Abstract
Rieske nonheme monooxygenase 3-ketosteroid 9α-hydroxylase (KSH) enzymes play a central role in bacterial steroid catabolism. KSH is a two-component iron-sulfur-containing enzyme, with KshA representing the terminal oxygenase component and KshB the reductase component. We previously reported that the KshA1 and KshA5 homologues of Rhodococcus rhodochrous DSM43269 have clearly different substrate preferences. KshA protein sequence alignments and three-dimensional crystal structure information for KshAH37Rv of Mycobacterium tuberculosis H37Rv served to identify a variable region of 58 amino acids organized in a β sheet that is part of the so-called helix-grip fold of the predicted KshA substrate binding pocket. Exchange of the β sheets between KshA1 and KshA5 resulted in active chimeric enzymes with substrate preferences clearly resembling those of the donor enzymes. Exchange of smaller parts of the KshA1 and KshA5 β-sheet regions revealed that a highly variable loop region located at the entrance of the active site strongly contributes to KSH substrate preference. This loop region may be subject to conformational changes, thereby affecting binding of different substrates in the active site. This study provides novel insights into KshA structure-function relationships and shows that KSH monooxygenase enzymes are amenable to protein engineering for the development of biocatalysts with improved substrate specificities.
INTRODUCTION
3Ketosteroid 9α-hydroxylase (KSH) is a Rieske-type nonheme oxygenase (RO) that plays a central role in bacterial steroid catabolism and is involved in opening of the steroid B ring (10). Hydroxylated steroids are of industrial and medical interest since many of them are bioactive compounds. 9α-Hydroxylated steroids are of particular interest for the synthesis of corticosteroids (14). KSH is a class IA monooxygenase, a two-component enzyme system comprised of the terminal oxygenase KshA, containing a Rieske Fe2S2 cluster and a nonheme Fe2+ located at the active site, and the reductase KshB, containing a plant-type Fe2S2 cluster and the flavin cofactor flavin adenine dinucleotide (2, 21, 25). Interestingly, KshA and KshB both have been identified to be essential factors in the pathogenicity of Mycobacterium tuberculosis H37Rv (11).
Previously, we reported on the molecular and biochemical characterization of five KshA homologues of Rhodococcus rhodochrous DSM43269. Each of these five kshA genes displayed a unique steroid induction pattern, coding for KshA enzymes with an identical reaction selectivity (they all introduce a hydroxyl moiety at the C-9 position of 3-ketosteroids), but interesting differences in their substrate preferences were observed (21, 22). Empowered with multiple kshA genes, R. rhodochrous DSM43269 may deal in an effective manner with the various sterol/steroid substrates present in its natural habitat (soil), with C9α-hydroxylation occurring at different levels during microbial steroid degradation (22). The KshA5 homologue of DSM43269 (GenBank accession number ADY18328) has a broad substrate range with no apparent preference for any of the tested steroids. In contrast, the KshA1 homologue (GenBank accession number ADY18310) has a narrow substrate range with a high preference for 23,24-bisnorcholesta-4-ene-22-oic acid (4-BNC) and 23,24-bisnorcholesta-1,4-diene-22-oic acid (1,4-BNC) and preference to a lesser extent for 1,4-androstadiene-3,17-dione (ADD) and 4-pregnene-3,20-dione (progesterone) (22) (Tables 1 and 2). In view of the relatively high protein sequence similarity of the KshA1 and KshA5 homologues (61% amino acid identity), their marked differences in substrate preferences may be based on subtle but clearly different structural features. This prompted us to study the structure-function relationships of these KshA enzymes.
Table 1.
Relative initial activities of wild-type and chimeric KshA1 with a range of steroid substrates
| Steroid substrated | Relative initial KSH activitye |
|||||
|---|---|---|---|---|---|---|
| A1c | A1A5β | A1A5loop | A1A5β201-210 | A1D242W | A1A5TG | |
| 4-Androstene-3,17-dione (AD) | 100 | 100 | 100 | 100 | 100 | 100 |
| 1,4-Androstadiene-3,17-dione (ADD) | 244 ± 24 | 132 ± 9a | 171 ± 10a | 149 ± 3a | 307 ± 28 | 274 ± 22 |
| 4-Androstene-17β-ol-3-one (testosterone) | 158 ± 14 | 144 ± 6 | 191 ± 20 | 117 ± 8a | 205 ± 26 | 193 ± 17a |
| 4-Pregnene-3,20-dione (progesterone) | 372 ± 59 | 213 ± 22a | 294 ± 30 | 231 ± 9a | 499 ± 91 | 468 ± 34a |
| 19-Nor-4-androstene-3,17-dione (nordion) | 24 ± 6 | 19 ± 5 | 24 ± 5 | 18 ± 5 | 23 ± 9 | 23 ± 5 |
| 1-(5α)-Androstene-3,17-dione | 23 ± 4 | 22 ± 3 | 25 ± 5 | 13 ± 1 | 19 ± 4 | 14 ± 5 |
| 5α-Androstane-3,17-dione | 12 ± 10 | — | 10 ± 1 | 5 ± 1 | 11 ± 2 | 12 ± 5 |
| 5β-Androstane-3,17-dione | 19 ± 4 | 8 ± 5 | 6 ± 1a | 5 ± 2a | 13 | 8 ± 2a |
| 5-Cholestene-3β-ol (cholesterol)b | — | — | — | — | — | — |
| 5α-Androstane-17β-ol-3-one (stanolon) | — | 7 ± 3a | 5a | 4 ± 1a | 17 ± 2a | 11 ± 4a |
| 3α-Hydroxy-5α-pregnane-20-oneb | — | — | — | — | — | — |
| 11β-Hydrocortisone | — | 7 ± 5a | 7 ± 1a | 15 ± 4a | 13 ± 3a | 10 ± 3a |
| 3β-Hydroxy-5α-androstane-17-one | — | — | — | — | — | — |
| 23,24-Bisnorcholesta-4-ene-22-oic acid (4-BNC) | 548 ± 63 | 177 ± 36a | 241 ± 12a | 229 ± 24a | 704 ± 141 | 953 ± 137a |
| 9α-Hydroxy-4-androstene-3,17-dione (9OHAD) | — | — | — | — | — | — |
P < 0.02 (analysis of variance).
The steroid concentration is 25 μM due to low solubility.
Data taken from Petrusma et al. (22).
For chemical structures of steroids used in this study, see Fig. S1 in the supplemental material.
Relative KSH activities (with standard deviations) compared to activity with AD, which is set at 100% (Table 4). Steroid substrates were tested at a concentration of 200 μM. —, no initial KSH activity detectable.
Table 2.
Relative activities of wild-type and chimeric KshA5 with a range of steroid substrates
| Steroid substrated | Relative KSH activitye |
||||
|---|---|---|---|---|---|
| A5c | A5A1β | A5A1loop | A5A1β207-216 | A5W248D | |
| 4-Androstene-3,17-dione (AD) | 100 | 100 | 100 | 100 | 100 |
| 1,4-Androstadiene-3,17-dione (ADD) | 51 ± 4 | 189 ± 35a | 114 ± 19a | 53 ± 4 | 66 ± 8a |
| 4-Androstene-17β-ol-3-one (testosterone) | 113 ± 20 | 155 ± 19a | 125 ± 13 | 95 ± 7 | 109 ± 7 |
| 4-Pregnene-3,20-dione (progesterone) | 65 ± 1 | 179 ± 37a | 106 ± 5a | 95 ± 21 | 74 ± 9 |
| 19-Nor-4-androstene-3,17-dione (nordion) | 111 ± 18 | 28 ± 5a | 27 ± 4a | 118 ± 7 | 102 ± 10 |
| 1-(5α)-Androstene-3,17-dione | 99 ± 9 | 28 ± 6a | 33 ± 3a | 94 ± 20 | 94 ± 12 |
| 5α-Androstane-3,17-dione | 73 ± 6 | 13 ± 1a | 14 ± 7a | 69 ± 9 | 58 ± 14 |
| 5β-Androstane-3,17-dione | 99 ± 12 | 20 ± 10a | 25 ± 5a | 50 ± 16a | 55 ± 3a |
| 5-Cholestene-3β-ol (cholesterol)b | — | — | — | — | — |
| 5α-Androstane-17β-ol-3-one (stanolon) | 100 ± 10 | — | 16 ± 3a | 95 ± 8 | 57 ± 19a |
| 3α-Hydroxy-5α-pregnane-20-oneb | — | — | — | — | — |
| 11β-Hydrocortisone | 93 ± 9 | 22 ± 5a | 24 ± 11a | 91 ± 10 | 94 ± 7 |
| 3β-Hydroxy-5α-androstane-17-one | — | — | — | — | — |
| 23,24-Bisnorcholesta-4-ene-22-oic acid (4-BNC) | 88 ± 11 | 216 ± 44a | 120 ± 29 | 89 ± 21 | 50 ± 37 |
| 9α-Hydroxy-4-androstene-3,17-dione (9OHAD) | — | — | — | — | — |
P < 0.02 (analysis of variance).
Steroid concentration is 25 μM due to low solubility.
Data taken from Petrusma et al. (22).
For chemical structures of steroids used in this study, see Fig. S1 in the supplemental material.
Relative KSH activities (with standard deviations) compared to activity with AD, which is set at 100% (Table 4). Steroid substrates were tested at a concentration of 200 μM. —, no initial KSH activity detectable.
Several three-dimensional (3D) structures of ROs are currently available (3, 4, 6, 7, 8, 9, 13, 15, 17, 19, 20), including a single 3D structure of KshA, namely, that of M. tuberculosis H37Rv (Rv3526; KshAH37Rv) (2). Although the protein sequences of ROs vary considerably, their tertiary structures are very similar overall. Characteristic for ROs, including KshAH37Rv, is the formation of trimers. These trimers either occur as α3 subunits (2, 4, 17, 19) or may additionally include a smaller β subunit of the oxygenase component, resulting in an (αβ)3 enzyme complex (3, 6, 7, 8, 9, 13, 15, 20). The active site of KshAH37Rv was shown to be composed of a β sheet, including a loop region located at the entrance of the active site, flanked by two α helices. The α helices that flank the β sheet contain residues that coordinate the Fe2+ atom at the core of the active site (2). This so-called helix-grip fold of the active site is a common feature of ROs and part of the StAR (steroidogenic acute regulatory protein)-related lipid transfer (START) domain superfamily (12). Several amino acid residues predicted to be involved in steroid substrate binding were identified in a docking experiment with KshAH37Rv (2). All these residues were located within the helix-grip fold, either in the aforementioned β-sheet or in the flanking α helices.
To gain insight into the structural features responsible for the observed differences in the substrate preference of the homologous KshA enzymes of R. rhodochrous DSM43269, chimeric enzymes of KshA1 (KshA1A5) and KshA5 (KshA5A1) were constructed (Table 3; Fig. 1), and their KSH activities with a range of steroid substrates were analyzed. The results show that a highly variable loop region, located within the β sheet at the entrance of the active site of KshA, strongly contributes to the substrate preference of these KSH enzymes.
Table 3.
Chimeric KshA enzymes used in this study
| Chimeric KshA | Template | Description | Mutation(s) | KSH activitya |
|---|---|---|---|---|
| KshA1A5β | KshA1 | KshA1 with β sheet of KshA5 | kshA1 E198 to V255 exchanged for kshA5 E204 to I261 | + |
| KshA1A5α | KshA1 | KshA1 with α helix of KshA5 | kshA1 I257 to I306 exchanged for kshA5 I263 to V312 | − |
| KshA5A1β | KshA5 | KshA5 with β sheet of KshA1 | kshA5 E204 to I261 exchanged for kshA1 E198 to V255 | + |
| KshA5A1α | KshA5 | KshA5 with α helix of KshA1 | kshA5 I263 to V312 exchanged for kshA1 I257 to I306 | +/− |
| KshA1A5loop | KshA1 | KshA1 with loop region of KshA5 | kshA1 209-QAREDTRPHANGQPKMIGS-227 exchanged for kshA5 215-TGREDVISGTNYDDPNAEL-233 | + |
| KshA1A5TG | KshA1 | KshA1 with 2 aac mutations at the start of the loop | Q209T, A210G | + |
| KshA5A1loop | KshA5 | KshA5 with loop region of KshA1 | kshA5 215-TGREDVISGTNYDDPNAEL-233 exchanged for kshA1 209-QAREDTRPHANGQPKMIGS-227 | + |
| KshA1A5β201-210 | KshA1 | KshA1 with mutations in β sheet | V201T, S203T, F205Y, R207H, G208S, Q209T, A210G | + |
| KshA5A1β207-216 | KshA5 | KshA5 with mutations in β sheet | T207V, T209S, Y211F, H213R, S214G, T215Q, G216A | + |
| KshA1A5β230-238b | KshA1 | KshA1 with mutations in β sheet | D230E, S232T, F238Y | − |
| KshA5A1β236-244 | KshA5 | KshA5 with mutations in β sheet | E236D, T238S, Y244F | − |
| KshA1D242W | KshA1 | KshA1 with 1 aa mutation in β sheet | D242W | + |
| KshA5W248D | KshA5 | KshA5 with 1 aa mutation in β sheet | W248D | + |
Initial KSH activity with AD; −, no initial KSH activity detectable; +, KSH activity detectable (Table 4).
Several cloning attempts were unsuccessful.
aa, amino acids.
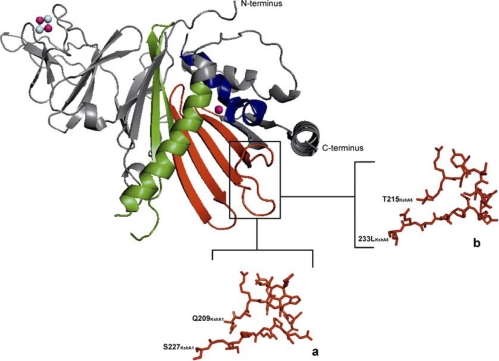
Fig 1.
3D model structure of KshA1DSM43269 depicted as a cartoon. Green, α-helix domain; red, β-sheet domain exchanged between KshA1 and KshA5; purple spheres, iron atoms; light blue, sulfur atoms. The first α helix (located above the nonheme iron indicated in blue), the β sheet (in red), and the second α helix (in green) form the helix-grip fold. The loop region swapped between KshA1DSM43269 (a) and KshA5DSM43269 (b) is enlarged, and the amino acids are shown in sticks.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli cloning strain DH5α (Stratagene) and expression strain E. coli C41(DE3) (18) were grown in Luria-Bertani (LB) medium at 37°C and 220 rpm unless stated otherwise. Ampicillin was added to a final concentration of 100 μg ml−1 when appropriate.
Steroids.
4-Androstene-3,17-dione (AD), 1,4-androstadiene-3,17-dione (ADD), 19-nor-4-androstene-3,17-dione (nordion), 3α-hydroxy-5α-pregnane-20-one, 5α-androstane-17β-ol-3-one (stanolon), 3β-hydroxy-5α-androstane-17-one, and 9α-hydroxy-4-androstene-3,17-dione were obtained from HSD (Oss, The Netherlands). 17β-Hydroxy-4-androstene-3-one (testosterone), 11β-hydrocortisone, 3α-, 7α-, and 12α-trihydroxy-5β-cholan-24-ioc acid (cholic acid), and 5-cholestene-3β-ol (cholesterol) were obtained from Sigma-Aldrich. 4-Pregnene-3,20-dione (progesterone) was obtained from ICN Biomedicals Inc. 1-(5α)-Androstene-3,17-dione (1-5α-AD), 5α-androstane-3,17-dione (5α-AD), 5β-androstane-3,17-dione (5β-AD), and 23,24-bisnorcholesta-4-ene-22-oic acid (4-BNC) were obtained from Steraloids. For chemical structures of the steroids used in this study, see Fig. S1 in the supplemental material.
Construction of chimers of kshA1 and kshA5.
Parts of the kshA1 (nucleotides 591 to 767; encoding peptides E198 to V255) and kshA5 (nucleotides 609 to 785; encoding peptides E204 to I261) genes were exchanged as BstBI/BclI DNA fragments, resulting in chimeric genes kshA1A5β and kshA5A1β. The QuikChange site-directed mutagenesis protocol (Stratagene) was used to introduce BclI sites in kshA1 and kshA5 by making the silent point mutations C768G in kshA1 and C786G in kshA5. The BstBI restriction site in kshA5 was introduced by a G612A silent mutation. For the exchange of the second α helix, BclI/BglII DNA fragments were exchanged between kshA1 (nucleotides 768 to 920; encoding peptides I257 to I306) and kshA5 (nucleotides 786 to 938 of kshA5; encoding peptides I263 to V312), resulting in chimeric genes kshA1A5α and kshA5A1α (Table 3). The PCR mixture contained 4 mM MgSO4, 2% dimethyl sulfoxide, and 2.5 units (50 μl−1) Pfu DNA polymerase (Fermentas) using pKSH814 and pA5rho5 (22) (see Table S1 in the supplemental material) as DNA templates.
The chimeric genes kshA1A5loop, kshA1A5TG, and kshA5A1loop (Table 3) were made via ligase-independent cloning (LIC) (23) using pKsh814 or pA5rho5 (see Table S1 in the supplemental material) as the DNA template for PCR amplification.
The chimeric genes kshA1A5β201-210 and kshA1A5β230-238 (pKSH814 as template) and kshA5A1β207-216 and kshA5A1β236-244 (pA5rho5 as template) were constructed by PCR techniques (Table 3). The forward primers (see Table S2 in the supplemental material) contain a stretch of DNA at the 5′ end (phosphorylated) corresponding to the βstretch donor gene. The PCR mixtures consisted of Tris HCl (10 mM, pH 8.0), 1× polymerase buffer with 1.5 mM MgCl2, MgCl2 (2.5 mM), deoxyribonucleotide triphosphates (0.2 mM), dimethyl sulfoxide (2%), primers (10 ng μl−1), High Fidelity DNA polymerase (0.5 units [25 μl−1]; Fermentas), and 1 pg DNA template under the following conditions: 5 min at 95°C and 30 cycles of 1 min at 95°C, 45 s at 68°C, and 4 min at 72°C, followed by 5 min at 72°C.
Site-directed mutagenesis (QuikChange protocol; see above) was used to generate kshA1D242W and kshA5W248D (Table 3). Primers used in the construction of the chimeric genes are shown in Table S2 in the supplemental material.
For uniformity, throughout the paper all mutant kshA genes are referred to as chimers.
Cloning, heterologous expression of KSH enzymes in E. coli, protein purification, and standard KSH enzyme activity assay.
The chimeric kshA genes were cloned into expression vector pET15b (Novagen) and then subcloned into pET15b containing kshBDSM43269, as described previously (21), to obtain constructs for coexpression of a kshA chimer with kshB. Details of plasmid vectors used are given in Table S1 in the supplemental material. Coexpression of the chimeric KshA enzymes with KshB, His tag purification of the enzymes yielding purified KSH enzymes, and KSH activity assays were performed as described previously (21, 22). The assay mixture (total volume, 500 μl) consisted of 50 mM Tris-HCl buffer (pH 7.0), NADH (105 μM), 25 to 40 μg of KSH enzyme, and 200 μM steroid substrate. NADH consumption over time was recorded with the Soft-max PRO4 (Life Science edition) program. Activities were calculated in nmol min−1 mg−1 of KSH enzyme. Km values (with standard deviations) for the 4-BNC substrate were estimated by determining KSH activity over a range of substrate concentrations (1 to 200 μM). Sigmaplot (version 10.0) was used to process the data using the Michaelis-Menten formula y = (a × x)/(b + x) (21). Total uncoupling of the oxygenase enzyme reaction, which would result in NADH oxidation but not product formation, was checked by high-pressure liquid chromatography–UV or gas chromatography (GC) analysis to confirm product formation (22).
KshA activity, as used in this paper, always refers to the activity of the KSH enzyme (made up of the KshA wild-type or chimeric protein and the KshB protein).
3D modeling of KshA1 and KshA5 of R. rhodochrous DSM43269.
The 3D crystal structure of KshAH37Rv (2) served as template (Protein Data Bank [PDB] accession number 2ZYL) to model the tertiary structures of KshA1 and KshA5 of R. rhodochrous DSM43269. 3D models of both KshA1DSM43269 and KshA5DSM43269 were generated on the SCWRL server (http://www1.jcsg.org/scripts/prod/scwrl/serve.cgi). Alignments were made using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2) and edited manually using Vim (version 7.3.46). PyMOL (The PyMOL Molecular Graphics System, version 1.1; Schrödinger, LLC) was used to produce images.
RESULTS
Construction of chimeric enzymes of KshA1 and KshA5 of R. rhodochrous DSM43269 by exchanging parts of the helix-fold grip.
Our previous work has shown that KshA1 and KshA5 of R. rhodochrous DSM43269 are homologues with highly diverse substrate preferences (22). KshA1 showed relatively high substrate preferences for 4-BNC, ADD, and progesterone (Table 1), whereas KshA5 displayed a broad substrate range (Table 2). We exploited these differences in order to identify KshA regions involved in substrate preference by constructing and characterizing chimeric enzymes of KshA1 and KshA5. To identify regions of interest within the putative substrate binding pocket as targets for exchange between KshA1 and KshA5, we first aligned protein sequences of the 5 KshA homologues of DSM43269 with all the known, e.g., KshAH37Rv (2), and putative KshA protein sequences from databases (a total of 71 KshA amino acid sequences). In addition, we constructed 3D structural models of KshA1 and KshA5 using the 3D crystal structure of KshAH37Rv (2) (Fig. 1). The KshA1 and KshA5 proteins share 60% amino acid identity with KshAH37Rv. The protein sequence alignment (data not shown) and the 3D models of KshA1 and KshA5 revealed that the second α helix of the helix-grip fold (Fig. 1) contains many amino acid residues that vary between KshA1 and KshA5. To investigate whether these variations determine the differences in substrate preference of the KshA homologues, the second α-helix region was exchanged between KshA1 (I257 to I306) and KshA5 (I263 to V312), resulting in the chimeric enzymes KshA1A5α and KshA5A1α (Fig. 1; Table 3).
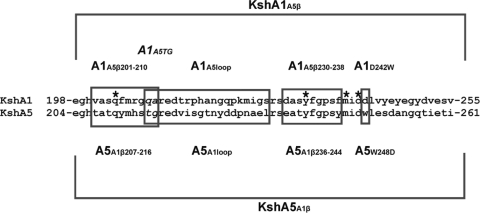
Several putative substrate-interacting residues of KshAH37Rv are located within the β sheet of the helix-grip fold (2). Indeed, this region contains highly conserved amino acid residues, but the alignment also revealed several residues that differ between KshA1 and KshA5 (Fig. 2). To investigate whether this β sheet contributes to the differences in substrate preference of KshA1 and KshA5, the β sheets (58 residues) of KshA1 (E198 to V255) and KshA5 (E204 to I261) were exchanged and chimeric enzymes KshA1A5β and KshA5A1β were constructed (Fig. 2; Table 3).
Fig 2.
Amino acid sequences of the β-sheet regions exchanged between KshA1 and KshA5 resulting in the chimeric enzymes KshA1A5β and KshA5A1β, respectively. The boxed amino acid residues were exchanged between KshA1 and KshA5, resulting in the chimeric enzymes KshA1A5loop, KshA1A5TG (italic), KshA5A1loop, KshA1A5β201-210, KshA5A1β207-216, KshA1A5β230-238, KshA5A1β236-244, KshA1D242W, and KshA5W248D. *, putative substrate-interacting residues (2).
Chimeric KshA enzymes with exchanged helix-grip fold β sheets retain KSH activity.
In order to determine whether the constructed KshA chimers retained KSH activity, the chimeric KshA enzymes were coexpressed with KshBDSM43269 in E. coli C41(DE3), copurified by nickel-nitrilotriacetic acid chromatography, and subsequently assayed for KSH activity using 4-androstene-3,17-dione (AD) as the steroid substrate (Table 3). KshA1A5β and KshA5A1β both clearly possessed KSH activity, indicating that functional chimeric KshA enzymes had been constructed (Tables 3 and 4). In contrast, the chimeric enzyme KshA1A5α did not show any detectable activity, and KshA5A1α showed very low activity only. The molar ratio of KshA5A1α/KshB was determined to be 1:8 ± 0.83, indicating low levels of expression of soluble KshA5A1α enzyme, providing a possible explanation for the low activity observed with KshA5A1α.
Table 4.
Molar ratios of KshA/KshBa and activity of (chimeric) enzymes with AD
| Chimeric KshA | Molar ratio of KshA/KshBb | Exptl Vmax ADb (nmol min−1 mg−1) |
|---|---|---|
| KshA1 | 1:1 ± 0.16 | 261 ± 11 |
| KshA5 | 1:0.59 ± 0.04 | 151 ± 30 |
| KshA1A5β | 1:3.16 ± 0.16 | 336 ± 27 |
| KshA5A1β | 1:1.43 ± 0.03 | 82 ± 11 |
| KshA1A5loop | 1:1.6 ± 0.1 | 366 ± 25 |
| KshA1A5TG | 1:1.27 ± 0.07 | 196 ± 24 |
| KshA5A1loop | 1:1 ± 0.01 | 248 ± 24 |
| KshA1A5β201-210 | 1:1.51 ± 0.11 | 415 ± 40 |
| KshA5A1β207-216 | 1:0.93 ± 0.04 | 421 ± 34 |
| KshA1D242W | 1:1.86 ± 0.04 | 125 ± 20 |
| KshA5W248D | 1:2.6 ± 0.3 | 231 ± 22 |
Determined by densitometry.
Error is standard error of the mean (SEM), n = 3.
Substrate preferences of KshA1A5β and KshA5A1β.
Next, we determined the effects of the exchanged β-sheet regions on the substrate preferences of both KshA1A5β and KshA5A1β by measuring their KSH activity with a range of steroid substrates (Tables 1 and 2). Their enzyme activity levels cannot be directly compared, however, because the KshA/KshB molar ratios in the various purified KSH samples differed (Table 4). KSH enzyme activities with the different substrates are therefore presented as percentages of the activity with AD (set at 100%) and can thus be compared between the different chimers (Table 4).
The exchange of the 58-amino-acid-residue β-sheet region (Fig. 2) had a substantial effect on the substrate preferences of both KshA1 and KshA5. Compared to wild-type KshA1, the preference of KshA1A5β for ADD, progesterone, and 4-BNC was significantly reduced relative to that for AD (Table 1). Interestingly, KshA1A5β activity with 4-BNC was reduced more than 3-fold compared to that of KshA1. Consistent with the effect on the substrate preference of KshA1, the KshA5A1β chimer displayed a higher preference for ADD, progesterone, and 4-BNC than wild-type KshA5, with a 2.5-fold increase in the activity with 4-BNC (Table 2). Also, the KSH activity of KshA5A1β on nordion, 1-5α-AD, 5α-AD, 5β-AD, and hydrocortisone was about 4-fold lower than that of KshA5 (Table 2), more closely resembling the substrate preference displayed by KshA1 (Table 1). Interestingly, KshA5A1β activity with stanolon, a relatively good substrate for KshA5, had become completely abolished. These results show that exchange of the β sheets of the helix-grip fold between different KshA homologues clearly affected their substrate preferences.
KshA1-KshA5 chimeric enzymes with exchanged fragments within the β-sheet region.
To further narrow down the residues potentially involved in KshA substrate preference, smaller regions within the β sheets of KshA1 and KshA5 were exchanged (Fig. 2). Several conserved residues putatively interacting with substrates are located within the β-sheet region (Fig. 2). Amino acid residues that are in close proximity to these putative substrate-interacting residues and which differ between KshA1 and KshA5 were targeted for exchange between these KshA homologues.
Striking is the large loop of 19 amino acid residues (loop region) that interrupts the β sheet and which appears to be poorly conserved in the KshA amino acid sequence (Fig. 1 and 2). KshA1A5loop and KshA5A1loop (Fig. 2) were found to retain KSH activity with AD (Table 4). KshA1A5β201-210 and KshA5A1β207-216 (Fig. 2) were also successfully expressed and displayed KSH activity with AD (Table 4). However, the construction of the plasmid for expression of KshA1A5β230-238 failed repeatedly for unknown reasons, and chimeric enzyme KshA5A1β236-244 did not show any detectable KSH activity.
Residues D242 (KshA1) and W248 were exchanged because these highly different amino acids are situated directly adjacent to the putative substrate-interacting amino acids D241 (KshA1) and D247 (KshA5) (2). The chimeric enzymes KshA1D242W and KshA5W248D (Table 3; Fig. 2) were successfully expressed and displayed KSH activity (Table 4).
The variable loop within the β sheet plays a role in the substrate preference of KshA enzymes.
The chimeric enzyme KshA1A5β201-210 (Fig. 2) displayed an approximately 2-fold decrease in activity with ADD, progesterone, and 4-BNC relative to KshA1 (Table 1). However, KshA5A1β207-216, the chimeric counterpart of KshA1A5β201-210, is comparable to KshA5 in terms of its substrate preference (Table 2). Interestingly, exchange of the 19-amino-acid-residue loop regions (Fig. 1 and 2) affected the substrate preferences in KshA1 and KshA5 in a way similar to that observed with the exchange of the β sheets between these enzymes. KSH activity of KshA1A5loop with ADD and 4-BNC had become reduced about 1.5-fold and >2-fold, respectively, compared to that of KshA1 (Table 1). Also, the KshA5A1loop chimer substrate preference more closely resembled that of KshA1 (Tables 1 and 2). The KSH activity of KshA5A1loop with the preferred substrates of KshA1 (ADD, progesterone) had increased approximately 2-fold. On the other hand, the activity of KshA5A1loop with nordion, 1-5α-AD, 5α-AD, 5β-AD, stanolon, and hydrocortisone was about 4-fold reduced compared to that of KshA1 (Tables 1 and 2). Surprisingly, there was no significant change in KSH activity of KshA5A1loop toward 4-BNC. Overall, these results indicate that the highly variable 19-amino-acid-residue loop region is an important determinant of the substrate preferences of KshA1 and KshA5.
When constructing KshA1A5loop, we accidently obtained a KshA1A5 chimer harboring point mutations Q209T and A210G, located at the entrance of the loop region (KshA1A5TG; Fig. 2). These two point mutations were shown to have intriguing effects on the substrate preference of KshA1. Moreover, KshA1A5TG displayed elevated activity with 4-BNC (almost 2-fold higher than that of wild-type KshA1), progesterone, and testosterone (Table 1).
Exchange of the amino acid residues D242 (KshA1) and W248 (KshA5), located directly adjacent to a putative substrate-interacting residue, did not have a substantial effect on substrate preference. The substrate range of KshA1D242W was not significantly different from that of KshA1, except that this chimeric enzyme had gained low activity with stanolon and hydrocortisone, in contrast to wild-type KshA1 (Table 1). Conversely, KshA5W248D displayed a strongly reduced activity with stanolon compared to wild-type KshA5 (Table 2). KshA5W248D also showed an approximately 2-fold reduced activity with 5β-AD. However, the activity of KshA1D242W with this substrate is similar to that of wild-type enzyme KshA1.
Kinetic analysis of the activity of chimeric KshA enzymes with 4-BNC.
Several chimeric KshA enzymes showed significant differences in KSH activity with 4-BNC compared to the wild-type enzymes (Tables 1 and 2). To investigate whether these differences were due to changes in 4-BNC substrate affinity, kinetic studies were performed to determine the 4-BNC affinity constants of all enzymes. KshA1 and KshA5 both have a Km value of <10 μM with 4-BNC. Similar results were obtained with most of the chimeric KshA enzymes. KshA1A5β and KshA5A1loop have Km values of 14 ± 6 μM and 12 ± 6 μM, respectively. KshA1A5loop shows an elevated Km value of 43 ± 10 μM relative to the wild-type enzyme KshA1. This is in agreement with the finding that the KSH activity of KshA1A5loop with 4-BNC was significantly lower than that of wild-type KshA1 (Table 1). The reduced activity of this chimer with 4-BNC may thus be due to a lowered substrate affinity.
DISCUSSION
Insights into structure-function relationships of KSH enzymes will aid in the engineering of KSH enzymes with higher activities and/or improved substrate specificities. KSH is an interesting biocatalyst for the biotechnological production of 9α-hydroxylated steroids. Moreover, both KshA and KshB are essential for pathogenesis of M. tuberculosis. Detailed insights into the identity of active-site residues interacting with substrates will provide a firm basis for structure-based design of KSH inhibitors.
Using the successful coexpression and copurification protocol previously developed for KshA and KshB of R. rhodochrous DSM43269 (21), we also succeeded in constructing and producing mutant KSH enzymes with chimers of 2 of the 5 KshA homologues (this study). The majority of these KSH enzymes with chimeric KshA proteins displayed KSH activity (Tables 3 and 4), providing a clear scope for engineering of KshA proteins to optimize desirable properties. Interestingly, several of the KSH enzymes with chimeric KshA proteins showed increased activities with AD compared to the native KshA1 or KshA5 proteins (Table 4). However, the KshA/KshB ratios vary between the different KshA chimers (Table 4). These different KshA/KshB ratios may at least partly account for the differences in KSH activities measured (21). Thus, the conclusion that KSH enzymes displaying higher activities indeed are made up of KshA chimers that are more active is not warranted at present. Nevertheless, KSH enzymes with KshA5A1β207-216 and KshA5A1loop have similar KshA/KshB ratios (1:1), but KshA5A1β207-216 has a nearly 2-fold higher activity with AD. Thus, KshA5A1β207-216 clearly provides a more active KSH enzyme than KshA5A1loop.
In order to investigate whether the helix-grip fold β sheet, which forms the basis of the catalytic domain, and the α helix that flanks the β sheet are determinants of the substrate preference in KshA enzymes, these regions were targeted for mutagenesis (Fig. 1). Exchange of the α helix between KshA1 and KshA5 resulted in poorly active or fully inactive chimeric enzymes. Since the α helix contains an Fe2+ binding amino acid (2), we speculate that exchange of the α helix changed the tertiary structure in such a way that the coordination of the Fe2+ atom was altered in these chimers, rendering them (virtually) inactive. Interestingly, exchange of the β sheets yielded active chimeric enzymes KshA1A5β and KshA5A1β (Table 3), displaying significantly altered substrate preferences resembling more the preferences displayed by the β-sheet donor enzyme. The data indicate that this specific helix-grip fold β sheet plays an important role in the substrate preferences of KshA enzymes.
To further pinpoint the identity of residues determining KshA substrate preference, smaller regions within the helix-grip fold β sheet were exchanged between KshA1 and KshA5 (Fig. 2; Table 3). Intriguingly, chimeric KSH enzymes KshA1A5loop and KshA5A1loop (Fig. 2) had significantly altered substrate preferences, similar to KshA1A5β and KshA5A1β (Tables 1 and 2). Similar loop regions have been found at the entrance of the active site of various ROs. Ferraro et al. (5) already speculated that such loops are involved in the accommodation of substrates in the active site. The loop regions are disordered and have high-temperature factors, indicating that they are very flexible. Dicamba monooxygenase (PDB accession number 3GKE) shows the highest structural similarity (3D) with KshA of M. tuberculosis (www.rcsb.org). On the basis of an analysis of the crystal structures of dicamba monooxygenase, D'Ordine et al. (4) suggested that entry of the substrate into the active site is enabled by movement of a helix region and a loop region, resulting in a more open or closed state of the active site. Characterization of the KshA chimeric proteins in the present study thus provides the first experimental evidence that the loop present at the entrance of the active site strongly affects the substrate preference of KSH enzymes.
Analysis of further chimers showed that amino acid differences between the β sheets of KshA1 and KshA5, apart from the variations within the loop region (Fig. 2), do not have strong effects on the substrate preference of KSH enzymes. Nevertheless, these chimeric enzymes also display some significant differences from the wild-type enzymes (Tables 1 and 2). Intriguing is the high substrate preference of KshA1A5TG for 4-BNC and progesterone (Table 1). KshA1 mutant enzymes KshA1A5loop and KshA1A5β201-210 possess the same mutations as KshA1A5TG, in addition to other mutations (Fig. 2; Table 3). However, both chimeric enzymes have a decreased preference for both 4-BNC and progesterone compared to KshA1 (Table 1). Thus, the additional mutations in KshA1A5loop and KshA1A5β201-210 abolish the increase of activity observed for KshA1A5TG with 4-BNC. Mutations Q209T and A210G in KshA1 apparently facilitate a spatial configuration preferred for the accommodation of 4-BNC and, to a lower extent, of progesterone.
In summary, the data presented in this paper show that a loop region at the entrance of the KshA active site, part of the β sheet of the helix-grip fold, strongly affects KSH substrate preference. This loop region may be subject to conformational changes, thereby affecting binding of different substrates in the active site. This study provides novel insights into KshA structure-function relationships and shows that KSH enzymes are amenable to protein engineering for the development of improved substrate specificities.
Supplementary Material
ACKNOWLEDGMENTS
This project was financially supported by The Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (http://www.b-basic.nl) through B-Basic, a public-private NWO (Netherlands Organization for Scientific Research)-ACTS (Advanced Chemical Technologies for Sustainability) program.
We gratefully acknowledge MSD (Oss, The Netherlands) for supporting this project. We thank C. C. van Oosterwijk (Biophysical Chemistry, University of Groningen) for assistance with 3D modeling of KshA1 and KshA5.
Footnotes
Published ahead of print 21 October 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Reference deleted.
- 2. Capyk JK, D'Angelo I, Strynadka NC, Eltis LD. 2009. Characterization of 3 ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J. Biol. Chem. 284:9937–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong X, et al. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J. Bacteriol. 187:2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Ordine RL, et al. 2009. Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation. J. Mol. Biol. 392:481–497 [DOI] [PubMed] [Google Scholar]
- 5. Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175–190 [DOI] [PubMed] [Google Scholar]
- 6. Ferraro DJ, et al. 2007. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct. Biol. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friemann R, et al. 2005. Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J. Mol. Biol. 348:1139–1151 [DOI] [PubMed] [Google Scholar]
- 8. Friemann R, et al. 2009. Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system. Acta Crystallogr. D Biol. Crystallogr. 65:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furusawa Y, et al. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041–1052 [DOI] [PubMed] [Google Scholar]
- 10. Gibson DT, Wang KC, Sih CJ, Whitlock H., Jr 1966. Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms.. J. Biol. Chem. 241:551–559 [PubMed] [Google Scholar]
- 11. Hu Y, et al. 2010. 3-Ketosteroid 9alpha-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 75:107–121 [DOI] [PubMed] [Google Scholar]
- 12. Iyer LM, Koonin EV, Aravind L. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43:134–144 [DOI] [PubMed] [Google Scholar]
- 13. Jakoncic J, Jouanneau Y, Meyer C, Stojanoff V. 2007. The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. FEBS J. 274:2470–2481 [DOI] [PubMed] [Google Scholar]
- 14. Kano S, Tanaka K, Hibino S, Shibuya S. 1979. New synthesis of cortico steroids from 17-ketosteroids: application and stereochemical study of the unsaturated sulfoxide-sulfenate rearrangement. J. Org. Chem. 44:1582–1584 [Google Scholar]
- 15. Kauppi B, et al. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571–586 [DOI] [PubMed] [Google Scholar]
- 16. Reference deleted.
- 17. Martins BM, Svetlitchnaia T, Dobbek H. 2005. 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure 13:817–824 [DOI] [PubMed] [Google Scholar]
- 18. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 19. Nojiri H, et al. 2005. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase. J. Mol. Biol. 351:355–370 [DOI] [PubMed] [Google Scholar]
- 20. Parales RE, et al. 2005. Purification, characterization, and crystallization of the components of the nitrobenzene and 2-nitrotoluene dioxygenase enzyme systems. Appl. Environ. Microbiol. 71:3806–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petrusma M, Dijkhuizen L, van der Geize R. 2009. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 75:5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petrusma M, Hessels GI, Dijkhuizen L, van der Geize R. 2011. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for the specific degradation of different classes of steroids. J. Bacteriol. 193:3931–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tillett D, Neilan BA. 1999. Enzyme-free cloning: a rapid method to clone PCR products independent of vector restriction enzyme sites. Nucleic Acids Res. 27:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reference deleted.
- 25. Van der Geize R, Hessels GI, Van Gerwen R, Van der Meijden P, Dijkhuizen L. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45:1007–1018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.