Abstract
We present the draft genome for the Rickettsia endosymbiont of Ixodes scapularis (REIS), a symbiont of the deer tick vector of Lyme disease in North America. Among Rickettsia species (Alphaproteobacteria: Rickettsiales), REIS has the largest genome sequenced to date (>2 Mb) and contains 2,309 genes across the chromosome and four plasmids (pREIS1 to pREIS4). The most remarkable finding within the REIS genome is the extraordinary proliferation of mobile genetic elements (MGEs), which contributes to a limited synteny with other Rickettsia genomes. In particular, an integrative conjugative element named RAGE (for Rickettsiales amplified genetic element), previously identified in scrub typhus rickettsiae (Orientia tsutsugamushi) genomes, is present on both the REIS chromosome and plasmids. Unlike the pseudogene-laden RAGEs of O. tsutsugamushi, REIS encodes nine conserved RAGEs that include F-like type IV secretion systems similar to that of the tra genes encoded in the Rickettsia bellii and R. massiliae genomes. An unparalleled abundance of encoded transposases (>650) relative to genome size, together with the RAGEs and other MGEs, comprise ∼35% of the total genome, making REIS one of the most plastic and repetitive bacterial genomes sequenced to date. We present evidence that conserved rickettsial genes associated with an intracellular lifestyle were acquired via MGEs, especially the RAGE, through a continuum of genomic invasions. Robust phylogeny estimation suggests REIS is ancestral to the virulent spotted fever group of rickettsiae. As REIS is not known to invade vertebrate cells and has no known pathogenic effects on I. scapularis, its genome sequence provides insight on the origin of mechanisms of rickettsial pathogenicity.
INTRODUCTION
The blacklegged tick Ixodes scapularis (also known as the deer tick) is an obligate blood-feeding vector of prominent medical importance (19, 32), as it propagates the causative agents of several mammalian diseases, such as babesiosis (Babesia microti), Lyme disease (Borrelia burgdorferi), and human granulocytic anaplasmosis (HGA) (Anaplasma phagocytophilum) (1, 27, 78, 80, 91, 96, 103, 110). In addition to transmitting pathogenic microorganisms, ticks (like many other arthropods) carry a wide range of symbiotic organisms (44, 83). Although the main disease transmitted by I. scapularis appears to be Lyme disease and not babesiosis and HGA (111, 129), the ecological relationships within the tick microbiome and the impacts of other microorganisms harbored by the tick, including Bartonella spp. and Rickettsia spp. (57, 80, 94, 99, 112), remain to be elucidated.
The sequence of the I. scapularis genome has been drafted recently (GenBank accession no. NZ_ABJB000000000 GI:255764735). The analysis of the tick genomic DNA revealed the presence of an uncharacterized species of Rickettsia that we describe in the manuscript. Although it was anticipated that I. scapularis genome sequencing and assembly could reveal the presence of new microbes, the uniqueness of the new Rickettsia species was unexpected, providing an invaluable tool for comparative genomics. Little is known about the relationship between I. scapularis and its resident species of Rickettsia in the context of pathogenicity or symbiosis. For that reason, comparative genomic analysis with more than a dozen complete rickettsial genomes (including human pathogens) is important for providing insights into the evolution of pathogenicity and symbiosis.
Rickettsia spp. (class Alphaproteobacteria) are obligate intracellular bacteria with both symbiotic and pathogenic lifestyles. It is well established that rickettsiae exploit a wide range of arthropod and vertebrate hosts, and recent studies have shown a broader metazoan host range for Rickettsia (92), with the host relationship (symbiosis versus pathogenicity) not determined. Robust phylogenomic analyses illustrate the wide diversity of Rickettsia species, which are classified into at least four groups: ancestral group (AG), typhus group (TG), transitional group (TRG), and spotted fever group (SFG) rickettsiae (47, 51). Topical studies of a larger number of rickettsiae based on partial sequence analysis (including one or a few gene regions) suggest further delineations within the Rickettsia tree (17, 122); however, it is widely agreed that the TG, TRG, and SFG rickettsiae are the most derived rickettsiae lineages. Given that these three lineages contain the only known pathogenic species of rickettsiae (45), it has been suggested that vertebrate pathogenicity is a derived condition in Rickettsia (92), a probable consequence of genome degradation tolerated by dependence on host resources (20).
During the past two decades, uncharacterized species of rickettsiae have been detected in various U.S. populations of I. scapularis (11, 78–80, 86, 112, 114). Cytosolic rickettsiae in ovarian tissues of Connecticut populations of I. scapularis were first observed being moderately destroyed by tick lysosomal-like organelles (78). The analysis of partial gene sequences of Rickettsia outer membrane protein A (RompA), citrate synthase (GltA), and 23S and 16S rRNA from ticks collected in Minnesota and Wisconsin suggested that this I. scapularis-associated species is a member of the SFG rickettsiae (86, 123). Not surprisingly, a species of Rickettsia detected in I. scapularis from Texas cross-reacted with an antibody generated from infection with another SFG rickettsia, R. rickettsii, and further sequence analysis (RompA-, GltA-, and 17-kDa antigen-encoding gene regions) prompted the elevation of a novel species epithet, R. cooleyi (11). Interestingly, additional partial GltA sequences generated from various central and eastern U.S. I. scapularis populations have since been deposited at GenBank under the name “R. midichlorii”. However, limited divergence across all available Rickettsia-like sequences generated from I. scapularis populations strongly suggests a single species, with geographic location accounting for this observed diversity (data not shown). Consistent with this observation, an uncharacterized species of rickettsiae collected in Slovakia from the closely related tick, I. ricinus, shares a similar phylogenetic position in the rickettsial tree based on the analysis of 16S rRNA, GltA, and RompA sequences (105).
In the manuscript, we present the first draft of the genome sequence of the Rickettsia endosymbiont of Ixodes scapularis (REIS) with the objective of providing insights into the biology of this poorly characterized species, particularly regarding its relationship with its acarine host. REIS is the most basal member of the SFG rickettsiae, sharing attributes of both pathogenic and symbiotic rickettsiae, and it is the largest Rickettsia genome sequenced to date, with a chromosome size of ∼1.8 Mb that contains 2,059 predicted open reading frames (ORFs). The REIS genome is extraordinary compared to other completed rickettsial genome sequences, in that 35% of its genome encodes products or facilitators of the microbial mobile gene pool, including a Rickettsiales amplified genetic element (RAGE), previously identified as proliferative elements in scrub typhus group (STG) rickettsiae (Orientia tsutsugamushi) genomes, as well as four distinct plasmids. The genetic profile of the REIS genome allows for an assessment of the evolution of pathogenesis across the derived rickettsiae lineages, particularly in light of the acquisition and proliferation of mobile genetic elements (MGEs).
MATERIALS AND METHODS
Genome reconstruction. (i) Summary.
The REIS genome is a hybrid assembly consisting of shotgun sequences from I. scapularis (Wikel colony) bacterial artificial chromosome (BAC) clones and recruited whole-genome shotgun (WGS) reads (accession number ABJB000000000; project 16232). The assembly consists of 20 scaffolds, including the main chromosome and four plasmids (CM000770 to CM000773) plus 16 unplaced (not annotated) scaffolds (GG688301 and GG688316), 9 of which are singletons. The draft chromosome of REIS is a scaffold containing 109 gaps of estimated sizes (derived from mate pair information distances), with the majority of gaps present in regions of multiple transposon insertions. Flanking regions of all gaps were compared to other Rickettsia genomes to determine genes possibly missed in the assembly. Since REIS is the product of 12 laboratory generations of transovarial transmission, wherein ticks may pass only one species of rickettsiae to progeny, it is unlikely that the observed difficulty in closing the genome stems from multiple Rickettsia species or any intraspecific variation within REIS of the Wikel colony. The evidence supporting a single rickettsial species within the I. scapularis Wikel colony is described in more detail in Document S1 in the supplemental material. The total genome coverage is 10.7×.
(ii) Fragment recruitment.
Rickettsia reads were extracted from the full I. scapularis read set by first recruiting reads to known Rickettsia sequences and then using mate pairs to iteratively pull in I. scapularis scaffolds with at least two mate pairs to the recruited read set. All I. scapularis reads were aligned to 10 Rickettsia BAC sequences (BURC, BURE, BURF, BURG, BURH, BURJ, BURQ, BURR, BURU, and BURW) and seven Rickettsia genomes (R. africae, R. bellii, R. conorii, R. felis [including the plasmid rfeplasmid1], R. massiliae, R. prowazekii, and R. typhi) (15) using the snapper aligner (http://kmer.sourceforge.net/) with the following parameters: -mersize 17, -minhitcoverage 0, -minhitlength 17, -minmatchidentity 90, and -minmatchcoverage 50. A total of 10,942 fragments were recruited, with 8,214 fragments recruited to the BAC sequences and 7,310 fragments recruited to the genomes.
(iii) Scaffold recruitment.
The fragment recruitment read set was expanded using an iterative scaffold recruitment process. First, all reads whose mates were in the set were added. Second, a list of I. scapularis scaffolds that contained at least one read in the set was generated. Third, scaffolds with fewer than two fragments in the set were removed from the list. Fourth, all fragments in the remaining list of scaffolds were added to the set. These four steps were repeated four times, after which there was no change in the number of reads in the set. This added 3,152 reads from 136 scaffolds to the set, for a total of 14,094 reads. Increasing the mate requirement for retaining a scaffold did not significantly change the number of reads recruited. The requirement of three mates recruited 88 scaffolds and 14,070 reads. The dramatic drop in scaffold count was caused by all but five of those scaffolds consisting of exactly two reads.
(iv) Assembly.
Reads (recruited WGS fragments and shotgunned BACs) were assembled with Celera Assembler v.4.0 (http://www.jcvi.org/cms/research/projects/cabog) using default parameters, except the unitig genome size (parameter utgGenomeSize) was set to 800 kb to overcome the depth-of-coverage biases resulting in unique sequences being treated as if they were repeats. WGS contigs were combined systematically with BAC shotgun contigs in jumpstart assemblies using the TIGR Assembler (113). WGS contigs were chosen over BAC shotgun contigs when conflicts in orientation occurred. BAC contigs were broken as needed to support the WGS contigs, which represent a better so-called average Rickettsia endosymbiont than individually shotgunned BACs. The assembly was electronically improved through making unrealized joins, correcting misassemblies, and recruiting small contigs and singletons to further extend contigs. PCR was performed to link two chromosome scaffolds, as well as to confirm the circularity of three plasmids. There are remaining ambiguities in the consensus caused by discrepancies in BAC shotgun versus WGS reads, between WGS reads, in repeat regions, and in areas with low quality or low coverage. The current assembly contains 109 contigs linked into one chromosome spanning 1.82 Mb and four confirmed circular plasmids. The chromosome contigs containing both BAC and WGS reads had an average depth of 14.3×, with WGS-only contigs having an average depth of 4.3×. Plasmids contain only WGS reads and range in average coverage from 7.0 to 10.5×.
(v) Consensus quality.
The preliminary annotation of the REIS genome indicated hundreds of potential framshifts and premature stop codons. In an effort to better understand the quality of the consensus, 45 genes with potential frameshifts were manually reviewed, with 27 sites (60%) confirmed as high quality. Additionally, 1 area represented a discrepancy between BAC shotgun and WGS reads, 11 areas were false and were resolved with editing, and 6 areas could not be resolved due to low quality. The low coverage of areas covered only by WGS reads likely causes many of the consensus quality problems, but the fact that 60% of gene defects were confirmed supports that this genome contains many psuedogenes. Support for this was further garnered by comparing all predicted ORFs to homologs in other Rickettsia genomes.
Annotation.
The REIS genome was annotated at the J. Craig Venter Institute (JCVI) using a standard set of processes as described previously (18). In brief, putative protein-encoding genes were identified using the GLIMMER algorithm (98). Predicted proteins were searched against a nonredundant amino acid database. Domains were identified using HMMer with the Pfam (37) and TIGRfam (55) databases. Frame shifts and point mutations that were manually verified were designated authentic. Gene annotation also was performed using the RAST (Rapid Annotation using Subsystems Technology) system (5). At PATRIC (PathoSystems Resource Integration Center) (50, 109), the annotation of all available bacterial genomes (2,865 bacterial genomes as of 1 July 2011) has been standardized using RAST, primarily to afford consistency in comparative genomic analysis. The REIS proteins are included within conserved clusters generated across a selected number of input genomes using FIGfams technology (82). All other automated methods that are standardized across bacterial genomes at PATRIC (e.g., metabolic pathway reconstruction, gene and protein phylogeny estimations, literature compilation, etc.) were applied to the REIS genome as recently described (50), with all pertinent information available at PATRIC (http://www.patricbrc.org/portal/portal/patric/Genome?cType=genome&cId=40569).
The JCVI and PATRIC annotation sets were merged with GSAC (Gene Structure Annotation Comparison; http://www.sequenceontology.org/gff3.shtml), with discrepancies between the two annotation schemes evaluated as previously described (77). The manual assessment of discrepancies also was influenced by referral to other Rickettsia genomes. In a few cases, manual adjustments were made to gene and protein annotations to maintain consistency with other rickettsial genomes. This final gene set was used for all comparative analyses and was submitted to GenBank (accession number NZ_ACLC00000000).
Comparative genomic analysis. (i) Genome alignments.
Six genome sequence alignments were performed using Mauve v.2.0.0 (28). Unmodified Fasta files for each rickettsial genome were used as the input, except that the R. sibirica genome sequence was reindexed using the reverse complement of its circular permutation from the original position 668301 as previously described (51). Repeat densities within and across genomes were evaluated with the Nucmer program in MUMmer v. 3.0 (71).
(ii) Protein family clustering.
OrthoMCL (74) was used to generate orthologous groups (OGs) of protein families from a total of 20,035 predicted proteins across 16 complete Rickettsiaceae genomes: O. tsutsugamushi strain Boryong (NC_009488), O. tsutsugamushi strain Ikeda (NC_010793), R. bellii strain RML369-C (NC_007940), R. bellii strain OSU 85-389 (NC_009883), R. canadensis strain McKiel (NC_009879), R. typhi strain Wilmington (NC_006142), R. prowazekii strain Madrid E (NC_000963), R. felis strain URRWXCal2 (NC_007109), R. akari strain Hartford (NC_009881), REIS, R. massilae strain MTU5 (NC_009897), R. rickettsii strain Sheila Smith (NC_009882), R. rickettsii strain Iowa (NC_010263), R. conorii strain Malish 7 (NC_003103), R. sibirica strain 246 (NZ_AABW01000001), and R. africae strain ESF-5 (NC_012633). From this analysis, proteins were designated unique, core, or nonconserved based on their distribution across these genomes. One genome (R. prowazekii strain P22; CP001584) released subsequently to our analyses was not included in the OG clustering but was included in phylogeny estimations and other comparative analyses. Given its high occurrence of pseudogenes (36), the genome of R. peacockii strain Rustic (NC_012730) also was not included in OG clustering but was included in separate analyses focused on pseudogenization and transposition. The R. peacockii strain Rustic genome also was included in all phylogeny estimations. Finally, sequences of plasmids pREIS1 to pREIS4 were analyzed in a separate OG clustering analysis with plasmid sequences from R. felis strain URRWXCal2 (pRF; NC_007110), R. monacensis strain IrR/Munich (pRM; NC_010927), R. massilae strain MTU5 (pRMA; NC_009900), R. peacockii strain Rustic (pRPR; NC_012732), and R. africae strain ESF-5 (pRAF; NC_012634).
(iii) Genome phylogeny.
For genome-based phylogeny estimation for 46 Rickettsiales taxa, an automated workflow for gene family selection and tree building was implemented through a set of Perl scripts (126). All protein sequences annotated by RAST (5) for 46 Rickettsiales genomes (plus two outgroup taxa) were downloaded from PATRIC. To estimate Rickettsiales phylogeny, BLAT (refined BLAST algorithm) (68) searches were performed to identify similar protein sequences between all genomes, including the two outgroup taxa. To predict initial homologous protein sets, mcl (119) was used to cluster BLAT results, with the subsequent refinement of these sets using in-house hidden Markov models (31). These protein families then were filtered to include only those with membership in >80% of the analyzed genomes (39 or more taxa included per protein family). Multiple-sequence alignment of each protein family was performed using MUSCLE (default parameters) (33, 34), with the masking of regions of poor alignment (length heterogeneous regions) done using Gblocks (default parameters) (21, 116). All modified alignments then were concatenated into one data set. Tree building was performed using FastTree (93). Support for generated lineages was estimated using a modified bootstrapping procedure, with 100 pseudoreplications sampling only half of the aligned protein sets per replication (standard bootstrapping tends to produce inflated support values for very large alignments). Local refinements to tree topology were attempted in instances where highly supported nodes have subnodes with low support. This refinement is executed by running the entire pipeline on only those genomes represented by the node being refined (with additional sister taxa for rooting purposes). The refined subtree then was spliced back into the full tree.
(iv) Additional analyses.
Various analyses were performed on selected protein families, with most analyses, including sequence alignment (as describe above) and phylogeny estimation. In all cases, trees were estimated under parsimony in PAUP v.4.10 (Altivec) (115) and heuristic searches with 500 random sequence additions holding 50 trees per replicate. Singlemost parsimonious trees or consensus trees of equally parsimonious topologies were generated, with branch support assessed using bootstrapping (a similar search strategy). In some cases, data sets also were analyzed under maximum likelihood using Bayesian inference with the program MrBayes v3.1.2 (59, 97). Two independent analyses from different starting seeds were run for three million generations, with samples taken every 1,000 generations throughout each analysis using four Markov chains. All chains were kept at the same temperature, and all branch lengths were saved throughout the analyses with flat priors implemented. For tree building, burn-in values were determined by plotting log likelihoods and tree lengths over sampled generations using the program Tracer v1.4 (http://beast.bio.ed.ac.uk/Tracer). Ultimately, the first 250,000 generations from both analyses were discarded. The estimated sample sizes for all likelihoods, trees, and model parameters from both analyses were determined using Tracer to ensure that the MCMC procedure was effectively sampling from the posterior distribution. Tree files from parsimony and Bayesian analyses were used to draw trees using PAUP.
All amino acid sequence comparisons were based on blastp results. The nr database (including the GenBank, RefSeq Nucleotides, EMBL, DDBJ, and Protein Data Bank databases) was used, coupled with a search against the Conserved Domains Database. Searches were performed across all organisms with composition-based statistics. No filter was used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. The taxonomic distributions of blastp hits were graphically depicted as previously described (46). In some instances, all sequences within alignments were screened for possible signal peptides using SignalP v.3.0 (10) and LipoP v.1.0 (65) servers. Potential transmembrane-spanning regions were predicted using the transmembrane hidden Markov model (TMHMM) v.2.0 server (92).
RESULTS AND DISCUSSION
Genome architecture.
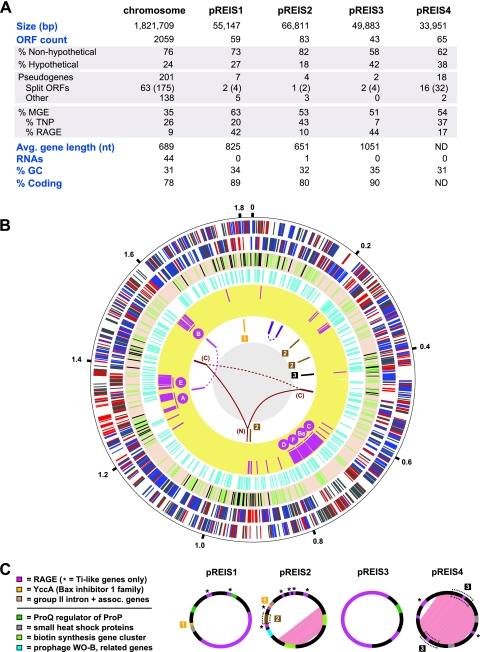
The assembled genome of REIS, consisting of one chromosome and four plasmids, is an estimated 2,027,501 nucleotides (nt) long, with a total of 2,309 predicted open reading frames (ORFs) (Fig. 1A). The proportion of ORFs with assigned functions versus annotations as hypothetical proteins (HP) is consistent with other Rickettsiales genomes. Split genes (84 genes comprised of 217 ORFs) and predicted pseudogenes (148) reflect genome decay that is typical of most Rickettsia genomes (see Table S1 in the supplemental material). However, unlike other Rickettsia genomes, the REIS genome is dominated by ORFs encoding members of the bacterial mobile gene pool, especially transposases (TNPs) and related insertion sequence elements. This characteristic of accessory genome plasticity is primarily what distinguishes REIS from other sequenced Rickettsia genomes, and intriguingly it establishes a novel evolutionary link between it and STG rickettsiae.
Fig 1.
Architecture of the REIS genome. (A) General statistics for the REIS chromosome and four plasmids (pREIS1 to pREIS4). For split genes, total genes are shown with split ORFs in parentheses. (B) Characteristics of the REIS chromosome. The putative origin of replication, positioned at 12 o'clock, was selected based on limited synteny with other SFG genomes (see Fig. S1 in the supplemental material). The outer black circle is a scale with coordinates (in Mb) listed at 200-kb intervals. Six rings inside the scale are the following: plus (1)- and minus (2)-strand genes with core Rickettsia genes colored blue, REIS singletons (not present in other Rickettsiaceae genomes) colored red, and all other genes colored gray; (3) 232 putative pseudogenes colored green (see Table S1 in the supplemental material) and gaps (109 total; see Table S3 in the supplemental material) in the assembly colored black; (4) mobile genetic elements, including transposases, integrases, phage-related ORFs, and other genes typically encoded on plasmids colored turquoise; (5) RAGE genes colored pink, with the location of the seven complete or nearly complete RAGE clusters illustrated (C, Be, F, D, A, E, and B); (6) regions of large-scale duplication and recombination (joined by lines) and regions resulting from plasmid integration (boxed numbers 1 to 3). The ring's six-color scheme is the following (clockwise from the top): purple, duplication of five ORFs and the 6S RNA gene; brown, group II intron and associated genes; black, six-gene insert from pREIS4; burgundy, N-terminal region of rickA (N) joined to the duplicated C-terminal regions (C) (see Fig. S5 in the supplemental material); magenta, four-gene duplication associated with RAGE-A and RAGE-B. (C) Main features of the four REIS plasmids. Duplicated regions within pREIS2 and pREIS4 are depicted with pink shading. Multigene regions (described above) that have been transferred to the chromosome are within dashed boxes. The color scheme depicts genes present on both the chromosome and plasmids (above line) and unique genes of the plasmids discussed in the text.
Regarding stable RNA genes, several features typical of Rickettsia genomes are conserved in REIS: (i) the 5S and 23S rRNA genes are separated from the 16S rRNA gene (2); (ii) the transfer-messenger RNA (tmRNA) gene, which is circularly permuted like other Alphaproteobacteria tmRNA genes (67), has a large intervening sequence between the coding (193 nt) and acceptor regions (74 nt); and (iii) the RNaseP gene has an insertion sequence in the region coding helix P12 of the mature RNA (54). The latter two insertion sequences contain Rickettsia palindromic elements (RPEs) (87), and these regions in REIS are highly divergent from comparable regions in other Rickettsia genomes (81). The genes encoding the signal recognition particle RNA (4.5S RNA) and transcription-associated 6S RNA are conserved as in other Rickettsia genomes, although the latter is present twice, as it is encoded within a duplicated region of the genome that contains five ORFs (coordinates 93946 to 101518 and 143906 to 148622) (Fig. 1B). The REIS genome carries 33 tRNA genes, with a novel duplication of tRNAPhe-GAA, a duplication of tRNAAsp-GTC that it shares only with R. bellii genomes, and deletions of tRNAAla-GGC and tRNATrp-CCA. The two missing tRNA genes likely are present within separate gaps of the assembly (see Table S2 in the supplemental material). Finally, we report the first occurrence of a group II intron in a Rickettsia genome (discussed below), bringing the total number of stable RNA coding genes to 45.
Chromosome.
The assembled REIS chromosome is an estimated 1,821,709 nt long, with a total of 2,059 predicted ORFs (Fig. 1B, first and second rings). Unexpectedly, 29% of the predicted ORFs on the chromosome do not have homologs within other Rickettsia genomes. Because of the nature of the generated bacterial DNA (see Materials and Methods), 109 gaps of estimated sizes averaging 790 nt are present throughout the final scaffold (Fig. 1B, third ring; also see Table S3 in the supplemental material). The effect of these gaps on the calculation of the core Rickettsiaceae and Rickettsia genomes and the determination of the degree of pseudogenization in REIS versus other Rickettsia genomes is discussed below.
An extraordinary 35% of the predicted ORFs on the chromosome can be classified as MGEs, comprised of TNPs, integrases (INTs), phage-related ORFs, and other genes typically located on plasmids (Fig. 1B, fourth ring). A specific MGE, named here RAGE and previously detected in large numbers within the genomes of O. tsutsugamushi strains Boryong (23) and Ikeda (85), alone accounts for 9% of the chromosome (Fig. 1B, fifth ring). While several RAGE components are found scattered throughout the chromosome and typically are associated with TNPs, seven complete (or nearly complete) modules, encoding an F-like type IV secretion system (F-T4SS) among other distinct features, are found on the chromosome in regions of high coverage (labeled in the fifth ring). The excessive proliferation of MGEs, particularly the TNPs and RAGEs, have caused an excess of recombination events throughout the REIS chromosome, resulting in minimal synteny with other Rickettsia genomes (see Fig. S1 in the supplemental material) and a lack of an identifiable GC-skew plot and trinucleotide skew typical of many bacterial genomes (data not shown). Aside from MGEs, several large recombination-mediated duplications occur throughout the chromosome, and duplicate regions across the chromosome and plasmids suggest multiple independent invasions into the former by the latter (Fig. 1B, sixth ring).
Plasmids.
The REIS genome contains four distinct plasmids (pREIS1 to pREIS4) (Fig. 1C), with 72% of encoded ORFs not present in other Rickettsia genomes. While all four of the plasmids contain RAGE components, notably genes encoding TraAITi and TraDTi (Ti indicates the plasmid pTi of Agrobacterium tumefaciens) that process plasmid DNA prior to conjugation, only pREIS1 and pREIS3 encode a complete cluster (RAGE p1 and p3), bringing the total number of complete (or nearly complete) RAGEs to nine in the REIS genome. pREIS1 and pREIS3 also encode ProQ, a regulator of ProP proline/betaine transporters, which are involved in osmoregularity and likely are critical for adaptation to intracellular environments (118, 128). While proP genes are widespread and proliferated in all Rickettsia genomes (51, 88, 95), this is the first occurrence of proQ regulators. Two adjacent ORFs encoding small heat shock proteins, which are found on most rickettsial plasmids (8), are present on all plasmids, except pREIS3, and are duplicated on pREIS4. Interestingly, a homolog of yccA, with an integral membrane gene product that inhibits the FtsH protease and may be involved in the regulation of apoptosis (120), is carried on pREIS1 and pREIS2 and has been recombined to a region on the chromosome (coordinates 1803800 to 1806030). The recombination event has deleted the majority of the yccA homolog on the chromosome, as well as the dihydrodipicolinate reductase gene (dapB) that is involved in the biosynthesis of diaminopimelate (DAP) and lysine. Given that yccA is conserved in all sequenced Rickettsiales genomes, the plasmid copies may be critical for REIS survival. The pseudogenization of dapB suggests that this gene is nonessential in REIS, which is consistent with incomplete pathways for lysine biosynthesis and the ability to synthesize DAP from asparate (without DapB) in other Rickettsia spp.
pREIS2 and pREIS4 encode several multigene regions, comprised mostly of pseudogenes, that have been transferred to various regions of the chromosome. A group II intron, coupled with an associated reverse transcriptase (RT), is present on pREIS2 and three disparate regions on the chromosome (Fig. 1B and C). Additional RTs (as well as genes encoding TraAITi, TraDTi, and TNPs) flank these group II introns, although none of the modules appear to be functional. The entire element shares little similarity with group II introns from STG rickettsiae and Wolbachia spp. and appears to be derived from distant nonproteobacterial genomes (data not shown). A stretch of three ORFs within the large duplication on pREIS4 also has been transferred to the chromosome (Fig. 1B and C). This element is flanked by TNPs and contains genes coding ProP, a stringent response regulator SpoT_H (hydrolase domain), and an S-adenosylmethionine (SAM)-dependent methyltransferase (type 11). Most of these ORFs are split genes and pseudogenes; however, multiple homologs are present throughout the REIS genome and the other Rickettsia genomes, suggesting a continual invasion of these ORFs via MGEs such as conjugative plasmids. It was previously reported that nearly half of the 68 ORFs encoded on the pRF plasmid of R. felis strain URRWXCal2 have homologs on the R. felis chromosome and the chromosomes of other Rickettsia genomes (47). The observations described here further support the incorporation of ORFs from rickettsial plasmids into their resident chromosomes.
pREIS2 LGT.
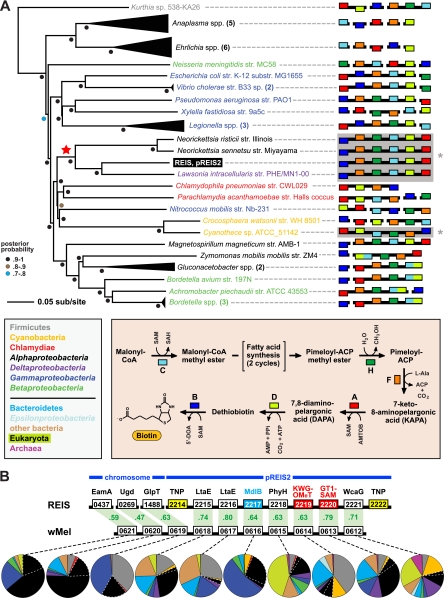
Plasmid pREIS2 contains two extraordinary regions of lateral gene transfer (LGT) that are unknown from other Rickettsia spp. but are recurrent among diverse obligate intracellular bacteria. First, two copies of a cluster of genes encoding all six enzymes involved in the conversion of malonyl-coenzyme A (CoA) to biotin (75) are present within an inverted repeat comprising >25% of pREIS2 (Fig. 1C). This is the first report of an entire biotin (bio) synthesis operon being encoded on a bacterial plasmid, and the orientation of the bio genes is similar only in the genomes of three obligate intracellular bacterial pathogens (Neorickettsia risticii strain Illinois, N. sennetsu strain Miyayama, and Lawsonia intracellularis strain PHE/MN1-00) and the divergent cyanobacterium Cyanothece sp. strain ATCC 51142 (Fig. 2A). Phylogeny estimation strongly supports a recent acquirement of this bio operon in the genomes of REIS, Neorickettsia spp., and L. intracellularis (also see Table S4 in the supplemental material). In the Rickettsiales, strategies for biotin synthesis/acquisition are quite variable (30, 49). Ehrlichia spp. and Anaplasma spp. encode five of the six enzymes (lacking bioH) and likely have an unknown mechanism for the hydrolysis of methyl ester from pimeloyl-ACP (Fig. 2A, inset). In contrast, Wolbachia spp. encode none and Rickettsia spp. encode only one (bioC) of the bio synthesis enzymes; however, a BioY importer is encoded by these genomes, suggesting that the import of host biotin has fostered the deletion of the bio genes. Interestingly, all of the bio synthesis genes, as well as the bioY gene, are absent from the STG rickettsial genomes, which is consistent with the lack of several genes coding for biotin-dependent enzymes (data not shown). Even the biotin ligase-encoding birA gene, which is universal in Rickettsiales genomes, is absent from STG rickettsial genomes. Thus, while a unique bio operon exists in the Rickettsiales mobilome and likely has been incorporated and stabilized into at least the Neorickettsia genomes, the necessity for biotin utilization may not be a defining characteristic of these bacteria. The presence of both bioY and two copies of the bio operon in REIS is staggering, indicating a potential functional redundancy for biotin utilization.
Fig 2.
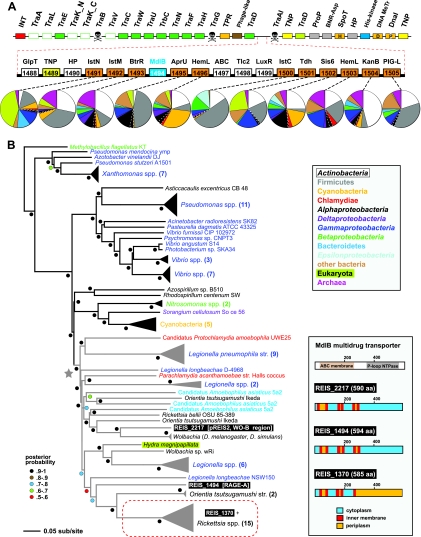
Two regions of lateral gene transfer (LGT) on pREIS2. (A) LGT of a biotin (bio) operon between the obligate intracellular bacteria REIS, Neorickettsia spp., and Lawsonia intracellularis (noted with a red star at the root). Phylogeny was estimated from the concatenation of six bio genes (bioC, bioH, bioF, bioA, bioD, and bioB; see Table S4 in the supplemental material) from 39 diverse bacteria across seven major taxonomic groups (see the color scheme in the inset at the bottom left; only taxonomic groups above the line are represented in the tree). The schema at the right show the gene order and coding strand for all bio genes per genome, with breaks in black bars denoting noncontiguous genes. The contiguous arrays of all six bio genes in REIS, Neorickettsia spp., Lawsonia intracellularis, and Cyanotheca sp. strain ATCC 51142 are shaded and denoted with an asterisk (see the text for further details). Gene color is described in the inset at the bottom right, which illustrates the recent amendment to the biotin synthesis pathway (75). (B) LGT of a 10-gene region between the prophage WO-B of the Wolbachia endosymbiont of Drosophila melanogaster (wMel) and REIS. EamA, S-adenosylmethionine (SAM) transporter; Ugd, UDP-glucose 6-dehydrogenase; GlpT, glycerol-3-phosphate transporter; LtaE, low-specificity l-threonine aldolase; MdlB, ATP-binding multidrug resistance transporter; PhyH, phytanoyl-CoA dioxygenase; KWG-OMeT, N-terminal KWG repeat domain fused to C-terminal O-methyltransferase (type 2) domain; GT1-SAM, N-terminal glycosyltransferase (type1) domain fused to C-terminal radical SAM domain; WcaG, nucleoside-diphosphate-sugar epimerase. The MdlB ORF is colored blue and is included in the estimated phylogeny shown in Fig. 6. The two domain fusion proteins are colored red and were detected as contiguous ORFs in only one other bacterial genome, Haliangium ochraceum (KWG-OMeT, YP_003265293; GT1-SAM, YP_003265292). The two transposases (TNPs) flanking the pREIS2-carried ORFs are colored yellow. Orthology across REIS and wMel genes is shown with green shading, with percent similarity listed for each comparison. Dashed lines connect each gene with a graphical depiction of the top 100 Blastp subjects using the REIS sequences as queries. The taxonomic color scheme is the same as that in the inset in panel A, with taxa listed from top to bottom (Firmicutes to Archeae) depicted clockwise starting at 12 o'clock on the graphs. Dashed white lines distinguish the Rickettsiales subjects from the remaining Alphaproteobacteria. Additional information pertaining to this region is found in Fig. S2 in the supplemental material.
The second LGT event on pREIS2 comprises a sequence of seven genes that are arrayed in the same configuration as in the WO-B prophage of several Wolbachia genomes (Fig. 2B). This region of WO-B originally was predicted to contribute to Wolbachia-host interactions (130). It encodes two divergent l-allo-threonine aldolases (LtaE) involved in glycine biosynthesis, an ATP-binding multidrug resistance transporter (MdlB), a phytanoyl-CoA dioxygenase (PhyH) involved in antibiotic synthesis, a nucleoside-diphosphate-sugar epimerase (WcaG) involved in lipopolysaccharide (LPS) biosynthesis, and two curious ORFs resulting from gene fusions (KWG-OMeT and GT1-SAM). KWG-OMeT consists of an N-terminal KWG repeat domain fused to a C-terminal SAM-dependent O-methyltransferase (type 2) domain, while GT1-SAM comprises an N-terminal glycosyltransferase (type 1) domain fused to a C-terminal radical SAM domain (see Fig. S2 in the supplemental material). LGT recently was proposed for the distribution of these seven genes in REIS and most sequenced WO-B-harboring Wolbachia genomes (60). Our analysis revealed two novel findings further strengthening this claim. First, three of the ORFs in this cluster (PhyH, KWG-OMeT, and GT1-SAM) share strong homology with the halophilic myxobacterium Haliangium ochraceum strain DSM 14365 (Deltaproteobacteria) (42). All three of these ORFs are contiguously arrayed in the H. ochraceum genome (61), and the gene fusions KWG-OMeT and GT1-SAM were not detected in any other organism (see Fig. S2). Second, three adjacent genes in the WO-B prophage also likely have been transferred between REIS and WO-B-harboring Wolbachia genomes: a SAM transporter (EamA), a UDP-glucose 6-dehydrogenase (Ugd) involved in outer membrane and LPS biosynthesis, and a glycerol-3-phosphate (G3P) transporter (GlpT). While these genes are scattered throughout the genomes of REIS and other Rickettsia spp., they share the same degree of homology and a phylogenetic signal with WO-B-harboring Wolbachia spp. similar to that of the genes described above (see Fig. S2). Collectively, the combined 10 genes of this MGE-facilitated cluster have functions in SAM acquisition (EamA) and utilization (KWG-OMeT and GT1-SAM), drug efflux (MdlB) and biosynthesis (PhyH), metabolite scavenging (GlpT) and biosynthesis (LtaE), and cell surface polysaccharide modification (WcaG and Ugd). Three of these LGT products (EamA, Ugd, and GlpT) have become mainstays of Rickettsia spp. genomes (discussed below).
Phylogenomics.
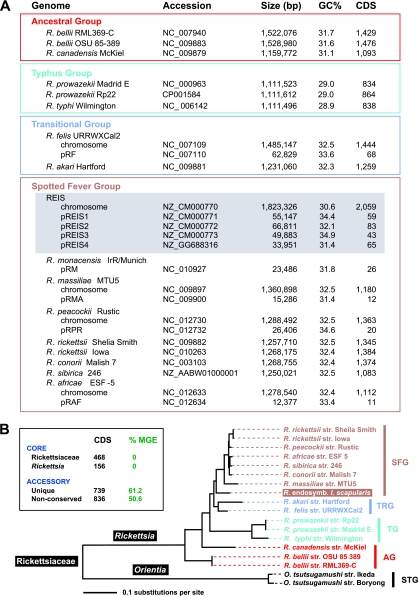
REIS has the largest sequenced genome of the 15 diverse Rickettsia taxa (Fig. 3A). The combined 2,309 ORFs found on the REIS chromosome and four plasmids are 797 more than the next largest Rickettsia genome, R. felis (1,512 ORFs). With the smallest Rickettsia genomes of the typhus group (TG) rickettsiae having an average of 836 ORFs, REIS is exceptional in having an accessory genome of nearly twice the size of TG rickettsial genomes. Despite a large portion of the accessory genome encoding singletons (32%) and MGEs (52%) (Fig. 3B, inset), the base composition of the REIS genome is typical of other Rickettsia genomes. This suggests that LGT products are mostly from organisms with AT biases similar to those of rickettsiae, or that rapid conversion to ∼31% GC has occurred in the majority of transferred MGEs. A slightly higher %GC of the pREIS plasmids is typical of other Rickettsia genomes harboring plasmids and can be attributed to the extreme AT bias of the intergenic regions of the chromosome (data not shown), which comprise an unusually large portion of Rickettsia genomes and likely are graveyards for pseudogenes (3).
Fig 3.
Rickettsia phylogenomic analysis. (A) Genome statistics for 15 complete Rickettsia genomes and the sequenced plasmid of R. monacensis (7). Statistics were computed from the PATRIC web site (50, 109). Rickettsia spp. are classified according to previous studies (47, 51). The REIS chromosome and plasmids are shaded gray. (B) Phylogeny estimated from 191 concatenated protein alignments (see Materials and Methods for details). STG, scrub typhus group; AG, ancestral group; TG, typhus group; TRG, transitional group; SFG, spotted fever group. The inset illustrates brief results of orthologous group (OG) clustering of 2,309 total predicted REIS ORFs across 16 Rickettsiaceae genomes (see Materials and Methods for details). In green, the percentage of mobile genetic elements per OG category is provided. The complete distribution of OGs across all genomes is provided in Fig. S3 in the supplemental material.
Core genome.
Despite this expanded accessory genome that is atypical of Rickettsia spp., REIS shares 468 protein families with 15 Rickettsiaceae genomes and an additional 166 protein families with 13 other Rickettsia genomes (Fig. 3B, inset; also see Fig. S3 in the supplemental material). Eighteen additional protein families were conserved in all sampled Rickettsia genomes except REIS (see Table S2 in the supplemental material). Several of these genes likely are important for rickettsial biology, encoding proteins involved in translation (GatB and RimJ), protein processing (SecE, ClpX, and HtrA), general metabolism (DapB, HemB, andFni), cell envelope biosynthesis (GpsA and RlpA), RNA regulation (HipB and Rbn), and DNA repair (UvrC). The function of the remaining ORFs is unknown (n2B ATPase, DUF2532, COG3671, DUF461, and DUF2674). For 12 of these 18 coding sequences (CDS; those except DUF461, ClpX, RimJ, Rbn, n2B, and Fni), ORFs of variable lengths were detected within the WGS reads using R. rickettsii strain Iowa homologs, suggesting that they were not included in the assembly. However, the insertion of TNPs and larger multigene MGEs, as well as TNP-mediated recombination, could account for the deletion or disruption of all but five of these conserved ORFs (GatB, SecE, HtrA, RlpA, and COG3671; see Table S2 in the supplemental material). If they are true pseudogenes, the determination of possible roles these genes play in the ability of other Rickettsia spp. to invade vertebrate cells and cause host pathogenesis is an important avenue for future research.
Phylogeny.
Robust phylogeny estimation suggests that REIS is the earliest branching lineage of the SFG rickettsiae (Fig. 3B). The REIS genome shares more protein families with R. felis of the TRG rickettsiae than with its closest relative in the SFG rickettsiae, R. massiliae (see Table S5 in the supplemental material), further supporting its phylogenetic position. Most of the duplicate REIS ORFs are MGEs and have homologs in the larger Rickettsia genomes (R. bellii, R. felis, and R. massiliae), suggesting the ongoing transfer of these MGEs or their gradual decay in relation to the more streamlined Rickettsia genomes. The presence of plasmids in these genomes is likely an indication that LGT is an active process sculpting genomic diversity in Rickettsia spp. (8).
Since the initial discovery of a rickettsial plasmid (pRF of R. felis) (90), a diversity of plasmids has been uncovered in various Rickettsia species (6–8). Plasmids also have been identified in many poorly characterized Rickettsia species associated primarily with phytophagous arthropods and nonarthropod hosts (121). However, prior to this report, only the genomes of R. bellii and R. massiliae were known to encode full F-T4SSs likely involved in the spread of rickettsial plasmids. Thus, rickettsial genomes harboring plasmids, yet without a complete F-T4SS, either are incapable of transferring plasmids or use an alternative mechanism for conjugation, such as transduction or the conserved Rickettsiales vir homolog (rvh) P-like T4SS (46, 48). The analysis of common protein families across all four pREIS plasmids, as well as five additional rickettsial plasmids (pRF of R. felis, pRM of R. monacensis, pRMA of R. massiliae, pRPR of R. peacockii, and pRFA of R. africae), identified 40 genes present at least once on multiple rickettsial plasmids (see Table S6 in the supplemental material). The most commonly occurring genes encode proteins involved in plasmid maintenance and stability (parA and dnaA), protein structure maturation (ibpA and hsp), conjugation (traAITi), gene regulation (HTH_Xre family), and gene mobilization (rve; several TNPs and DNA INTs). Of note, pREIS1 and pREIS3 contain two additional RAGEs, suggesting that this MGE is a member of the integrative conjugative element (ICE) family spread by diverse plasmids of the bacterial mobile gene pool (16, 24). Importantly, because several of the plasmid genes are present on the chromosomes of rickettsial genomes, the use of single genes to estimate rickettsial phylogeny requires the careful consideration of the evolutionary trajectory of said genes. The phylogeny estimate we present is robust, in that vertically transmitted genes shared with sampled outgroup proteobacteria were selected for inference (see Fig. S4 in the supplemental material).
REIS shares two signature characteristics of SFG rickettsiae, including a pseudogenized RalF gene and a RickA gene. RalF encodes a eukaryotic Sec7 domain that is known in prokaryotes only from some Rickettsia and Legionella spp. (25). In Legionella, RalF is a characterized Dot/Icm I-like T4SS effector, functioning as a guanine nucleotide exchange factor that recruits ADP ribosylation factor to occupied phagosomes. This process aids Legionella spp. in the ability to replicate free from the host immune system and subvert vesicular trafficking (84). Like other SFG genomes (52), the REIS RalF ORF lacks the N-terminal Sec7 domain and associated central Sec-7-capping domain, containing only the Pro-rich C-terminal tail (data not shown). RickA, a protein considered to activate host Arp2/3 complexes resulting in actin nucleation (52, 64), has been implicated in the intercellular spread of some rickettsiae (58). The protein lacks predicted transmembrane regions and a sec secretion signal (data not shown), yet it is localized to the bacterial surface (53), suggesting that it is secreted via the rvh T4SS (52). The rickA gene in REIS is interrupted by TNP ISPg3 insertion at the C-terminal region of the ORF (see Fig. S5 in the supplemental material). Homologous recombination between similar ISPg3 sequences resulted in a large genome rearrangement separating the C-terminal region from the remaining ORF. This C-terminal fragment was duplicated along with five additional gene remnants. Thus, rickA of REIS likely is inactive, possibly preventing host cell actin polymerization. However, recent studies suggest that rickA expression alone does not suffice for actin polymerization (9, 35), and evidence from host cell actin tail composition suggests that other bacterial factors are involved in intercellular mobilization (56, 106). One of these, rickettsial surface cell antigen 2 (Sca2), recently has been characterized as a formin-like mediator of actin-based motility in R. parkeri (SFG rickettsiae) (56). Like rickA, sca2 of REIS is a pseudogene (K. T. Sears et al., submitted for publication).
Comparison to a nonpathogen.
The recently sequenced genome of R. peacockii strain Rustic provided insight into possible factors involved in rickettsial colonization and pathogenesis of vertebrate host cells (36). R. peacockii, with a lineage close to that of R. rickettsii in the SFG rickettsia (Fig. 3B), causes no known pathogenicity in mammals or its tick (Dermacentor spp.) hosts, and its genome contains 42 copies of the ISRpe1 transposon that have facilitated a loss of synteny with other SFG rickettsiae via extensive ISRpe1-mediated homologous recombination. The ISRpe1 transposon was originally identified in R. peacockii as the possible factor abolishing actin polymerization, as its insertion point truncates rickA (108). We analyzed the characteristics of 50 genes across 15 Rickettsia genomes that were reported as deleted or mutated in the R. peacockii genome (see Fig. S6 in the supplemental material). Less than half of these genes (22) likely were vertically inherited from Alphaproteobacteria ancestors, and only five of the genes are conserved in all other Rickettsia genomes (SAM-methyltransferase [COG0500], SAM-dependent methyltransferase with a tetratricopeptide repeat domain, HP with a COG3264 N-terminal domain, coproporphyrinogen III oxidase, and thioredoxin DsbA). This suggests that the majority of these pseudogenized ORFs in R. peacockii are constituents of the hypervariable Rickettsia accessory genome that is dominated by MGEs and genes for cell surface structures, most of which are either dispensable or involved in species-specific functions with their hosts (43).
Three of the R. peacockii pseudogenes were conserved in all genomes except REIS: n2B ATPase, HP with an N-terminal alanyl-tRNA synthetase domain (HP_N-AlaS), and phosphatidylethanolamine transferase (YhbX) (see Fig. S6 in the supplemental material). In REIS, n2B is deleted by a TNP (mutator family)-mediated recombination event, while a premature stop codon truncated the gene in R. peacockii. HP_N-AlaS is interrupted by IS200 transposon (TNP_17 superfamily) insertion in REIS, while an ISRpe1 transposon insertion deletes part of the gene in R. peacockii. Finally, YhbX, which was suggested to play a role in LPS assembly in Rickettsia (36), contains frameshift mutations in both REIS and R. peacockii genomes. These shared mutations between two bona fide nonpathogenic Rickettsia species lend insight into potential factors involved in rickettsial virulence. Importantly, we note that mutational events affecting these genes have occurred as independent events. Indeed, the analysis of the distribution of the ISRpe1 transposon across all Rickettsia genomes suggests that it is a minor player in the rampant proliferation of MGEs in the REIS genome (see Fig. S7 in the supplemental material). Despite footprints of at least seven transposition events, only one full-length ISRpe1 transposon is encoded in the REIS genome, and the flanking terminal inverted repeat sequence is degraded (data not shown). Collectively, the mechanisms underlying genome shuffling and gene degradation in REIS and R. peacockii likely are evolutionarily independent, although several shared pseudogenes hint at a common inability to establish infection in vertebrate hosts.
Accessory genome.
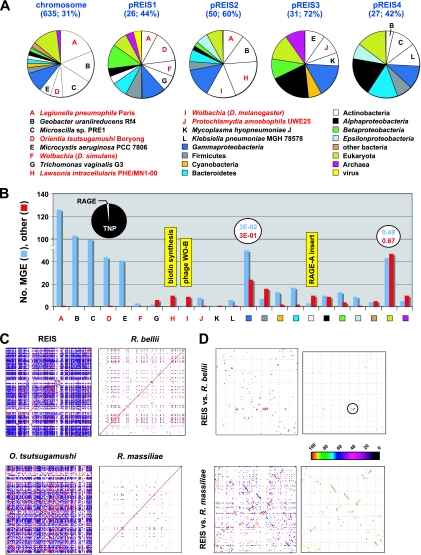
A substantial degree of LGT has shaped the REIS accessory genome (Fig. 4). Of 1,664 REIS proteins of the accessory genome, 46% have closest homologs in non-Rickettsia genomes (Fig. 4A). More than half (54%) of these proteins are most similar to counterparts in one of five distantly related bacterial species, with two of these species having an intracellular lifestyle: Legionella pneumophila strain Paris and O. tsutsugamushi strain Boryong. The larger Rickettsia genomes, namely, R. bellii and R. felis, were demonstrated previously to share genes with Legionella spp. (89, 90), although the amount of similar genes (127 ORFs) shared between REIS and L. pneumophila strain Paris is far greater. The common accessory genes shared between STG rickettsial genomes and REIS is not as unexpected given the close phylogenetic relationship of species from Orientia and Rickettsia; however, the lack of this amount of shared MGEs between Orientia spp. and any other Rickettsia genomes suggests novel LGT events between these genomes (discussed below). The three other bacteria species with a significant number of best blastp hits to REIS proteins of the accessory genome are distantly related taxa: Geobacter uraniireducens strain Rf4 (Deltaproteobacteria), Microscilla sp. strain PRE1 (Bacteroidetes), and Microcystis aeruginosa strain PCC 7806 (Cyanobacteria). Of note, Microcystis aeruginosa genomes are highly plastic and TNP enriched compared to other cyanobacterial genomes (39, 66).
Fig 4.
REIS genome plasticity. (A) Taxonomic breakdown of the best non-Rickettsia Blastp hits (n = 769). Homologs from other Rickettsia genomes were either undetected or had a significantly lower E value than the top five non-Rickettsia subjects. For the chromosome and plasmids, the number of total non-Rickettsia hits is provided, followed by the percentage of these sequences per total ORF count. Taxa A to L depict sequences from genomes that are overrepresented on the chromosome and each plasmid, with taxa colored red depicting intracellular species. General taxonomic divisions are colored accordingly, with other bacteria depicting Aquificae (1), Chloroflexi (3), Deinococcus-Thermus (1), Fusobacteria (1), Spirochaetes (1), and Tenericutes (2). (B) Number of molecular genetic elements (MGEs) and other CDS per taxon or taxonomic category listed in panel A. For taxa A to E, the 414 total MGEs are further divided into transposases and related elements (TNPs, 405) and components of the RAGE (7) as illustrated in the pie chart. Average E values are shown for both the MGEs and other CDS for the Gammaproteobacteria and Eukarya hits, illustrating the low average significance of the latter. Three regions of lateral gene transfer in the REIS genome are mapped to their respective taxon as illustrated in yellow (see the text for details). (C) Plots of sequence repeat density in four select Rickettsiales genomes. Each dot represents a repeat (stretches of similar sequences located in multiple regions of a genome), with blue depicting direct repeats and red depicting reverse repeats. (D) Genome synteny and comparative analysis of repeat density for REIS versus R. bellii strain RML369-C (top) and REIS versus R. massiliae strain MTU5 (bottom). The color scheme for plots at the left is the same as that for panel C, and the color scheme at the right follows the color spectrum depicting percent sequence identity. The high similarity between the RAGE of R. bellii and REIS RAGE-Be is encircled, with the monophyly of these two elements relative to the remaining RAGEs being shown in Fig. 5C.
Distinguishing between accessory genes that encode MGEs and those encoding other proteins reveals a substantial bias toward MGEs, particularly TNPs (Fig. 4B; also see Table S7 in the supplemental material). Nearly 80% of the REIS accessory genome encodes MGEs, and 68% of these MGEs have best blastp hits to the five taxa described above. Only 2 of 414 sequences from these five taxa are non-MGE, and 98% are TNPs (and related derivatives) as opposed to being components of the RAGE (Fig. 4B, pie chart at the left). The most commonly occurring TNPs form four major groups, IS4 (n = 49), IS200-TNP17 (n = 86), IS4-TNP11 (n = 54), and rve-COG3335 (n = 43), with a substantial amount of similarity between most members within each group (see Fig. S8 in the supplemental material). Indeed, roughly half of the REIS TNPs are singletons or duplicated singletons (see Table S8 in the supplemental material), yet given the similarity of these ORFs to homologs in other diverse bacterial species, it is likely that multiple independent invasions occurred in the REIS genome, and that subsequent homologous recombination across proliferated TNP families has facilitated the scrambled gene order in REIS relative to that in other Rickettsia genomes. This is clear in visualizing the density of repetitive sequences per genome, as REIS is much more similar to STG rickettsial genomes in this capacity than to its closer relatives R. bellii and R. massiliae (Fig. 4C).
Of the remaining TNPs with homologs present in other Rickettsiaceae genomes (311), none of the REIS TNPs are found in a conserved distribution (see Table S8 in the supplemental material). These MGEs represent 40% of the total REIS genes in the nonconserved protein families, with genomes of STG rickettsiae, TRG rickettsiae, and R. massiliae encoding substantially more homologs than the remaining Rickettsiaceae genomes (see Fig. S9 in the supplemental material). Thus, there is some phylogenetic correlation, as well as correlation with genome size, underlying TNP abundance in REIS and other rickettsial genomes. This is supported by a greater number of shared syntenic regions between the genomes of REIS and R. massiliae, as opposed to REIS versus R. bellii, for example (Fig. 4D). However, the predominance of shared TNPs between REIS and the STG rickettsial genomes, as well as a recent exchange of RAGEs between REIS and R. bellii (Fig. 4D, encircled region), strongly allude to a highly plastic REIS accessory genome overlaying a typical core rickettsial genome.
RAGE.
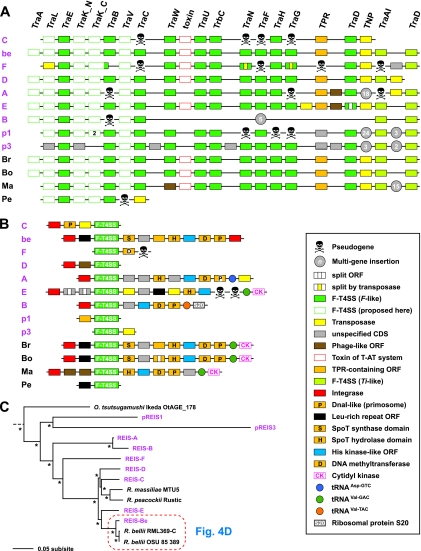
The most unexpected discovery within the REIS genome was the presence of numerous RAGEs. These MGEs originally were identified in the O. tsutsugamushi strain Boryong genome as being essentially two separate units, one comprised of F-T4SS genes (tra) and another composed of tra-associated genes, including those encoding histidine kinase (HK), ankyrin repeat (AR), and tetratricopeptide (TPR) domains (23). While highly proliferated, no complete copy of either unit was identified, and many genes within these units were indicative of pseudogenization. A similar pattern was observed in the O. tsutsugamushi strain Ikeda genome, although the authors used the proliferated elements to reconstruct a hypothetical complete element called OtAGE (O. tsutsugamushi amplified genetic element) (85). The sequence and gene order of the tra genes were similar to those of the F-T4SS clusters encoded by the genomes of R. bellii and R. massiliae; however, the tra-associated genes were noted as being variable across the Orientia and Rickettsia elements. This, combined with several different families of INTs associated with the OtAGE, led Nakayama et al. to hypothesize that multiple invasions of these elements occurred in the Rickettsiaceae genomes, and that these elements are best characterized as members of the ICE family (16, 24).
F-T4SS.
Unlike STG genomes, the REIS genome encodes multiple intact RAGEs (Fig. 5). The structure of the F-T4SS genes throughout the nine largest RAGEs is highly similar to the elements of the STG genomes and R. bellii and R. massiliae (Fig. 5A). The RAGE T4SS genes can best be described as an amalgamation of tra-like genes similar to those encoded on the F plasmid (73) and two genes (traAITi and traDTi) encoded on plasmid Ti of Agrobacterium tumefaciens (127). TraAITi is a multidomain protein containing a TraA-like MobA/MobL DNA mobilization domain coupled with the TraITi-like helicase/nickase domains. TraAITi is thus a relaxase and is likely associated with plasmid transfer in Rickettsia spp., an observation supported by the presence of traAITi on several of the Rickettsia plasmids (see Table S6 in the supplemental material). This large ORF corresponds to two smaller genes in the RAGEs of STG genomes, which to date have not been shown to harbor plasmids. The junction of the F-like and Ti-like conjugative T4SS genes across all RAGEs is highly variable, with the insertion of phage-like, TNPs, and other cassettes (discussed below) being common. The presence on pREIS1 and pREIS3 of two additional RAGE clusters suggests that this MGE is a member of the ICE family (16, 24) and likely utilizes diverse plasmids of the bacterial mobile gene pool for dispersal. Importantly, this is the first report that the whole RAGE can insert and mobilize on rickettsial plasmids.
Fig 5.
Characteristics of the RAGE. (A and B) Schema of gene organization within nine complete (or nearly complete) RAGEs in the REIS genome (pink) and their comparison to the single-copy RAGEs encoded in four other Rickettsia genomes: Br, R. bellii strain RML369-C; Bo, R. bellii strain OSU 85-389; Ma, R. massiliae strain MTU5; Pe, R. peacockii strain Rustic. The genome coordinates of the REIS RAGEs are illustrated in Fig. 1B. Gene color and symbols are described in the inset. (A) Illustration of the RAGE-encoded F-like type IV secretion system (F-T4SS) genes. (B) Illustration of the regions flanking the RAGE F-T4SS genes, including several proposed tRNA insertion sites. (C) Phylogeny estimation of the RAGE genes illustrated in panel A. Nineteen protein families were included in the analysis (excluding TraA pilins and the conserved transposase flanked by TraDF and TraAITi), with pseudogenes not included. Branch support (posterior probabilities) is from Bayesian analysis (see the text for details). Phylogeny estimates of the individual RAGE genes are provided in Fig. S10 in the supplemental material.
F-T4SS associated genes.
Like their STG rickettsial counterparts, the REIS RAGEs contain a variable stretch of genes associated with the F-T4SS genes that flank traDTi (Fig. 5B). Similar genes in this region across all Rickettsiaceae RAGEs include those encoding a DNA methyltransferase (D12 class N6 adenine specific), a PolC-like DNA helicase, stringent response hydrolases (SpoT_H) and synthetases (SpoT_S), and proteins containing HK and TPR domains. Unlike STG genomes, this region in the Rickettsia RAGEs does not encode AR domain-containing genes, and the HK domain-encoding genes are not duplicated in cassettes. Along with RAGE-E of REIS, the RAGEs of R. bellii and R. massiliae inserted into the tRNAVal-GAC gene that flanks the cytidyl kinase-encoding gene (cmk). cmk is deleted from the genomes of STG rickettsiae, and the proliferated nature of the RAGE in these genomes prevented an accurate location of the insertion foci (85). Other tRNA genes of REIS, including tRNAVal-TAC and tRNAAsp-GTC, have served as insertion sites for RAGEs (Fig. 5B), and the presence of at least seven divergent RAGE-associated INT genes suggests multiple invasions of these elements into the genome. The lack of INTs in the two plasmid-encoded RAGEs suggests that these particular elements are not capable of further transmission but likely can contribute to recombination with other chromosomally encoded RAGEs.
RAGE evolution.
The phylogeny estimation of the F-T4SS proteins from 13 Rickettsia RAGEs suggests that the REIS RAGEs are highly divergent and are the products of multiple genomic invasions (Fig. 5C). This is exemplified by REIS RAGE-Be and the RAGEs of R. bellii, which are extraordinarily similar despite the limited colinearity of genes across these genomes (Fig. 4D). A similar pattern of high similarity across the RAGEs of R. massiliae and R. bellii was previously observed and attributed to LGT between these diverse Rickettsia species (14); however, it is evident from our analysis that RAGE-C of REIS is the closest element to R. massiliae, with several other elements (RAGE-D and RAGE-E) separating this group from the highly similar RAGE-Be and the RAGEs of R. bellii (Fig. 5C). Phylogeny estimation of the individual RAGE proteins further suggests multiple genomic invasions of divergent RAGEs (see Fig. S10 in the supplemental material). Counter to this observed divergence across the F-T4SS genes of REIS RAGEs, the F-T4SS genes of the STG rickettsial RAGEs were demonstrated to be highly similar to one another, prompting Cho et al. to propose that gene conversion has kept the proliferated RAGEs static in these genomes (23). These discordant evolutionary pressures on RAGEs encoded in STG genomes versus Rickettsia genomes may reflect their neofunctionalization in the former versus the latter, with a possible role in shaping antigenic variation in scrub typhus populations. There also may be a drive for gene conversion in STG RAGEs to ensure the formation of at least one functional F-T4SS, although the lack of any identified plasmids in O. tsutsugamushi strains doesn't support the need for a functioning conjugative T4SS. Despite the possibility that the REIS RAGEs are mosaics resulting from recombination across the duplicate copies, the observed divergence between different elements and the presence of complete copies on two plasmids leads us to conclude that multiple genomic invasions have occurred (Fig. 5C).
Evolutionary implications.
Surprisingly, the proliferation of MGEs in the REIS genome has had a minimal impact on the inactivation of genes conserved in other Rickettsia genomes (see Table S9 in the supplemental material). Aside from rickA (discussed above), a few exceptions are noteworthy. Several genes encoding components of the rvh P-T4SS, which is highly conserved in Rickettasiales (48), are split or truncated relative to their counterparts in other genomes. Of the 18 rvh T4SS genes conserved in Rickettsia genomes (46), eight are problematic in REIS (rvhB1, rvhB6a, rvhB6b, rvhB6c, rvhB6e, rvhB9b, rvhB10, and rvhD4). Despite being a mainstay in Rickettsia genomes, no substrates of the rvh T4SS have been identified to date, thus the effects of these possible pseudogenes are difficult to predict. gppA (encoding guanosine pentaphosphate phosphohydrolase), a gene flanking the rvh substrate recognition particle (rvhD4) in all Rickettsia genomes, also is pseudogenized in REIS. This likely affects the efficiency of the stringent response in REIS. Two genes encoding metabolic enzymes involved in phospholipid biosynthesis (GpsA) and SAM formation from methionine and ATP (MetK) are pseudogenized in REIS, yet as described above, the genes glpT and eamA (both LGT products) encode transporters for the uptake of these metabolites from the host. Finally, REIS does not encode the repertoire of rickettsial surface cell antigens typical of SFG rickettsial genomes (13). Specifically, relative to other SFG genomes, sca0 (rompA), sca2, and sca5 (rompB) all are pseudogenes, sca4 is truncated with additional copies on plasmids pREIS1 and pREIS3, and various other rickettsial surface cell antigens are partially or entirely encoded or are completely absent (see Table S9 in the supplemental material) (104). Interestingly, a complete Sca5 protein is available in GenBank from a “rickettsial endosymbiont of I. scapularis” (ABQ57416), suggesting either misassembly due to low sequencing coverage or natural variation within the REIS population at the sca5 locus. The generation of additional sequences will be needed to validate these putative pseudogenes, as well as to fortify the species limits and pangenome of REIS.
In addition to the variability in gene content flanking their F-T4SS components, the RAGEs can serve as hotspots for multigene insertions. This was shown originally for the RAGE within the R. massiliae genome, which contains a 15-gene insert between traAITi and traDTi (14). Thus, the RAGE can serve a piggybacking role, acting as a vehicle for the dissemination of the bacterial mobilome. A staggering 18-gene insert in RAGE-A of REIS illustrates this point (Fig. 6). In RAGE-A, the region between the F plasmid-like and Ti plasmid-like F-T4SS genes served as a hot spot for the integration of an element dominated by components typical of Gram-positive aminoglycoside antibiotic biosynthesis (AGAB) gene clusters (107) (Fig. 6A). Eleven genes in this insert are highly similar to those of kanamycin and gentamicin biosynthetic gene clusters (69) found in some Actinobacteria and Firmicutes genomes (see Table S10 in the supplemental material). To our knowledge, this is the first report of such genes within a Rickettsiales genome, and the significance of their insertion into a nonpathogenic Rickettsia species awaits experimental determination. This anomalous insert also contains three internally arrayed genes uncharacteristic of AGAB gene clusters that encode (i) an uncharacterized component of the ABC-type transport system, (ii) one of two divergent type 2-like nucleotide transporters (Tlc-2-like), and (iii) a LuxR-like transcriptional activator (see Fig. S11 in the supplemental material). Importantly, this is the first demonstration in Rickettsia spp. of tlc genes associated with MGEs, and it fortifies the dogma for the dispersal of these genes across diverse intracellular bacteria (101). Also located internally within the AGAB cluster is a gene encoding an uncharacterized multidrug resistance transporter (MdlB) highly homologous to the mdlB gene located within the prophage WO-B of plasmid pREIS2 (Fig. 2B). Including a third mdlB gene conserved in all Rickettsia genomes, this multigene family is present in a diverse array of intracellular bacterial species and has a common origin relative to homologs from extracellular species (Fig. 6B). This strongly suggests that these transporters are important components of the intracellular lifestyle. Finally, a second copy of the G3P importer gene (glpT), which is also a component of WO-B (Fig. 2B), is located upstream of the AGAB genes. This illustrates a stunning commonality in gene content across diverse MGEs of the intracellular mobilome.
Fig 6.
Eighteen-gene insert piggybacking on RAGE-A. (A) The schema at the top depicts RAGE-A, and the color scheme and symbols are explained in Fig. 5. Red dashed lines illustrate an 18-gene insert between traDF (F plasmid-like) and traAITi (Ti plasmid-like). ORFs colored orange have the closest homology to components typical of aminoglycoside antibiotic biosynthesis gene clusters: IstN, SAM-methyltransferase; IstM, NDP-d-glucosaminyltransferase; BtrR, l-glutamine:deoxy-scyllo-inosose (DOI) aminotransferase; AprU, apramycin kinase; HemL, glutamate-1-semialdehyde aminotransferase; IstC, 2-deoxy-scyllo-inosose synthase; Tdh, l-threonine 3-dehydrogenase; Sis6, sugar-alcohol dehydrogenase; TbmB, l-glutamine:DOI aminotransferase; PIG-L, GlcNAc-PI de-N-acetylase. Dashed lines connect each gene with a graphical depiction of the top 100 Blastp subjects using the REIS sequences as queries. The taxonomic color scheme follows the inset at the bottom, with taxa listed from top to bottom (Actinobacteria to Archeae) depicted clockwise starting at 12 o'clock on the graphs. Dashed white lines distinguish the Rickettsiales subjects from the remaining Alphaproteobacteria. The top five Blastp hits for each of these ORFs are listed in Table S10 in the supplemental material. The MdlB ORF is colored blue and included in the estimated phylogeny in panel B. An associated transposase (TNP) is colored yellow. Additional ORFs are GlpT, glycerol-3-phosphate transporter (see Fig. 2B); HP, hypothetical protein; ABC, uncharacterized ABC transporter (COG4178); Tlc, ADP/ATP translocase (rickettsial type 2 Tlc); and LuxR, HTH transcriptional activator. Additional information pertaining to ABC, Tlc, and LuxR is provided in Fig. S11 in the supplemental material. (B) Estimated phylogeny of an uncharacterized multidrug resistance transporter, MdlB (EC 3.6.3.44). A gray star depicts the monophyly of mdlB genes within diverse intracellular bacterial species, with these lineages colored gray (except for the eukaryote Hydra magnipapilata). The three REIS ORFs are boxed, with a red dashed box enclosing the ortholog conserved in all Rickettsia genomes. The predicted membrane association of all three REIS orthologs is shown at the right, with TMS regions depicted using TMHMM v.2.0 (70).
While rampant invasion by MGEs has equipped REIS with dozens of genes unknown from other Rickettsia genomes, it is likely that similar elements seeded the ancestral rickettsiae with genes that have become mainstays, functioning primarily in aiding these bacteria in survival within a eukaryotic cellular niche (Table 1). All of these genes have strong support as products of LGT and are present in unrelated intracellular bacterial species. For example, many of these genes were recently demonstrated to have highly similar homologs in the genome of the distantly related deltaproteobacterium “Candidatus Amoebophilus asiaticus” (102) (see Table S11 in the supplemental material). In Rickettsia spp., several of these LGT products have become duplicated and proliferated, including those encoding Tlc nucleotide transporters (see Fig. S11 in the supplemental material), SpoT stringent response regulators (see Fig. S12 in the supplemental material), ProP osmoregulatory proteins (see Fig. S13 in the supplemental material), and AmpG permeases involved in peptidoglycan recycling and beta-lactamase induction (see Fig. S14 in the supplemental material). Their expansion into multigene families suggests a strong selection on their retention in Rickettsia genomes, and their association with MGEs implies that a rich source for further diversification via LGT is present in the mobilome. Importantly, while REIS shows evidence of pseudogenization for several putative virulence factors characterized for pathogenic species of Rickettsia (discussed above), there is strict conservation in the genes identified here as being essential for an intracellular lifestyle.
Table 1.
Conserved Rickettsia genes putatively seeded by MGEs
| Protein | Function | MGE | Details |
|---|---|---|---|
| Tlc | Nucleotide translocase | RAGE | Antiporter that provides the microbial cell with host ATP in exchange for ADP. Tlc1 of R. prowazekii is a typical Tlc protein (ATP/ADP); Tlc4 (CTP, UTP, GDP) and Tlc5 (GTP, GDP) function as ribonucleotide importers, while the substrates of Tlc2 and Tlc3 have not been experimentally determined (4). All sequenced Rickettsia genomes encode all five Tlc transporters (see Fig. S11 in the supplemental material). |
| SpoT | Global regulator of stringent response | RAGE, plasmid | Full-length eubacterial SpoT proteins catalyze both the synthesis and degradation of ppGpp (alarmone), which is a mediator of cellular activities in response to changes in nutrient abundance (100). Rickettsia spoT genes are truncated and encode proteins that contain only the synthetase or hydrolase domain (see Fig. S12 in the supplemental material). Genomes contain from 4 (TG) to 14 (REIS; R. felis) spoT genes, with pseudogenes present in most genomes. |
| ProP | Osmoregulation, amino acid import | RAGE, plasmid | Proton symporter (sensitive to osmotic shifts) importing osmolytes, i.e., proline, glycine betaine, stachydrine, pipecolic acid, ectoine, and taurine (26). Rickettsia genomes encode from six (R. massiliae and R. peacockii) to 17 (R. felis) proP genes, with seven highly conserved groups and an additional clade comprised of less conserved genes (see Fig. S13 in the supplemental material). The proQ regulator of proP is found only in REIS (plasmids pREIS1 and pREIS3). |
| AmpG | IM GlcNAc-1,6-anhydro-muropeptide import and beta-lactamase inducer | NDa | IM permease involved in the translocation of peptidoglycan (PG) subunits from the periplasm to the cytoplasm (PG recycling) (62). It also acts as a permease in the beta-lactamase induction system (76). Four conserved groups are present in Rickettsia genomes, with each genome encoding at least three ampG genes (see Fig. S14 in the supplemental material). Three ampG3 pseudogenes in REIS are located in regions indicative of LGT. |
| RhlE | ATP-dependent RNA helicase | NDa | DEAD-box helicase (12) and putative ribosome assembly factor with a role in the interconversion of rRNA folding intermediates (63). Flanks ampG-2 in Rickettsia genomes and known only from Wolbachia genomes in the Anaplasmataceae. |
| EamA | SAM importer | WO-B | Possible export pump for several amino acids and their metabolites, especially cysteine (29). Demonstrated to uptake host SAM in R. prowazekii (117). eamA is highly conserved in all sequenced Rickettsiaceae genomes, illustrating the importance for SAM uptake (see Fig. S2 in the supplemental material). The pseudogenization of metK (encoding SAM synthase) in several genomes suggests that the acquirement of eamA has made the retention of metK obsolete. |
| Ugd | UDP-glucose 6-dehydrogenase | WO-B | Catalyzes the oxidation of UDP-glucose to yield UDP-glucuronic acid (40) and functioning in the biosynthesis of LPS and other polysaccharides, such as colanic acid and K-antigens (124). Associated with virulence in Xanthomonas campestris (22). Known in Rickettsiales only from some genomes of Wolbachia spp. and all sequenced Rickettsia genomes (see Fig. S2 in the supplemental material). Located near rvhB4b and ampG-3 in Rickettsia genomes. |
| GlpT | G3P importer | RAGE, WO-B | IM antiporter of the major facilitator superfamily, exchanging host G3P for bacterial cytosolic phosphate (Pi) (72). G3P import has been demonstrated in R. prowazekii, although it is not directly associated with GlpT (41). REIS encodes two glpT genes: one conserved across all Rickettsia genomes and a second encoded within RAGE-A and similar to the WO-B-related glpT from Wolbachia spp. (see Fig. S2 in the supplemental material). In Rickettsia genomes, the conserved glpT gene is flanked by tlc1 and ndk, the latter encoding a nucleoside diphosphate kinase functioning in nucleotide metabolism as well as the stringent response. |
| MdlB | ATP-binding multidrug resistance transporter | RAGE, WO-B | Uncharacterized transporter. Highly conserved in Rickettsiaceae genomes and multicopy in REIS and several others. Present in WO-B prophage (Fig. 2) and RAGE-A (Fig. 6) and part of the intracellular bacteria mobilome. |
| PlD | Phospholipase D | RAGE | Phospholipase involved in phagosomal escape (125). Conserved across Rickettsiales genomes with highly similar homologs in other intracellular bacterial species. Multiple pld copies are associated with RAGEs and TNPs in the REIS genome. |
| HP | DUF2608; HAD-like superfamily | rvh T4SS | Two divergent duplicate genes encoding putative haloacid dehalogenase-like hydrolases. The genes are adjacent and usually flank gppA, which itself flanks the gene encoding the rvh T4SS coupling protein (rvhD4). The closest homologs are proteins encoded in Parachlamydia genomes. |
Associated with other duplicate genes or TNPs.
Conclusions.
The analysis of the genome sequencing of I. scapularis revealed the presence of a rickettsial endosymbiont which was detected previously in various tick populations throughout the United States. We used the available sequence reads, in combination with information from 18 complete Rickettsiaceae genomes, to assemble a draft version of the REIS genome. Despite a conserved core genome shared with other Rickettsia species, the massive accessory genome (nearly twice the size of the entire R. prowazekii genome) makes REIS exceptional among Rickettsia genomes. The extreme proliferation of MGEs is reminiscent of the STG rickettsiae genomes, yet REIS encodes far more TNPs than RAGEs compared to those of Orientia. Our study is the first to identify plasmid-carried RAGEs, and it indicates that multiple invasions of these MGEs have occurred in the REIS genome. The ability of large gene clusters to insert and piggyback on the RAGEs suggests that these MGEs are efficient facilitators of the bacterial mobilome. Indeed, our analysis strongly suggests that the RAGE and other MGEs have seeded the Rickettsia genomes with genes typical of other intracellular bacterial species.
Unlike its sister lineage that diverged into the mitochondria, rickettsiae have evolved diverse strategies for life inside eukaryotic cells (49). Genome reduction and the acquisition of genes involved in exploiting host resources have concomitantly shaped the genomes of extant symbiotic and pathogenic rickettsial species. For Rickettsia spp., several scenarios have been proposed regarding the evolution of pathogenesis, ranging from gene loss leading to virulence (38) to the acquisition of virulence factors from LGT (90) to initial host virulence (stress) followed by gradual modification to symbiosis (17). Given that the degree of pathogenicity and the variation in host ranges does not directly correlate with rickettsial genome size and content, it is likely that no single scenario best explains the evolution of pathogenesis from symbiosis or vice versa for Rickettsia spp. Rather, through the avoidance of being trapped by a single host in an inextricable dependency, Rickettsia spp. have continually evolved strategies for manipulating multiple hosts for their survival. The observed diversification across the genus perhaps best reflects evolutionary continuums that likely keep genome size and composition in relative flux, allowing for the emergence of novel traits to offset reductive evolution. Although it is becoming more understood through the compilation of diverse genome sequences, the role of LGT in shaping the diversity of Rickettsia has remained largely unknown. To this end, the nature of the REIS genome and its impressive assortment of MGEs provide new insight.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Azad laboratory (University of Maryland, School of Medicine) and Roderick Felsheim, Timothy Kurtti, and Ulrike Munderloh (University of Minnesota, Department of Entomology) for invaluable feedback and discussion throughout the duration of this project. We thank Sean Daugherty (Institute for Genome Sciences, University of Maryland, School of Medicine) for advice regarding computational analysis of RAGE clusters.
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and Department of Health and Human Services under the following contract numbers: N01-AI-33071, HHSN266200400038C, and HHSN272200900040C (awarded to B.W.S.) and R01AI017828 and R01AI59118 (awarded to A.F.A.).
J.J.G., V.J., A.F.A., B.W.S., and E.C. conceived and designed the experiments, J.J.G., V.J., K.P.W., J.B.H., E.N., M.S., B.W., and E.C. performed the experiments, J.J.G., V.J., K.P.W., T.D., and E.C. analyzed the data, and J.J.G., V.J., K.P.W., T.D., A.F.A., C.A.H., B.W.S., and E.C. wrote the paper.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adelson ME, et al. 2004. Prevalence of Borrelia burgdorferi, Bartonella spp., Babesia microti, and Anaplasma phagocytophila in Ixodes scapularis ticks collected in Northern New Jersey. J. Clin. Microbiol. 42: 2799–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson SG, Stothard DR, Fuerst P, Kurland CG. 1999. Molecular phylogeny and rearrangement of rRNA genes in Rickettsia species. Mol. Biol. Evol. 16: 987–995 [DOI] [PubMed] [Google Scholar]
- 3. Andersson SG, et al. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140 [DOI] [PubMed] [Google Scholar]
- 4. Audia JP, Winkler HH. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 188: 6261–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldridge GD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. 2008. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl. Environ. Microbiol. 74: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldridge GD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. 2007. Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis. Appl. Environ. Microbiol. 73: 4984–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baldridge GD, et al. 2010. Wide dispersal and possible multiple origins of low-copy-number plasmids in rickettsia species associated with blood-feeding arthropods. Appl. Environ. Microbiol. 76: 1718–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balraj P, et al. 2008. RickA expression is not sufficient to promote actin-based motility of Rickettsia raoultii. PLoS One 3: e2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- 11. Billings AN, Teltow GJ, Weaver SC, Walker DH. 1998. Molecular characterization of a novel Rickettsia species from Ixodes scapularis in Texas. Emerg. Infect. Dis. 4: 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bizebard T, Ferlenghi I, Iost I, Dreyfus M. 2004. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43: 7857–7866 [DOI] [PubMed] [Google Scholar]
- 13. Blanc G, et al. 2005. Molecular evolution of rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 22: 2073–2083 [DOI] [PubMed] [Google Scholar]
- 14. Blanc G, et al. 2007. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 17: 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanc G, et al. 2007. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 3: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boyd EF, Almagro-Moreno S, Parent MA. 2009. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17: 47–53 [DOI] [PubMed] [Google Scholar]
- 17. Braig HR, Turner BD, Perotti MA. 2008. Symbiotic rickettsia, p. 221–249 In Bourtzis K, Miller TA. (ed.), Insect symbiosis, vol. 3. CRC Press, Boca Raton, FL [Google Scholar]
- 18. Buell CR, et al. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burgdorfer W, Gage KL. 1986. Susceptibility of the black-legged tick, Ixodes scapularis, to the Lyme disease spirochete, Borrelia burgdorferi. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 263: 15–20 [DOI] [PubMed] [Google Scholar]
- 20. Casadevall A. 2008. Evolution of intracellular pathogens. Annu. Rev. Microbiol. 62: 19–33 [DOI] [PubMed] [Google Scholar]
- 21. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552 [DOI] [PubMed] [Google Scholar]
- 22. Chang KW, Weng SF, Tseng YH. 2001. UDP-glucose dehydrogenase gene of Xanthomonas campestris is required for virulence. Biochem. Biophys. Res. Commun. 287: 550–555 [DOI] [PubMed] [Google Scholar]
- 23. Cho NH, et al. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U. S. A. 104: 7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Churchward G. 2008. Back to the future: the new ICE age. Mol. Microbiol. 70: 554–556 [DOI] [PubMed] [Google Scholar]
- 25. Cox R, Mason-Gamer RJ, Jackson CL, Segev N. 2004. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15: 1487–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Culham DE, et al. 1993. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229: 268–276 [DOI] [PubMed] [Google Scholar]
- 27. Curran KL, Kidd JB, Vassallo J, Van Meter VL. 2000. Borrelia burgdorferi and the causative agent of human granulocytic ehrlichiosis in deer ticks, Delaware. Emerg. Infect. Dis. 6: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dassler T, Maier T, Winterhalter C, Bock A. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36: 1101–1112 [DOI] [PubMed] [Google Scholar]
- 30. Dunning Hotopp JC, et al. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Durbin R, Eddy S, Krogh A, Mitchison G. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, MA [Google Scholar]
- 32. Ebel GD, Campbell EN, Goethert HK, Spielman A, Telford SRIII. 2000. Enzootic transmission of deer tick virus in New England and Wisconsin sites. Am. J. Trop. Med. Hyg. 63: 36–42 [DOI] [PubMed] [Google Scholar]
- 33. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellison DW, et al. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 76: 542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4: e8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fournier PE, et al. 2009. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frangeul L, et al. 2008. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Franzen JS, Ashcom J, Marchetti P, Cardamone JJ, Jr., Feingold DS. 1980. Induced versus pre-existing asymmetry models for the half-of-the-sites reactivity effect in bovine liver uridine diphosphoglucose dehydrogenase. Biochim. Biophys. Acta 614: 242–255 [DOI] [PubMed] [Google Scholar]
- 41. Frohlich KM, Roberts RA, Housley NA, Audia JP. 2010. Rickettsia prowazekii uses an sn-glycerol-3-phosphate dehydrogenase and a novel dihydroxyacetone phosphate transport system to supply triose phosphate for phospholipid biosynthesis. J. Bacteriol. 192: 4281–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fudou R, Jojima Y, Iizuka T, Yamanaka S. 2002. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J. Gen. Appl. Microbiol. 48: 109–116 [DOI] [PubMed] [Google Scholar]
- 43. Fuxelius HH, Darby AC, Cho NH, Andersson SG. 2008. Visualization of pseudogenes in intracellular bacteria reveals the different tracks to gene destruction. Genome Biol. 9: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gibson CM, Hunter MS. 2010. Extraordinarily widespread and fantastically complex: comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 13: 223–234 [DOI] [PubMed] [Google Scholar]
- 45. Gillespie JJ, Ammerman NC, Beier-Sexton M, Sobral BS, Azad AF. 2009. Louse- and flea-borne rickettsioses: biological and genomic analyses. Vet. Res. 40: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gillespie JJ, et al. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4: e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gillespie JJ, et al. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One 2: e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gillespie JJ, et al. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 78: 1809–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Phylogeny and comparative genomics: the shifting landscape in the genomics era. In Azad AF, Palmer GH. (ed), Intracellular pathogens II: Rickettsiales, in press. American Society for Microbiology, Washington, DC. [Google Scholar]
- 50. Gillespie JJ, et al. 2011. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 79: 4286–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gillespie JJ, et al. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3: e2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gouin E, et al. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427: 457–461 [DOI] [PubMed] [Google Scholar]
- 53. Gouin E, et al. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112: 1697–1708 [DOI] [PubMed] [Google Scholar]
- 54. Haas ES, Brown JW, Pitulle C, Pace NR. 1994. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. U. S. A. 91: 2527–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31: 371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. 2010. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12: 1057–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Halos L, et al. 2005. Evidence of Bartonella sp. in questing adult and nymphal Ixodes ricinus ticks from France and co-infection with Borrelia burgdorferi sensu lato and Babesia sp. Vet. Res. 36: 79–87 [DOI] [PubMed] [Google Scholar]
- 58. Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. 1993. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 61: 1926–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- 60. Ishmael N, et al. 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155: 2211–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ivanova N, et al. 2010. Complete genome sequence of Haliangium ochraceum type strain (SMP-2). Stand. Genomic Sci. 2: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13: 4684–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jain C. 2008. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 14: 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jeng RL, et al. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 6: 761–769 [DOI] [PubMed] [Google Scholar]
- 65. Juncker AS, et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12: 1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kaneko T, et al. 2007. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 14: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Keiler KC, Shapiro L, Williams KP. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 97: 7778–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kharel MK, et al. 2004. A gene cluster for biosynthesis of kanamycin from Streptomyces kanamyceticus: comparison with gentamicin biosynthetic gene cluster. Arch. Biochem. Biophys. 429: 204–214 [DOI] [PubMed] [Google Scholar]
- 70. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 71. Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62: 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224: 1–15 [DOI] [PubMed] [Google Scholar]
- 74. Li L, Stoeckert CJ, Jr., Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lin S, Hanson RE, Cronan JE. 2010. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat. Chem. Biol. 6: 682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lindquist S, et al. 1993. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol. Microbiol. 9: 703–715 [DOI] [PubMed] [Google Scholar]
- 77. Lorenzi HA, et al. 2010. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl. Trop. Dis. 4: e716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Magnarelli LA, Andreadis TG, Stafford KCIII, Holland CJ. 1991. Rickettsiae and Borrelia burgdorferi in ixodid ticks. J. Clin. Microbiol. 29: 2798–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Magnarelli LA, et al. 1995. Hemocytic rickettsia-like organisms in ticks: serologic reactivity with antisera to Ehrlichiae and detection of DNA of agent of human granulocytic ehrlichiosis by PCR. J. Clin. Microbiol. 33: 2710–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Magnarelli LA, Swihart RK. 1991. Spotted fever group rickettsiae or Borrelia burgdorferi in Ixodes cookei (Ixodidae) in Connecticut. J. Clin. Microbiol. 29: 1520–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mao C, et al. 2009. Variations on the tmRNA gene. RNA Biol. 6: 355–361 [DOI] [PubMed] [Google Scholar]
- 82. Meyer F, Overbeek R, Rodriguez A. 2009. FIGfams: yet another set of protein families. Nucleic Acids Res. 37: 6643–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42: 165–190 [DOI] [PubMed] [Google Scholar]
- 84. Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295: 679–682 [DOI] [PubMed] [Google Scholar]
- 85. Nakayama K, et al. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Noda H, Munderloh UG, Kurtti TJ. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63: 3926–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ogata H, Audic S, Abergel C, Fournier PE, Claverie JM. 2002. Protein coding palindromes are a unique but recurrent feature in Rickettsia. Genome Res. 12: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ogata H, et al. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293: 2093–2098 [DOI] [PubMed] [Google Scholar]
- 89. Ogata H, et al. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ogata H, et al. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3: e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pancholi P, et al. 1995. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J. Infect. Dis. 172: 1007–1012 [DOI] [PubMed] [Google Scholar]
- 92. Perlman SJ, Hunter MS, Zchori-Fein E. 2006. The emerging diversity of Rickettsia. Proc. Biol. Sci. 273: 2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Raoult D, Roux V. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10: 694–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Renesto P, Ogata H, Audic S, Claverie JM, Raoult D. 2005. Some lessons from Rickettsia genomics. FEMS Microbiol. Rev. 29: 99–117 [DOI] [PubMed] [Google Scholar]
- 96. Richter PJ, Jr, et al. 1996. Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae). J. Med. Entomol. 33: 1–5 [DOI] [PubMed] [Google Scholar]
- 97. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- 98. Salzberg SL, Delcher AL, Kasif S, White O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26: 544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sanogo YO, et al. 2003. Bartonella henselae in Ixodes ricinus ticks (Acari: Ixodida) removed from humans, Belluno province, Italy. Emerg. Infect. Dis. 9: 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sarubbi E, et al. 1989. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 264: 15074–15082 [PubMed] [Google Scholar]
- 101. Schmitz-Esser S, et al. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J. Bacteriol. 186: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schmitz-Esser S, et al. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. 1999. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37: 2215–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reference deleted. [Google Scholar]
- 105. Sekeyová Z, Fournier PE, Rehacek J, Raoult D. 2000. Characterization of a new spotted fever group rickettsia detected in Ixodes ricinus (Acari: Ixodidae) collected in Slovakia. J. Med. Entomol. 37: 707–713 [DOI] [PubMed] [Google Scholar]
- 106. Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. 2010. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe 7: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57: 138–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Simser JA, Rahman MS, Dreher-Lesnick SM, Azad AF. 2005. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol. Microbiol. 58: 71–79 [DOI] [PubMed] [Google Scholar]
- 109. Snyder EE, et al. 2007. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35: D401–D406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Spielman A. 1976. Human babesiosis on Nantucket Island: transmission by nymphal Ixodes ticks. Am. J. Trop. Med. Hyg. 25: 784–787 [DOI] [PubMed] [Google Scholar]
- 111. Steere AC. 2001. Lyme disease. N. Engl. J. Med. 345: 115–125 [DOI] [PubMed] [Google Scholar]
- 112. Steiner FE, et al. 2008. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J. Med. Entomol. 45: 289–297 [DOI] [PubMed] [Google Scholar]
- 113. Sutton GG, White O, Adams MD, Kerlavage AR. 1995. TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1: 9–19 [Google Scholar]
- 114. Swanson KI, Norris DE. 2007. Co-circulating microorganisms in questing Ixodes scapularis nymphs in Maryland. J. Vector Ecol. 32: 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Swofford D. 1999. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed Sinauer, Sunderland, MA [Google Scholar]
- 116. Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577 [DOI] [PubMed] [Google Scholar]
- 117. Tucker AM, Winkler HH, Driskell LO, Wood DO. 2003. S-adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185: 3031–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. van der Heide T, Stuart MC, Poolman B. 2001. On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20: 7022–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Van Dongen S. 2008. Graph clustering via a discrete uncoupling process. SIAM J. Matrix Anal. Appl. 30: 121–141 [Google Scholar]
- 120. van Stelten J, Silva F, Belin D, Silhavy TJ. 2009. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325: 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Weinert LA, Welch JJ, Jiggins FM. 2009. Conjugation genes are common throughout the genus Rickettsia and are transmitted horizontally. Proc. Biol. Sci. 276: 3619–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Weller SJ, et al. 1998. Phylogenetic placement of rickettsiae from the ticks Amblyomma americanum and Ixodes scapularis. J. Clin. Microbiol. 36: 1305–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75: 39–68 [DOI] [PubMed] [Google Scholar]
- 125. Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 73: 6668–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Williams KP, et al. 2010. Phylogeny of gammaproteobacteria. J. Bacteriol. 192: 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wood DW, et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294: 2317–2323 [DOI] [PubMed] [Google Scholar]
- 128. Wood JM. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63: 230–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wormser GP. 2006. Clinical practice. Early Lyme disease. N. Engl. J. Med. 354: 2794–2801 [DOI] [PubMed] [Google Scholar]
- 130. Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.