ, Graumann PL. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916–1920 [DOI] [PubMed] [Google Scholar]
- 14. Defeu Soufo HJ, Graumann PL. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Demchick P, Koch AL. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. den Blaauwen T, Aarsman ME, Vischer NO, Nanninga N. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539–547 [DOI] [PubMed] [Google Scholar]
- 17. Desvaux M, Dumas E, Chafsey I, Hébraud M. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1–15 [DOI] [PubMed] [Google Scholar]
- 18. Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 102:18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Domínguez-Escobar J, et al. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228 [DOI] [PubMed] [Google Scholar]
- 20. Dramsi S, Trieu-Cuot P, Bierne H. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289–297 [DOI] [PubMed] [Google Scholar]
- 21. Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 102:18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fasehee H, et al. 2011. Functional analysis of the sortase YhcS in Bacillus subtilis. Proteomics 11:3905–3913 [DOI] [PubMed] [Google Scholar]
- 23. Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 25. Formstone A, Carballido-López R, Noirot P, Errington J, Scheffers DJ. 2008. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J. Bacteriol. 190:1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garner EC, et al. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 105:14603–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Höltje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu P, Bian Z, Fan M, Huang M, Zhang P. 2008. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch. Oral Biol. 53:150–154 [DOI] [PubMed] [Google Scholar]
- 31. Jones LJ, Carballido-López R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 32. Kim CJ, Schumann W. 2009. Display of proteins on Bacillus subtilis endospores. Cell. Mol. Life Sci. 66:3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobayashi G, et al. 2000. Accumulation of an artificial cell wall-binding lipase by Bacillus subtilis wprA and/or sigD mutants. FEMS Microbiol. Lett. 188:165–169 [DOI] [PubMed] [Google Scholar]
- 34. Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57:1196–1209 [DOI] [PubMed] [Google Scholar]
- 35. Lee SG, Fischetti VA. 2006. Purification and characterization of LPXTGase from Staphylococcus aureus: the amino acid composition mirrors that found in the peptidoglycan. J. Bacteriol. 188:389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee SG, Pancholi V, Fischetti VA. 2002. Characterization of a unique glycosylated anchor endopeptidase that cleaves the LPXTG sequence motif of cell surface proteins of Gram-positive bacteria. J. Biol. Chem. 277:46912–46922 [DOI] [PubMed] [Google Scholar]
- 37. Lee SY, Choi JH, Xu ZH. 2003. Microbial cell-surface display. Trends Microbiol. 21:45–52 [DOI] [PubMed] [Google Scholar]
- 38. Maresso AW, Schneewind O. 2008. Sortase as a target of anti-infective therapy. Pharmacol. Rev. 60:128–141 [DOI] [PubMed] [Google Scholar]
- 39. Marraffini LA, Dedent AC, Schneewind O. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763 [DOI] [PubMed] [Google Scholar]
- 41. Meissner D, Vollstedt A, van Dijl JM, Freudl R. 2007. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl. Microbiol. Biotechnol. 76:633–642 [DOI] [PubMed] [Google Scholar]
- 42. Mesnage S, et al. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narita J, et al. 2006. Display of alpha-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein, and production of lactic acid from starch. Appl. Environ. Microbiol. 72:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen HD, Schumann W. 2006. Establishment of an experimental system allowing immobilization of proteins on the surface of Bacillus subtilis cells. J. Biotechnol. 122:473–482 [DOI] [PubMed] [Google Scholar]
- 45. Oussenko IA, Sanchez R, Bechhofer DH. 2004. Bacillus subtilis YhcR, a high-molecular-weight, nonspecific endonuclease with a unique domain structure. J. Bacteriol. 186:5376–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pallen MJ, Lam AC, Antonio M, Dunbar K. 2001. An embarrassment of sortases - a richness of substrates? Trends Microbiol. 9:97–102 [DOI] [PubMed] [Google Scholar]
- 47. Raz A, Fischetti VA. 2008. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. U. S. A. 105:18549–18554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Samuelson P, Gunneriusson E, Nygren PA, Ståhl S. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129–154 [DOI] [PubMed] [Google Scholar]
- 49. Scheffers DJ, Jones LJF, Errington J. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749–764 [DOI] [PubMed] [Google Scholar]
- 50. Schirner K, Marles-Wright J, Lewis RJ, Errington J. 2009. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28:830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shaner NC, et al. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shiomi D, Yoshimoto M, Homma M, Kawagishi I. 2006. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 60:894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shoham Y, Lamed R, Bayer EA. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275–281 [DOI] [PubMed] [Google Scholar]
- 54. Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262 [DOI] [PubMed] [Google Scholar]
- 55. Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swulius MT, et al. 2011. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem. Biophys. Res. Commun. 407:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van den Ent F, et al. 2006. Dimeric structure of the cell shape protein MreC and its functional implications. Mol. Microbiol. 62:1631. [DOI] [PubMed] [Google Scholar]
- 58. Wong SL, Doi M. 1986. Determination of the signal peptidase cleavage site in the preprosubtilisin of Bacillus subtilis. J. Biol. Chem. 261:10176–10181 [PubMed] [Google Scholar]
- 59. Wong SL, Kawamura F, Doi RH. 1986. Use of the Bacillus subtilis subtilisin signal peptide for efficient secretion of TEM beta-lactamase during growth. J. Bacteriol. 168:1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu CH, Mulchandani A, Chen W. 2008. Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol. 16:181–188 [DOI] [PubMed] [Google Scholar]
- 61. Wu SC, Wong SL. 1999. Development of improved pUB110-based vectors for expression and secretion studies in Bacillus subtilis. J. Biotechnol. 72:185–195 [DOI] [PubMed] [Google Scholar]
- 62. Wu SC, et al. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yan X, Yu HJ, Hong Q, Li SP. 2008. Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl. Environ. Microbiol. 74:5556–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang TT, Cheng L, Kain SR. 1996. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 24:4592–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.